Abstract
In filamentous fungi, a programmed cell death (PCD) reaction occurs when cells of unlike genotype fuse. This reaction is caused by genetic differences at specific loci termed het loci (for heterokaryon incompatibility). Although several het genes have been characterized, the mechanism of this cell death reaction and its relation to PCD in higher eukaryotes remains largely unknown. In Podospora anserina, genes induced during the cell death reaction triggered by the het-R het-V interaction have been identified and termed idi genes. Herein, we describe the functional characterization of one idi gene (idi-1) and explore the connection between incompatibility and the response to nutrient starvation. We show that IDI-1 is a cell wall protein which localizes at the septum during normal growth. We found that induction of idi-1 and of the other known idi genes is not specific of the incompatibility reaction. The idi genes are induced upon nitrogen and carbon starvation and by rapamycin, a specific inhibitor of the TOR kinase pathway. The cytological hallmarks of het-R het-V incompatibility (increased septation, vacuolization, coalescence of lipid droplets, induction of autophagy, and cell death) are also observed during rapamycin treatment. Globally the cytological alterations and modifications in gene expression occurring during the incompatibility reaction are similar to those observed during starvation or rapamycin treatment.
In filamentous fungi cell fusion is frequent within a single colony or between cells from different strains. In the latter case, the resulting cells are heterokaryotic, since they contain a mixture of genetically different nuclei. Most generally, such heterokaryotic cells undergo a cell death reaction. This is referred to as heterokaryon incompatibility (for a review, see reference 12). This cell death reaction results from genetic differences at specific loci, termed het loci. It has been proposed that heterokaryon incompatibility constitutes a self-versus-nonself discrimination system limiting cytoplasmic mixing between genetically unlike individuals to prevent horizontal transfer of deleterious replicons (9).
Podospora anserina is one of the model fungal species in which this phenomenon has been analyzed. Nine het loci which define allelic or nonallelic systems have been identified in this species (3). Incompatibility is triggered by the coexpression of different het alleles from the same gene in allelic systems or from distinct loci in nonallelic systems. A number of these loci have been cloned, but the molecular characterization of their products has not given insights into the cell death mechanism that they trigger (for a review, see reference 33). In order to characterize this fungal cell death reaction, we are using the P. anserina het-R het-V self-incompatible strain as a model. In that strain, the two incompatible het genes (het-R and het-V) are reunited in the same haploid nucleus but incompatibility is conditional (19). A het-R het-V strain displays a normal growth at 32°C, but at 26°C the incompatibility reaction is triggered throughout the mycelium. Growth stops and cell lysis occurs. At the molecular level, major modifications in gene expression occur. Two-dimensional gel electrophoresis experiments have shown that synthesis of most proteins stops while expression of at least 20 specific proteins is induced (4). Several genes induced during incompatibility (idi genes) have been identified using a differential hybridization strategy (5). idi-1, idi-2, and idi-3 encode small proteins predicted to be either localized in the cell wall or secreted. idi-4 encodes a bZIP transcription factor whose characterization will be described elsewhere (K. Dementhon and C. Clavé, unpublished results). idi-6 and idi-7 are both autophagy genes (27, 28). Autophagy corresponds to the bulk degradation of cytoplasmic content by vacuolar enzymes, to provide new pools of nutrients under starvation conditions (18). idi-6 (also termed pspA) encodes a vacuolar protease involved in degradation of autophagic bodies, and idi-7 encodes the ortholog of the yeast AUT7 gene that is involved in the formation of autophagosomes, the vesicles that target cytoplasmic material to the vacuole. We have shown recently that autophagy is induced during cell death by incompatibility in Podospora (28). Autophagy is induced upon starvation and is under the control of the TOR signaling pathway. The TOR (for target of rapamycin) protein kinases are conserved from yeast to humans and control cell growth in response to nutrient availability (30, 34). Upon nutritional starvation, inactivation of TOR leads to induction of autophagy, expression of starvation-induced genes, and inhibition of translation. TOR is specifically inhibited by the natural product rapamycin (14). Thus, rapamycin treatment mimics nutrient deprivation in yeast, Drosophila melanogaster, and mammals (1, 8, 13, 40) and triggers autophagy (25).
Herein, we explore the connection between the incompatibility reaction and the response to nutrient starvation. We show that idi-1 encodes a cell wall protein localized in the septum during normal growth and that expression of idi-1 is not only induced by incompatibility but also by nutrient limitation. We found that in addition to idi-1 the other known idi genes are induced by nutrient starvation and by rapamycin, a specific inhibitor of the TOR kinase. We show also that rapamycin treatment phenocopies the incompatibility reaction at the cytological level.
MATERIALS AND METHODS
Strains and media.
The het-r and het-v loci of P. anserina each display two distinct alleles termed het-r and het-R and het-V and het-V1, respectively. The het-R and het-V alleles are incompatible. Self-incompatible het-R het-V strains were obtained in the progeny of a cross between het-R het-V1 and het-r het-V. Self-incompatible het-R het-V strains grow normally at permissive temperature (32°C), but the incompatibility reaction is triggered when the strain is transferred at nonpermissive temperature (26°C) (19). Preparation and transformation of protoplasts were performed as described previously (2). For RNA extraction mycelia were grown on solid SA medium (dextrin, 20 g/liter; ammonium acetate, 4 g/liter; NaCl, 0.05 g/liter; CaCl2, 7.5 mg/liter; K2HPO4, 0.17 g/liter; MgSO4, 0.1 g/liter; biotin, 5 μg/liter; thiamine, 100 μg/liter; Bacto Agar, 25 g/liter; trace element concentrate solution, 0.1 ml/liter). The trace-element concentrate solution contains citric acid (50 g/liter), ZnSO4 heptahydrate (50 g/liter), Fe(NH4)2(SO4)2 hexahydrate (10 g/liter), CuSO4 pentahydrate (2.5 g/liter), MnSO4 monohydrate (0.5 g/liter), boric acid (0.5 g/liter), and MoNa2SO4 dihydrate (0.5 g/liter). Nitrogen-deprived medium is SA medium lacking ammonium acetate; carbon-deprived medium is SA lacking dextrin.
Plasmids.
For construction of the plasmid allowing expression of the IDI-1-green fluorescent protein (GFP) fusion protein, a 3-kbp SmaI-XhoI fragment (position 727 to 3686, AF505658) bearing the idi-1 gene was inserted into the Litmus 28 vector (Biolabs New England) restricted with EcoRV and XhoI. SmaI and NotI restriction sites were introduced upstream of the stop codon of the idi-1 open reading frame (ORF) by site-directed mutagenesis (Transformer site-directed mutagenesis kit; Clontech) and a SmaI-NotI fragment containing the enhanced GFP gene isolated from the pEGFP-1 plasmid (Clontech) was cloned into the newly created SmaI and NotI sites. For construction of the pAN52.1-idi-1 vector used to overexpress idi-1, the idi-1 ORF was amplified by PCR using oligonucleotides introducing BspHI and BamHI sites (underlined) at the 5′ and 3′ end of the ORF, respectively (5′-TGCTAGTCATGAAGTACACCACCGCCTC-3′ and 5′-CGCGGATCCTTAACAAGCAGACTGATTCCG-3′). The amplified fragment was then introduced into the pAN52.1 vector under control of the Aspergillus nidulans GPD promoter (29).
DNA and RNA procedures.
General methods for manipulation of nucleic acids were as described in Sambrook et al. (31). Genomic DNA and RNA from P. anserina were extracted as previously described (5, 22). Total RNA samples were quantified by measuring absorbance at 260 nm, and their concentration was adjusted to 3 mg/ml. A 28S ribosomal DNA (rDNA) probe (20) was used as a control in Northern blot analyses. The different idi gene probes were prepared from PCR products corresponding to the entire ORF of the corresponding genes. The GenBank accession numbers for idi-1, idi-2, idi-3, pspA, and idi-7 are, respectively, AF505658, AF500213, AF502253, AF047689, and AF502254. The idi-4 probe is a 1.1-kbp XhoI-XbaI fragment containing the entire idi-4 ORF (Dementhon and Clavé, unpublished data). Probes were labeled with 32P using the Prime it II random labeling kit (Stratagene). Visualization and quantification of radioactive signals were performed with a PhosphorImager using the ImageQuant program (Molecular Dynamics, Sunnyvale, Calif.).
Gene disruption of idi-1.
Inactivation of idi-1 by gene replacement was achieved by a two-marker strategy. idi-1 was cloned as a 4.5-kbp EcoRI-HindIII fragment into the pSelect-1 vector (Promega). The ura5 gene was introduced as an EcoRI fragment into the unique EcoRI site of this vector (37). An internal 2.5-kbp fragment containing the entire idi-1 gene (ORF and promoter region) was replaced by a 1.4-kbp XbaI fragment containing the hph gene conferring hygromycin resistance. This vector containing about 1 kbp of idi-1 flanking sequences on either side of hph was used to transform a ura5-6 auxotrophic strain (37). The expected double homologous crossing-over event leading to idi-1 disruption yields hygR auxotrophic strains. Among 1,600 hygR transformants, 64 were auxotrophic. These 64 transformants were analyzed as 16 pools of 4 transformants each by Southern blotting using a 2.4-kb EcoRI-EcoRV fragment corresponding to the 5′ flank of idi-1 as a probe. In one pool a transformant displaying the expected profile by Southern blot hybridization was obtained. The inactivation of idi-1 was also confirmed by Northern blot by showing that the idi-1 mRNA is undetectable in a Δidi-1 strain.
Light and fluorescence microscopy.
Synthetic medium containing 2% (wt/vol) agarose was poured as two 10-ml layers of medium. P. anserina hyphae were inoculated on this medium and cultivated for 16 to 24 h at 26°C. The top layer of the medium was then cut out and the mycelium was examined with a Leica DMRXA microscope equipped with a Micromax charge-coupled device (Princeton Instruments). For Congo red staining of cell wall material, the mycelium was overlaid with a 1-mg/ml Congo red solution in 150 mM NaCl for 2 min and rinsed with 150 mM NaCl. Nile Red staining of lipid droplets was performed as described elsewhere (36). Evans blue dye staining to identify dead cells was performed as described previously (15).
RESULTS
idi-1 is not an essential effector of cell death by incompatibility.
We have previously shown that the expression of idi-1 is strongly induced during het-R het-V incompatibility (5). In order to gain insight into the role of idi-1 in the incompatibility reaction, we have inactivated this gene by gene replacement using a two-marker strategy and identified idi-1 disruptants by Southern blot analysis (see Materials and Methods section). This strain was designated Δidi-1. The Δidi-1 strain displayed normal vegetative growth and normal fertility both as male and female parent. Thus, idi-1 is a nonessential gene. We then constructed a het-R het-V Δidi-1 self-incompatible strain by crossing the het-r het-V Δidi-1 strain with het-R het-V1. The het-R het-V Δidi-1 strain was grown at 32°C and transferred to 26°C to trigger the incompatibility reaction. The cell death reaction occurred as in a wild-type het-R het-V strain. We conclude that idi-1 is not required for cell death by incompatibility. Thus, IDI-1 is not an essential effector of this cell death reaction. We then analyzed the effect of the constitutive overexpression of idi-1. The idi-1 ORF was cloned under control of the strong constitutive GPD promoter (pGPD) of A. nidulans (29). Transformants containing the GPD-idi-1 construct expressed the idi-1 mRNA at very high levels. The amount of the idi-1 mRNA detected by Northern blot was at least fivefold higher than that detected during het-R het-V incompatibility (data not shown). These pGPD-idi-1 transformants displayed a normal growth rate, pigmentation, and fertility and did not display any detectable growth alteration. This observation indicates that overexpression of idi-1 alone is not sufficient to lead to the incompatibility reaction, further suggesting that idi-1 is not an effector of cell death by incompatibility. Apparently, induction of idi-1 is symptomatically rather than causally linked to cell death by incompatibility.
IDI-1 is a septal protein which accumulates during incompatibility.
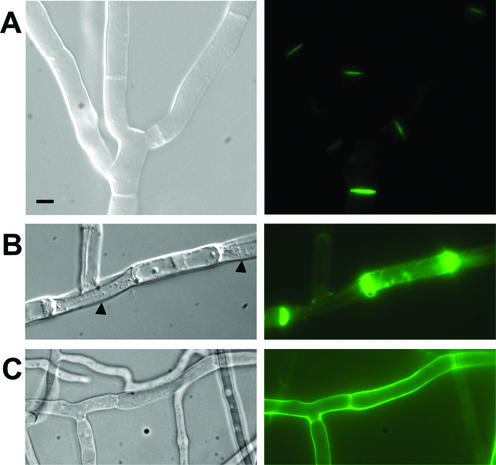
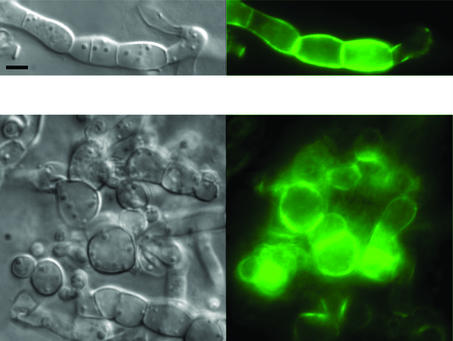
We have analyzed the cellular localization of IDI-1 by constructing a plasmid allowing expression of an IDI-1-GFP fusion protein under control of its own promoter. During vegetative growth on rich medium, the fusion protein was localized to the septa, the cross walls of the hyphae (Fig. 1A). We then analyzed IDI-1-GFP localization during the incompatibility reaction. We crossed the het-r het-V idi-1-gfp strain with the het-R het-V1 strain and selected the self-incompatible het-R het-V idi-1-gfp progeny. After transfer to 26°C, the IDI-1-GFP fluorescence signal was strongly increased and was detected at the septa but also in the rest of the cell wall compartment. In late stages of the incompatibility reaction, surviving cells often display cell wall thickening at their extremities when they are bordered by dead fungal cells. A very strong IDI-1-GFP fluorescent signal was observed at such cell wall thickenings (Fig. 1B). The same experiments were also performed in a Δidi-1 background and gave similar results (data not shown).
FIG. 1.
Localization of the IDI-1-GFP fusion protein. (A) Strain expressing the IDI-1-GFP fusion protein and grown on rich medium. (B) Incompatible het-R het-V strain expressing the IDI-1-GFP fusion protein and transferred for 4 h at 26°C (incompatibility conditions). The black arrowheads mark lysed cells surrounding a surviving cell. (C) Strain expressing the IDI-1-GFP fusion protein grown on nitrogen-deprived medium. For each panel, the Nomarski view is given on the left and the GFP fluorescence view is given on the right. Note that image acquisition time was 10 times longer for panel A than for panels B and C. The scale bar represents 2 μm.
During the course of these experiments, we observed that in vegetative cultures of the wild-type, non-self-incompatible, strain, the IDI-1-GFP signal was very strong in the older parts of the mycelium, suggesting that nutrient starvation might increase expression of idi-1. On nitrogen-starved medium, the IDI-1-GFP signal was indeed strongly increased. As observed during incompatibility, the signal was localized in the entire cell wall compartment and not exclusively to the septum (Fig. 1C). This observation prompted us to analyze expression of idi-1 and of the other idi genes during nutrient starvation.
idi genes are induced upon nitrogen and carbon starvation.
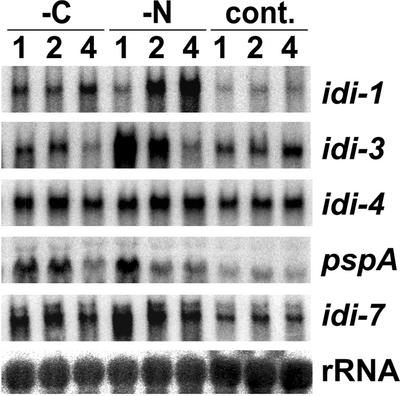
Expression of the idi-1-gfp fusion is apparently induced by starvation. In addition, in yeast the orthologs of idi-6/pspA and idi-7 are induced upon starvation (17, 24). We have therefore investigated the expression level of idi genes under carbon or nitrogen limitation. A wild-type Podospora strain was grown on synthetic (SA) medium and transferred onto SA medium deprived of carbon source or SA medium deprived of nitrogen source. As a control, the cultures were also transferred onto fresh SA medium containing both carbon and nitrogen sources. At different times after transfer, idi gene expression was analyzed by Northern blotting (Fig. 2). The mRNA of the different idi genes accumulated under nitrogen limitation. Peak expression levels were observed after 1 h for idi-3, pspA, and idi-7; after 2 h for idi-4; and after 4 h for idi-1. Compared to basal levels, expression increased about 10-fold for idi-3, 8-fold for pspA, 6-fold for idi-1, and 4-fold for idi-7. Induction of idi-4 was only two-fold. pspA was also induced to a same extent upon carbon starvation. The other idi genes were only slightly induced upon carbon starvation. Induction was about 2-fold for idi-1 and idi-7 and 1.5-fold for idi-3 and idi-4. We thus considered only idi-1, pspA, and idi-7 to be significantly induced upon carbon starvation. Expression of idi-2 was detected under none of the experimental conditions (data not shown).
FIG. 2.
idi gene expression under starvation conditions. A wild-type, non-self-incompatible, strain was grown on synthetic SA medium for 40 h and then transferred for 4 h onto fresh SA medium to rule out any preexisting starvation. Cultures were then transferred onto SA medium lacking dextrin (-C), SA medium lacking ammonium acetate (-N), or control SA medium (cont.). Total RNAs were extracted after 1, 2, or 4 h. Total RNA samples (30 μg per lane) were analyzed by Northern blotting and probed successively with the different idi genes and a control 28S rDNA probe.
Together these results indicate that induction of idi genes is not limited to incompatibility but also occurs upon starvation. Induction is more robust and generalized for nitrogen than carbon starvation.
idi genes are induced by rapamycin treatment.
In yeast, the TOR kinase pathway is the central regulator of the cellular response to nutrient limitation, in particular, nitrogen limitation. Rapamycin treatment that inactivates TOR leads to expression of various starvation-induced genes. This prompted us to investigate the effect of a rapamycin treatment on idi gene expression in Podospora. We first analyzed the effect of various amounts of rapamycin on the growth rate of Podospora. At 20 ng · ml−1, rapamycin had no effect on growth. At 200 ng · ml−1, the radial growth rate was reduced by 30%, and at 500 ng · ml−1 it was reduced by 90%. With rapamycin at a concentration of 1 μg · ml−1, growth was completely inhibited. idi gene expression experiments were performed with rapamycin at a concentration of 200 ng · ml−1. A wild-type strain was grown for 40 h on synthetic medium. Then, the mycelium was transferred onto fresh medium containing rapamycin. Mycelia were collected 2, 4, 6, 9, and 27 h after transfer, and idi gene expression was assessed by Northern blot analysis (Fig. 3). Rapamycin treatment induced the expression of all idi genes. Timing of induction and peak expression levels varied for the different genes. For instance, expression of idi-3 peaked after 2 h, while expression of idi-2 was found to be maximal after 27 h. Induction levels were at least 50-fold for idi-1 and idi-2, 25-fold for idi-3, and 20-fold for idi-4. pspA and idi-7 showed a more modest increase in expression, with a four- and a threefold increase, respectively.
FIG. 3.
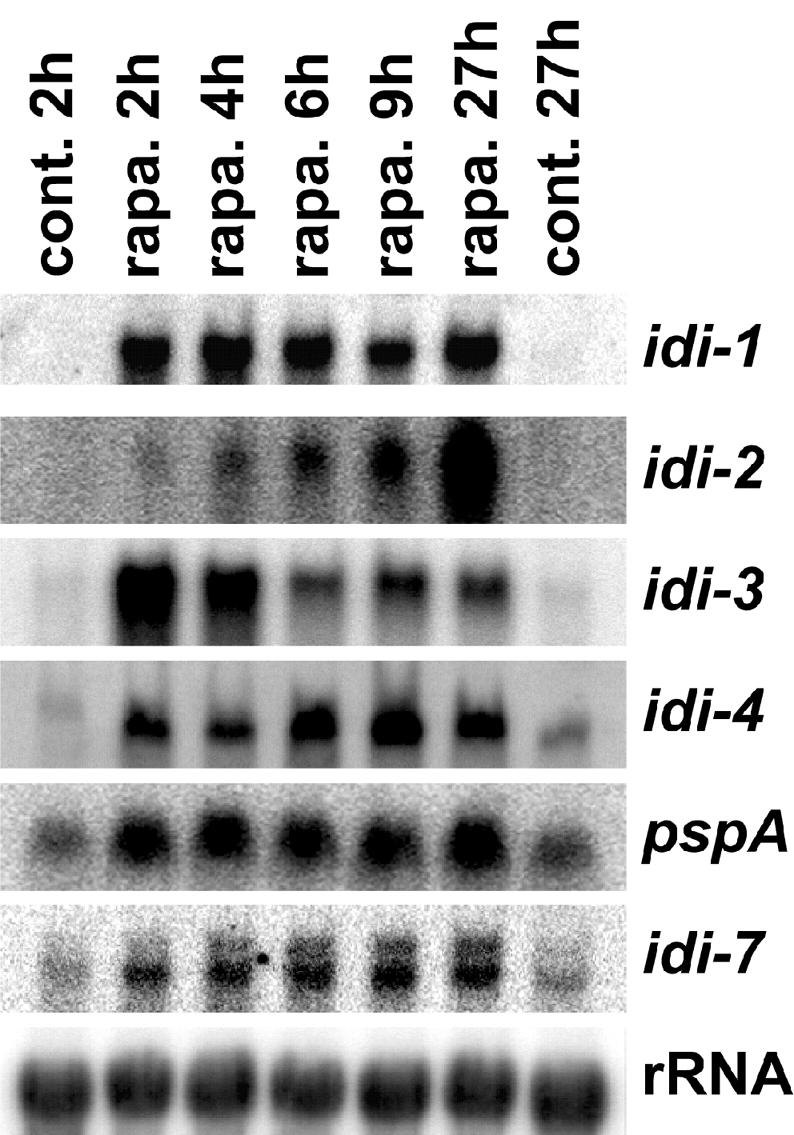
Effect of rapamycin on idi gene expression. A wild-type, non-self-incompatible, strain was grown on synthetic SA medium for 40 h and then transferred for 4 h onto fresh synthetic medium. Cultures were transferred onto SA medium supplemented or not with rapamycin (200 ng · ml−1). Total RNAs were extracted 2, 4, 6, 9, and 27 h after transfer onto rapamycin (rapa.). As a control (cont.), the strain was also transferred onto fresh SA medium lacking rapamycin for 2 or 27 h. Total RNA samples (30 μg per lane) were analyzed by Northern blotting and probed successively with the different idi genes and a control 28S rDNA probe.
Together, these results indicate that all known idi genes are induced by rapamycin.
Incompatibility and rapamycin trigger vacuolization, increased septation, coalescence of lipid droplets, morphological alterations, and cell death.
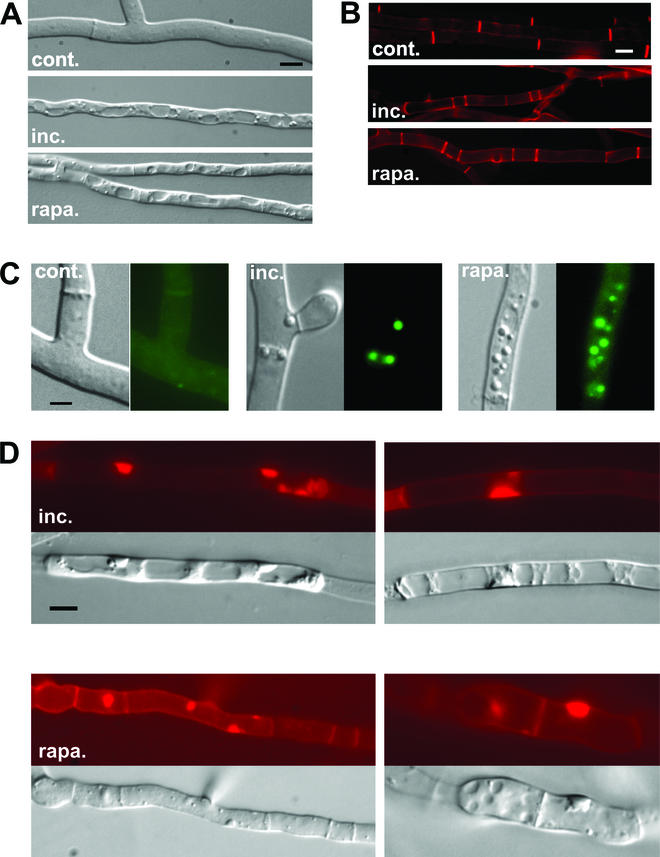
Based on the observation that rapamycin is able to strongly induce expression of all known idi genes, we chose to compare the cytological modifications occurring during incompatibility and rapamycin treatment. A het-R het-V self-incompatible strain was cultivated at 32°C and transferred at 26°C. The het-R het-V incompatibility reaction was found to be characterized by an extensive vacuolization. Spherical vacuoles formed and ultimately filled the entire fungal cell (Fig. 4A). In addition, deposition of cross walls (septa) was more frequent, thus giving rise to short cells (Fig. 4B). Moreover, during het-R het-V incompatibility, refringent spherical structures accumulated in the cytoplasm (Fig. 4C). Based on observation in the fungus Magnaporthe grisea, we hypothesize that these structures could correspond to lipid bodies which were found to form during appressorium formation in this phytopathogenic fungus (36). Such lipid bodies can be identified with the lipophilic dye Nile red. The spherical structures formed during het-R het-V incompatibility stained with Nile red; we therefore conclude that they correspond to lipid bodies (Fig. 4C). In the late stages of het-R het-V incompatibility, such lipid bodies accumulated in the vacuolar compartment (not shown). In a number of cells abnormal deposition of cell wall material took place (Fig. 4D). Cell wall thickenings, which could be labeled with Congo red, appear either on the lateral cell walls or at the end of hyphal cells. Finally, after about 4 h of transfer at restrictive temperature (26°C), the het-R het-V incompatibility reaction culminated in generalized cell death. Cell death was monitored using Evans blue dye, which stains dead cells (Fig. 5A).
FIG.4.
het-R het-V incompatibility and rapamycin induce the same cytological alterations. (A, B, and C) The normal growth control (cont.), the het-R het-V incompatible strain (inc.), and the strains treated with rapamycin (at 200 ng · ml−1) (rapa.) are shown. (A) The intense vacuolization observed both during incompatibility and rapamycin treatment is shown. (B) Septa are labeled with Congo red. Note that septation is increased during incompatibility and rapamycin treatment. (C) Lipid body formation during incompatibility and rapamycin (200 ng · ml−1) treatment is shown. For each panel, the Nomarski view is given on the left and the fluorescence of the Nile red lipophilic dye is given on the right. (D) Abnormal deposition of cell wall induced by het-R het-V incompatibility and rapamycin (at 500 ng · ml−1) is shown. The two upper panels correspond to het-R het-V incompatibility. The lower panels correspond to rapamycin treatment. In each case the Nomarski view and the Congo red fluorescence specific of cell wall material are given. The scale bar represents 2 μm.
FIG. 5.
Rapamycin induces cell death in Podospora. (A) het-R het-V incompatible strain transferred for 4 h at 26°C (incompatibility conditions). (B) Wild-type strain transferred for 4 h on medium containing rapamycin at a concentration of 1 μg · ml−1. Mycelia were stained with Evans blue dye, which stains dead cells. For each panel the Nomarski view and the Evans blue dye fluorescence are given.
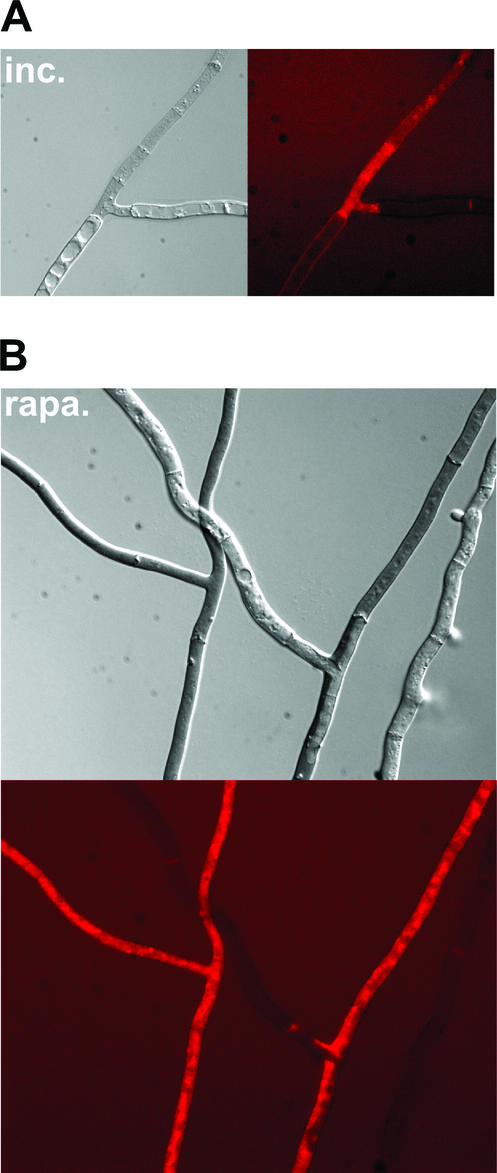
When a wild-type strain was cultivated on synthetic medium containing rapamycin at a concentration of 200 ng · ml−1, similar cytological alterations were observed. In particular numerous large spherical vacuoles were found in most filaments (Fig. 4A). Short cells resulting from increased septum deposition were also frequent (Fig. 4B). Spherical lipid bodies were numerous (Fig. 4C). At a higher rapamycin concentration (500 ng · ml−1), abnormal cell wall thickenings similar to those formed during incompatibility appeared (Fig. 4D). Moreover, morphological alterations could be detected. Filaments lost their cylindrical shape and became round and displayed constrictions at the septa (Fig. 6). In the apical region of the mycelium, hyphal branching was increased and growth polarity was affected (not shown). When cultures were transferred to a medium containing rapamycin at a concentration of 1 μg · ml−1, after a few hours, generalized cell death took place (Fig. 5B). Similar to what was observed during het-R het-V incompatibility, in dead cells, detected as cells taking up the Evans blue dye, the vacuoles were no longer visible, suggesting that cell death might results from rupture of the vacuolar membrane leading to coagulation of the cytoplasm.
FIG. 6.
Rapamycin induces morphological defects and expression of the IDI-1-GFP fusion protein. A strain expressing the IDI-1-GFP fusion protein was grown on medium containing rapamycin (500 ng · ml−1). The Nomarski view is given on the left, and the GFP fluorescence view is given on the right. The scale bar represents 2 μm. Note the abnormal cellular morphology induced by rapamycin. Image acquisition time was 50 times shorter than that for Fig. 1A.
To confirm that rapamycin is able to induce idi-1 not only at the mRNA level but also at the protein level, a strain expressing the IDI-1-GFP fusion protein was cultivated on rapamycin (at 500 ng · ml−1). The fluorescence signal was very strong and localized at the septum and in the rest of the cell wall compartment (Fig. 6).
Together these observations indicate that rapamycin treatment mimics the cytological manifestations of the het-R het-V incompatibility reaction and is able to induce cell death in Podospora.
Rapamycin treatment triggers autophagy in Podospora.
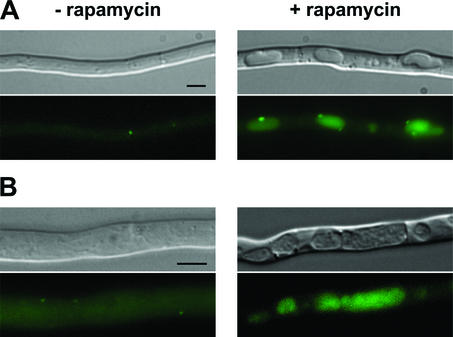
We have reported that autophagy is induced during the incompatibility reaction in Podospora (28). In order to further compare the cellular manifestations of incompatibility to those induced by rapamycin treatment, we have determined whether rapamycin treatment triggers autophagy in Podospora. IDI-7 is the ortholog of the yeast Aut7p protein involved in autophagosome formation, and relocalization of the GFP-IDI-7 fusion protein from the cytoplasm to the vacuole constitutes a cytological marker of the induction of autophagy. A wild-type strain expressing the GFP-IDI-7 fusion protein (28) was grown on SA medium for 16 h and transferred to SA medium supplemented with rapamycin at a concentration of 200 ng · ml−1. As a control, the same strain was transferred to SA medium without rapamycin. As already described (28), in the absence of rapamycin, the GFP-IDI-7 fusion protein had a diffuse cytoplasmic distribution and was associated with dot-like structures distinct from the vacuolar compartment. Upon transfer on rapamycin, the GFP-IDI-7 fusion protein localized to large punctate perivacuolar structures and to the lumen of the vacuole (Fig. 7A). Inactivation of the PSPA vacuolar protease prevents degradation of autophagic bodies in the vacuole (28). Induction of autophagy can thus be readily detected in a ΔpspA background by the accumulation of autophagic bodies within the vacuolar lumen. To confirm induction of autophagy upon rapamycin treatment, the same experiment was repeated in a ΔpspA background. As described above, in the presence of rapamycin relocalization of GFP-IDI-7 was observed together with the accumulation of autophagic bodies inside the vacuole (Fig. 7B). Detection of autophagic bodies upon rapamycin treatment in a ΔpspA strain further evidences induction of autophagy.
FIG. 7.
Cytological evidences of induction of autophagy by rapamycin: relocalization of the GFP-IDI7 marker and detection of autophagic bodies. A gfp-idi7 strain (A) and a gfp-idi7 ΔpspA strain (B) were grown for 16 h on synthetic SA medium. The strains were then transferred on fresh SA medium (− rapamycin) or SA medium containing rapamycin at 200 ng · ml−1 (+ rapamycin). The strains were further incubated for 1 h at 26°C before observation by light microscopy (top) and by fluorescence microscopy (bottom) (scale bar = 3 μm). Note the granular aspect of the vacuoles of the ΔpspA strain in the presence of rapamycin.
We conclude that rapamycin treatment induces autophagy in Podospora. This observation further illustrates the similarity between the cellular manifestations occurring during incompatibility and rapamycin treatment.
DISCUSSION
Programmed cell death (PCD) has been described in plants and animals as a normal developmental process or as a defense mechanism against pathogens (21, 38). The most widely described form of PCD in metazoans is apoptosis. Plants and fungi lack caspases; therefore, PCD does not occur via classical apoptosis in these organisms (21). In filamentous fungi, a genetically controlled cell death reaction (incompatibility reaction) occurs when cells of unlike het genotype fuse (12). We have proposed that this cell death reaction involves autophagy (27, 28). Interestingly, autophagy has been described as a PCD mechanism in metazoans (7). Therefore, cell death by incompatibility appears to employ similar mechanisms as certain forms of PCD in higher eukaryotes.
The present study suggests that idi-1 is not an effector of the cell death reaction by incompatibility. Neither did inactivation of idi-1 suppress incompatibility, nor did overexpression of idi-1 lead to cell death. Apparently, incompatibility not only induces genes involved in the degradative process like idi-6 and idi-7. IDI-1 is a septal protein, and septation is increased during starvation, by incompatibility, and by rapamycin. This might explain why idi-1 is induced under these conditions. During incompatible cell fusion, the cell death reaction is generally restricted to the fusion cell because the septa to the adjacent cells are rapidly sealed. While this phenomenon is likely to involve septal sealing by Woronin bodies (16), it is possible that deposition of cell wall material further compartimentalizes the cell death reaction. IDI-1 might therefore have a protective rather than cell death-promoting function. Septum deposition is unaffected in a Δidi-1 mutant; this might be due to the fact that other proteins (such as IDI-2 or IDI-3 which displays the same amino acid content bias as IDI-1) are functionally redundant with IDI-1.
The main conclusion of the present work is that there is a clear connection between the incompatibility reaction and the cellular response to nutrient starvation. In all eukaryotes, the TOR kinase is one of the central regulators that coordinates cell growth and metabolism with nutrient availability. Rapamycin, a specific inhibitor of TOR, mimics starvation in yeast and metazoans. The present work shows that the incompatible het gene interactions and rapamycin treatment lead to similar cellular responses. Both conditions strongly induce idi gene expression and trigger autophagy and cell death. This analogy between rapamycin treatment and the incompatibility reaction is further illustrated by earlier analyses of het-R het-V incompatibility. Namely, it was shown that transfer of a het-R het-V strain from 32 to 26°C leads to a growth arrest, a drastic decrease in global transcription, and translation. In yeast and metazoan, inactivation of the TOR pathway by rapamycin shuts down translation and transcription and leads to a cell cycle arrest.
The similarity between the incompatibility reaction and response to rapamycin could be explained by hypothesizing that like rapamycin, the incompatible het gene interactions affect the TOR pathway. Two alternate models can be proposed. First, the interaction of the antagonistic het gene products might affect the general cellular metabolism, and this perturbation might then be sensed by the TOR pathway. The TOR pathway can integrate a number of cellular cues such as nutrient availability but also mitochondrial integrity and ATP levels (10, 11). Thus, various cellular dysfunctions might lead to inactivation of the TOR pathway and induction of autophagy. Although idi genes have been identified in the het-r/het-v system, they are also induced in the het-c/het-e system (5, 27, 28). The hypothesis of metabolic perturbations might explain how different het systems can induce the same set of genes. In that hypothesis, inactivation of the TOR pathway would represent an indirect consequence of the deleterious effect of het gene interaction on cellular integrity. An alternate hypothesis would be that the het gene products belong to a signaling cascade feeding into the TOR pathway. The het-c gene product of Podospora encodes a glycolipid transfer protein (32), and it has been proposed that the Arabidopsis thaliana ortholog of Podospora het-c, involved in PCD in plants, is part of a sphingolipid signaling pathway (6). Sphingolipid-based signaling is involved in stress response both in yeast and in higher eukaryotes (23, 26, 35, 39). One might hypothesize that the incompatible HET-C HET-E interaction would directly signal inactivation of TOR.
Clearly the connection established here between the rapamycin-sensitive pathway and incompatibility in Podospora is correlative. To demonstrate that het gene interactions affect the TOR pathway it will be necessary to analyze TOR activity during incompatibility. We have now isolated the TOR ortholog in Podospora (I. Iraqui and C. Clavé, unpublished data) and will be able to determine whether the TOR kinase activity is modified by incompatible het gene interactions and whether the TOR pathway participates in the execution of cell death program.
idi genes are not specific to cell death by incompatibility (28). Apparently the cellular program leading to PCD in Podospora employs components of the cellular machinery required for adaptation to starvation. It was shown previously that the het genes which genetically control this PCD reaction can display cellular functions in addition to their role in heterokaryon incompatibility (32). Together these results suggest that neither the genetic triggers of PCD nor the actual executers are specific to cell death by incompatibility, but rather that the incompatibility function has recruited a preexisting cellular pathway.
Acknowledgments
We express our gratitude to J. Bégueret for his critical reading of the manuscript and for valuable suggestions.
This work was supported by a grant from the Association pour la Recherche sur le Cancer (ARC). K.D. was funded by the Ministère de la Recherche et de l'Education (MRE).
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergès, T., and C. Barreau. 1989. Heat-shock at elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J. Gen. Microbiol. 135:601-604. [DOI] [PubMed] [Google Scholar]
- 3.Bernet, J. 1965. Mode d'action des gènes de barrage et relation entre l'incompatibilité cellulaire et l'incompatibilité sexuelle chez le Podospora anserina. Ann. Sci. Nat. Bot. 6:611-768. [Google Scholar]
- 4.Boucherie, H., C. H. Dupont, and J. Bernet. 1981. Polypeptide synthesis during protoplasmic incompatibility in the fungus Podospora anserina. Biochim. Biophys. Acta 653:18-26. [DOI] [PubMed] [Google Scholar]
- 5.Bourges, N., A. Groppi, C. Barreau, C. Clavé, and J. Bégueret. 1998. Regulation of gene expression during the vegetative incompatibility reaction in Podospora anserina: characterization of three induced genes. Genetics 150:633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodersen, P., M. Petersen, H. M. Pike, B. Olszak, S. Skov, N. Odum, L. B. Jorgensen, R. E. Brown, and J. Mundy. 2002. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bursch, W., A. Ellinger, C. Gerner, U. Frohwein, and R. Schulte-Hermann. 2000. Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann. N. Y. Acad. Sci. 926:1-12. [DOI] [PubMed] [Google Scholar]
- 8.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caten, C. E. 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72:221-229. [DOI] [PubMed] [Google Scholar]
- 10.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 11.Desai, B. N., B. R. Myers, and S. L. Schreiber. 2002. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 99:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, N. L., D. J. Jacobson, and P. K. Shiu. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34:165-186. [DOI] [PubMed] [Google Scholar]
- 13.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 14.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, D. J., K. Beurkens, and K. L. Klomparens. 1998. Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet. Biol. 23:45-56. [DOI] [PubMed] [Google Scholar]
- 16.Jedd, G., and N. H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226-231. [DOI] [PubMed] [Google Scholar]
- 17.Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi, T. Yoshimori, T. Noda, and Y. Ohsumi. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky, D. J., and S. D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labarère, J. 1973. Properties of an incompatibility system in Podospora anserina fungus and value of this system for the study of incompatibility. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D 276:1301-1304. (In French.) [PubMed]
- 20.Labat, N., M. Perrot, and J. Bégueret. 1985. Cloning and characterization of the rDNA repeat unit of Podospora anserina. Mol. Gen. Genet. 199:154-157. [DOI] [PubMed] [Google Scholar]
- 21.Lam, E., N. Kato, and M. Lawton. 2001. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411:848-853. [DOI] [PubMed] [Google Scholar]
- 22.Lecellier, G., and P. Silar. 1994. Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr. Genet. 25:122-123. [DOI] [PubMed] [Google Scholar]
- 23.Mathias, S., L. A. Pena, and R. N. Kolesnick. 1998. Signal transduction of stress via ceramide. Biochem. J. 335:465-480. [DOI] [PMC free article] [PubMed]
- 24.Naik, R. R., V. Nebes, and E. W. Jones. 1997. Regulation of the proteinase B structural gene PRB1 in Saccharomyces cerevisiae. J. Bacteriol. 179:1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 26.Ohanian, J., and V. Ohanian. 2001. Sphingolipids in mammalian cell signalling. Cell Mol. Life Sci. 58:2053-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paoletti, M., M. Castroviejo, J. Bégueret, and C. Clavé. 2001. Identification and characterization of a gene encoding a subtilisin-like serine protease induced during the vegetative incompatibility reaction in Podospora anserina. Curr. Genet. 39:244-252. [DOI] [PubMed] [Google Scholar]
- 28.Pinan-Lucarré, B., M. Paoletti, K. Dementhon, B. Coulary-Salin, and C. Clavé. 2002. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47:321-333. [DOI] [PubMed]
- 29.Punt, P. J., M. A. Dingemanse, B. J. Jacobs-Meijsing, P. H. Pouwels, and C. A. van den Hondel. 1988. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene 69:49-57. [DOI] [PubMed] [Google Scholar]
- 30.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276:9583-9586. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Saupe, S., C. Descamps, B. Turcq, and J. Bégueret. 1994. Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc. Natl. Acad. Sci. USA 91:5927-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saupe, S. J. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64:489-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 35.Schneiter, R. 1999. Brave little yeast, please guide us to thebes: sphingolipid function in S. cerevisiae. Bioessays 21:1004-1010. [DOI] [PubMed] [Google Scholar]
- 36.Thines, E., R. W. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turcq, B., and J. Bégueret. 1987. The ura5 gene of the filamentous fungus Podospora anserina: nucleotide sequence and expression in transformed strains. Gene 53:201-209. [DOI] [PubMed] [Google Scholar]
- 38.Vaux, D. L., and S. J. Korsmeyer. 1999. Cell death in development. Cell 96:245-254. [DOI] [PubMed] [Google Scholar]
- 39.von Haefen, C., T. Wieder, B. Gillissen, L. Starck, V. Graupner, B. Dorken, and P. T. Daniel. 2002. Ceramide induces mitochondrial activation and apoptosis via a Bax-dependent pathway in human carcinoma cells. Oncogene 21:4009-4019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]