Abstract
A genetic approach utilizing the yeast Saccharomyces cerevisiae was used to identify the target of antifungal compounds. This analysis led to the identification of small molecule inhibitors of RNA polymerase (Pol) III from Saccharomyces cerevisiae. Three lines of evidence show that UK-118005 inhibits cell growth by targeting RNA Pol III in yeast. First, a dominant mutation in the g domain of Rpo31p, the largest subunit of RNA Pol III, confers resistance to the compound. Second, UK-118005 rapidly inhibits tRNA synthesis in wild-type cells but not in UK-118005 resistant mutants. Third, in biochemical assays, UK-118005 inhibits tRNA gene transcription in vitro by the wild-type but not the mutant Pol III enzyme. By testing analogs of UK-118005 in a template-specific RNA Pol III transcription assay, an inhibitor with significantly higher potency, ML-60218, was identified. Further examination showed that both compounds are broad-spectrum inhibitors, displaying activity against RNA Pol III transcription systems derived from Candida albicans and human cells. The identification of these inhibitors demonstrates that RNA Pol III can be targeted by small synthetic molecules.
Defining the mechanism of action of small molecules in higher eukaryotes is hindered by their complexity, limited genetic methods, and protracted life cycles. In contrast, model eukaryotes are amenable to extensive genetic analyses in relatively short time frames. For example, unicellular eukaryotes, such as Saccharomyces cerevisiae and Aspergillus nidulans, have been used to rapidly elucidate the mechanism of action of many compounds, including drugs that are relevant to human therapeutics. The targets of the immunosuppressive compounds cyclosporine, FK506, and rapamycin were discovered in studies with yeast (9, 24). Resistance to cerulenin in S. cerevisiae was mapped to FAS2, and inhibition of the encoded enzyme, fatty acid synthase, was demonstrated biochemically (21). Similarly, isolation of an A. nidulans mutant resistant to a novel agricultural antifungal compound led to identification of dihydroorotate dehydrogenase as the target (15).
In the present report, we extend the utility of S. cerevisiae for discovering and characterizing antifungal compounds. Our initial studies focused on UK-118005, a compound that has broad-spectrum antifungal activity. Using classical genetics, molecular biology, and biochemistry, we show that UK-118005 inhibits RNA polymerase (Pol) III. Although natural products have been identified that inhibit different RNA Pols, such as α-amanitin (32, 33) and tagetitoxin (34, 35), this is the first example of a synthetic small molecule inhibiting RNA Pol. This finding demonstrates that cell-based screening can be a powerful method for identifying novel druggable targets.
Additional antifungal structural analogs of UK-118005 were identified and further characterized. These results showed that whereas some analogs inhibit RNA Pol III as expected, others caused growth inhibition by an entirely different mechanism. Thus, yeast can be used to monitor changes in the mechanism of action that occur during lead compound optimization.
MATERIALS AND METHODS
Strains and media.
Plasmid pJCP1 was derived from pcDNAII (Invitrogen) and contains an ∼1.45-kb mini-Tn7 stuffer fragment conferring kanamycin resistance cloned into the PstI site (30). A URA3 fragment was inserted into pJCP1 with a 2-micron origin to create pBM601. The plasmids pRPO31 and pRPO31-G1101Sare derived from pBM601 and contain the wild-type RPO31 gene and the mutant Rpo31 gene conferring resistance to UK-118005, respectively. The plasmids pSUP4 and pSUP53 are derived from pCR-TOPO (Invitrogen) and contain the SUP4 and SUP53 tRNA genes, respectively. Strains used in the present study are listed in Table 1 and were grown in YPD medium (1), Sabouraud dextrose broth (SDB; Difco) or synthetic dextrose (SD; yeast nitrogen base; Invitrogen) with required supplements.
TABLE 1.
Description of strains
| Strain | Genotype | Note(s) | Reference or source |
|---|---|---|---|
| BY4741 | MATahis3Δ1. ura3Δ0. leu2Δ0 met15Δ0 | 4 | |
| BY4742 | MATα his3Δ1. ura3Δ0 leu2Δ0 lys2Δ0 | 4 | |
| BY4743 | MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 MET15/met15Δ0 LYS2/lys2Δ0 | 4 | |
| MF37 | MATaade1::URA3 ura3-52 | Millennium strain collection | |
| MMB2175 | MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 MET15/met15Δ0 LYS2/lys2Δ0 RPO31-G3301A | UK-118005-resistant mutant isolated by EMS mutagenesis of BY4743 | This study |
| MMB2177 | MATα his3Δ1 ura3Δ0 leu2Δ0 RPO31-G3301A | UK-118005-resistant haploid from mutant MMB2175 | This study |
| MMB2154 | MATa/α his3Δ1/his3Δ1 ura3Δ0/ura3-52 LEU2/leu2Δ0 ADE1/ade1::URA3 RPO31/RPO31-G3301A | UK-118005-resistant diploid obtained from MMB2177 × MF37 | This study |
| MMB2404 | MATα ura3 leu2Δ0 RPO31-G3301A | UK-118005-resistant haploid from MMB2154 | This study |
| MMB1487 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 pdr5Δ::HIS3 snq2Δ::HIS3 | Millennium strain collection | |
| MMB1489 | MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 pdr5Δ::HIS3/pdr5Δ::HIS3 snq2Δ::HIS3/snq2Δ::HIS3 | 12 | |
| MMB1576 | MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 pdr5Δ::HIS3/pdr5Δ::HIS3 snq2Δ::HIS3/snq2Δ::HIS3 gcn4Δ::KanMX/gcn4Δ::KanMX | This study | |
| Y01.06 (C. albicans) | Clinical isolate | This study |
MIC test and IC50 determination.
The MIC of antifungal compounds was determined by a whole-cell assay in a 96-well plate format. Yeast cells with an initial cell optical density at 600 nm (OD600) of 0.001 in SDB medium were inoculated with serial dilutions of compounds in the SDB medium. Growth inhibition was measured by determining the OD600 at 48 h. The lowest concentration at which a compound led to an OD600 of ≤0.010 was determined as the MIC of the compound; the concentration that causes 50% growth inhibition (IC50) of the compound was also determined. The MIC for UK-118005 increases with increasing starting inoculum, as has been reported for other compounds (reviewed in reference 31). However, the MIC remains consistent at 24 and 48 h.
Mutant isolation and genetic analysis.
S. cerevisiae BY4743 (Table 1) was grown to log phase in YPD, washed twice, and resuspended in 0.1 M sodium phosphate buffer (pH 7.0) to 2 × 108 cells/ml. For mutagenesis, ethyl methanesulfonate (EMS; Sigma) was added to 1 ml of cells to a final concentration of 3%, and the cells were shaken at 30°C for 1 h. This treatment resulted in 50 to 70% killing of the cells. The EMS treatment was terminated by the addition of 5 volumes of freshly prepared sterile 5% sodium thiosulfate (Sigma). Mutagenized cells were washed twice with 5% sodium thiosulfate and once with sterile water and were then plated onto SDB agar plates containing UK-118005 at concentrations of 117 and 234 μM (four and eight times the MIC). The plates were then incubated at 30°C to allow resistant colonies to form. Resistance was confirmed by streaking onto fresh SDB agar plates with the compound at the concentration of 117 μM. Standard genetic methods (1, 16) were used to evaluate the number of genes responsible for conferring resistance and to determine the dominance.
Construction of genomic DNA library from UK-118005 resistant mutant cells.
Genomic DNA prepared from the UK-118005 resistant mutant, MMB2404 (Table 1), was partially digested separately with AluI, HaeIII, and EcoRV. The digested DNAs were pooled and size fractionated on a 1% agarose gel. DNA fragments of 4 to 7 kb were isolated from the gel, ligated with the BstXI linkers (5′-CTCTAAG-3′ and 5′-ACACGAGATTC-3′) and inserted into the pBM601 vector at the BstXI site. The library, with a 20-fold coverage of the yeast genome, was amplified in DH10B E. coli cells (Invitrogen) and then transformed into yeast strain BY4741 (Table 1) by using the lithium acetate yeast transformation method as described previously (1). Transformants were selected on synthetic medium minus uracil agar plates containing 176 μM UK-118005.
Subcloning of the wild-type and mutant RPO31 genes.
The wild-type and the mutant RPO31 genes, each with a 700-bp upstream region, were cloned by PCR, by using the primer pair of RPO31-f (5′-CTGCAGAACCAGTGTGCTGGAGACAAACTCCTGATGTGCC-3′) and RPO31-r (5′-ATGCATCCAGTGTGATGGTTATCTTCCAACTTTATAACCG-3′) and inserted into the 2-micron pBM601 vector at the BstXI site. The constructs containing either insert were sequence verified and transformed into strain BY4741 (Table 1).
RNA analysis.
To isolate RNA for northern analysis, wild-type yeast cells were grown in SDB media until the culture reached an OD600 of 0.05. This culture was then divided into three subcultures, and the UK-118005 compound was added to the final concentrations of 0, 15, and 30 μM, which are equivalent to nontreatment, 0.13× MIC, and 0.25× MIC, respectively, under this inoculation condition. The three subcultures were incubated at 30°C with shaking. Samples were taken from each subculture at 0, 1, 2, and 4 h, and the cells were harvested by centrifugation. Total RNA was isolated from each culture by using Trizol reagent (Invitrogen) according to the manufacturer's directions.
For Northern analysis, 15 μg of RNA samples in loading buffer (90% formamide, 1% bromphenol blue, 1% xylene cyanole) were heated 10 min at 75°C and run on 8% polyacrylamide-8 M urea-1× Tris-borate-EDTA gels. Separated RNA samples were transferred electrophoretically onto positive charged nylon membrane (Amersham) in 0.5× Tris-borate-EDTA and immobilized onto the membrane by UV cross-linking. Hybridization was performed as previously described (5, 6) by using a radiolabeled oligonucleotide complementary to the 5′ sequence of the initiator methionyl-tRNA (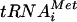 ): 5′-TCGTTTCGATCCCGACATCAGGGTTATGA-3′. The detected tRNA level was quantified by analysis on a Fujifilm BAS-2500 phosphorimager (Fuji Medical Systems, Stamford, Conn.).
): 5′-TCGTTTCGATCCCGACATCAGGGTTATGA-3′. The detected tRNA level was quantified by analysis on a Fujifilm BAS-2500 phosphorimager (Fuji Medical Systems, Stamford, Conn.).
To isolate RNA for transcript profiling, overnight cultures of cells were grown in SDB medium supplemented with 200 mg of histidine, 200 mg of uracil, and 300 mg of leucine/liter to mid-log phase (OD600 = 0.3). Cells were pelleted in 50-ml conical tubes and resuspended in fresh medium at an OD600 of 0.05. After the addition of 3 or 6 μg of UK-118005 or 0.75 μg of ML-22952 (dissolved in dimethyl sulfoxide [DMSO])/ml, all cultures (including untreated controls) were brought to 1% DMSO. Cells were cultured at 30°C with shaking (250 rpm) for 3 h before being pelleted and then frozen on dry ice.
DNA microarray production and transcript profiling were performed as described previously (12). For DNA microarray production, 6,144 DNAs representing different yeast open reading frames (19) were obtained from Research Genetics (Huntsville, Ala.) and amplified by PCR. Approximately 20 nl of each was spotted onto nylon membranes (Biodyne B; Invitrogen, Carlsbad, Calif.) at a density of ∼64/cm2. After being spotted, arrays were treated in 0.4 M NaOH, neutralized in 0.1 M Tris-HCl (pH 7.5), rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and dried to completion.
For transcript profiling, yeast total RNA was isolated by using Trizol (Invitrogen) according to manufacturer's directions. A total of 15 μg of total RNA and 1.5 μg of oligo(dT)12-18 were incubated with SuperScript II (Invitrogen) at 42°C for 1 h in the presence of 160 μM concentrations of dATP, dGTP, and dTTP; 1.6 μM dCTP; and 50 μCi of [α-33P]dCTP (2,000 to 4,000 Ci/mmol). Labeled cDNA was purified (Micro Bio-Spin 6 column; Bio-Rad, Hercules, Calif.), treated with 50 mM NaOH at 68°C for 15 min, and split into separate tubes containing duplicate nylon arrays. The hybridization and washing conditions were done essentially as previously described (8). Hybridization solutions contained 7% sodium dodecyl sulfate (SDS), 0.25 M sodium phosphate (pH 7.2), 1 mM EDTA, and 0.5% casein (Hammerstein grade). After overnight hybridization at 68°C, the filters were washed once for 15 min at 68°C in 4% SDS-0.02 M sodium phosphate (pH 7.2)-1 mM EDTA, three times for 15 min at 68°C in 1% SDS-0.02 M sodium phosphate (pH 7.2)-1 mM EDTA, and briefly at 22°C in 2× SSC. Dried filters were exposed to phosphorimager screens overnight. The hybridization signals were captured by a Fuji BAS-2500 phosphorimager and quantified by Grid Guru software (Millennium Pharmaceuticals, Cambridge, Mass.). For each array, the distribution of intensities across all yeast genes was normalized to a median of 1. Intensity values for duplicate filters were then averaged and analyzed with Spotfire software (Somerville, Mass.). Ratios were determined from data from drug-treated samples on a per-gene basis compared to the appropriate control sample.
Preparation of yeast nuclear extract.
Yeast subcellular extract was prepared as described previously (11). Cells were grown in YPD medium at 30°C to late log phase, harvested by centrifugation, and resuspended in sorbitol buffer (1 M sorbitol, 50 mM Tris [pH 7.9], 10 mM MgCl2) containing 30 mM dithiothreitol (DTT). After incubation at room temperature for 15 min, the suspension was spun at 1,500 × g for 5 min. The cell pellet was resuspended in sorbitol buffer containing 3 mM DTT. 60T zymolyase was added to the suspension to a final concentration of 0.15 mg/ml, and the lysis reaction was carried out at 30°C with gentle swirling for 40 min. Cells were collected by centrifugation at 1,500 × g for 5 min, washed with sorbitol buffer containing 3 mM DTT, and resuspended in ice-cold hypotonic buffer (15 mM KCl, 10 mM HEPES [pH 7.9], 5 mM MgCl2, 0.1 mM EDTA, 3 mM DTT) for 20 min after gentle homogenization. The nuclear pellet was obtained by centrifugation at 10,000 × g for 20 min, resuspended in hypotonic buffer, and extracted with one-fifth volume of 4 M (NH4)2SO4 (pH 7.9). The tube was set on ice for 30 min and then centrifuged at 100,000 × g for 60 min. The supernatant was filtered through cheesecloth, and solid (NH4)2SO4 (Sigma) was added to a final concentration of 0.25 g/ml. After centrifugation (30 min at 10,000 × g), the protein pellet was resuspended in one-half of the measured “nuclear pellet” volume with KBC 100 buffer (20 mM HEPES-KOH [pH 7.9], 0.2 mM EDTA, 20% glycerol, 100 mM KCl, 1 mM DTT) containing protease inhibitors at a concentration of one EDTA-free protease inhibitor cocktail tablet (Boehringer Mannheim) per 50 ml of KBC100 buffer and then aliquoted for storage at −80°C.
The extract from Candida albicans was prepared as described above. The human HEK293 cytosolic extract used for testing the Pol III inhibitors in the in vitro Pol III transcription assays was obtained from R. H. Lambalot (Pfizer Global Research and Development, Cambridge, Mass.), which was prepared as previously described (25).
In vitro yeast RNA Pol III transcription assay.
The reaction was carried out in a total volume of 50 μl. In each reaction tube 1.2 mM ATP, 0.6 mM CTP, 0.6 mM UTP, 25 μM GTP, 160 mM KCl, 10 mM MgCl2, 2 mM DTT, 10 μCi of [α-33P]GTP, and 0.04 to 1 μg of DNA template (e.g., pSUP4 or pSUP53 plasmids) were added, and the volume was adjusted to 25 μl (29). The reactions were initiated by addition of 25 μl of nuclear extract or whole-cell extract mixture; the wild-type and mutant subcellular extracts were adjusted to 50 μg of total protein (in KBC100) per reaction in the in vitro assays as determined by Bradford assay (Bio-Rad). The reactions were incubated at 15°C with constant shaking at 150 rpm for 1 h unless stated otherwise. The reaction was terminated by the addition of 70 μl of proteinase K (1 mg/ml in 10 mM Tris-HCl [pH 7.5]-0.1% SDS) and 5 μl of 10% SDS, followed by incubation at 37°C for 30 min. Phenol-chloroform-isoamyl alcohol (24:24:1) extraction was then performed. A total of 43 μl of 7.5 M ammonium acetate (pH 7.0) and 3 volumes of ethanol were added to the aqueous phase to precipitate the RNA. The RNA pellet was washed with 70% ethanol, dried in a Speed-Vac, and dissolved in 7 μl of RNA loading buffer (see above) for acrylamide gel electrophoresis. The activity of the RNA Pol III in vitro transcription was quantitated by measuring the photostimulated luminescence units from radiolabeled tRNA products on a Fujifilm BAS-2500 phosphorimager.
RESULTS
Identification of UK-118005 as an antifungal compound.
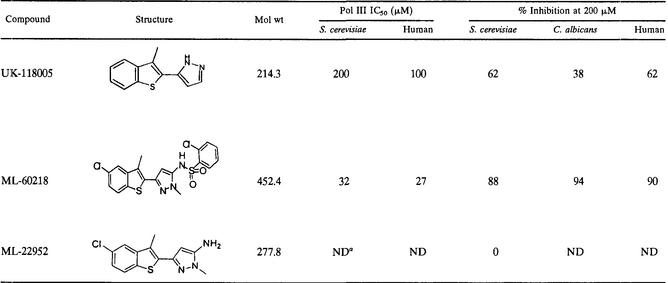
To identify antifungal compounds, a screen was carried out by using growth inhibition as the endpoint. The yeast C. albicans Y01.06 was grown in the presence of a library of compounds with the OD as an indicator for growth. UK-118005 (Table 2) was identified as a chemically acceptable, broad-spectrum antifungal compound, as determined by MIC testing, and was subjected to mechanism-of-action studies. UK-118005 inhibits the growth of both the wild-type BY4743 S. cerevisiae strain and an isogenic strain, MMB1489, deficient in two drug efflux pumps (pdr5 and snq2) that is more susceptible to some antifungal agents since export of the agents is compromised. Both strains have an MIC of 58 μM (12.5 μg/ml) and an IC50 of ∼10 μM.
TABLE 2.
Structures and IC50s of UK-118005 and two analogs
Note: nd = not determined
Transcript profiling of S. cerevisiae treated with UK-118005.
To gain insight into the pathways targeted by UK-118005, transcript profiling was performed to measure the transcriptional response to drug treatment. RNA was recovered from cultures treated for 3 h and compared to samples from untreated cultures. The most striking feature of the drug-treated samples was the upregulation of many genes associated with amino acid biosynthesis (Fig. 1A). This response is reminiscent of the well-characterized general amino acid control response, which is mediated by the transcription factor Gcn4p and results from amino acid or purine starvation or an imbalance of charged tRNAs (17). To determine whether UK-118005 was inducing the general control response, we profiled the mRNA from a gcn4Δ strain treated with UK-118005. In contrast to the wild-type strain, the amino acid pathway genes were not induced in the gcn4Δ mutant (Fig. 1B), showing that GCN4 is required for this response to UK-118005. If the induction of the general amino acid control response is due to amino acid or purine depletion, then addition of these nutrients to the medium is expected to bypass the effect of UK-118005. However, the MIC for UK-118005 is unaffected by amino acid or purine supplementation. Thus, the antifungal activity of UK-118005 is not due to inhibition of amino acid or purine biosynthesis. The possible relevance of the GCN4-dependent response to UK-118005 is discussed below.
FIG. 1.
Transcript profiling reveals that UK-118005 treatment induces a GCN4 response. (A and B) Transcript profiling of cultures of MMB1489 (GCN4+) (A) and MMB1576 (gcn4Δ) (B). In each panel, normalized intensity values from untreated cultures (x axis) are compared to treated samples (y axis). Only genes whose normalized intensity is >0.1 are shown, since this value represents the approximate intensity at which the signal-to-noise ratio decreases sharply. (A) A set of known Gcn4p target genes (blue squares) (28) involved in amino acid biosynthesis is significantly upregulated in UK-118005-treated (3 μg/ml) cells. These target genes comprise the bulk of the highly induced genes in this data set. (B) In gcn4Δ cells, a similar treatment (6 μg/ml) produces few highly induced genes. For comparison, the set of genes encoding the subunits of the 26S proteasome (red triangles) (12) are unchanged by the treatment in either genetic background. For the relevance of this finding, please refer to Fig. 8.
Isolation of UK-118005 resistant mutants.
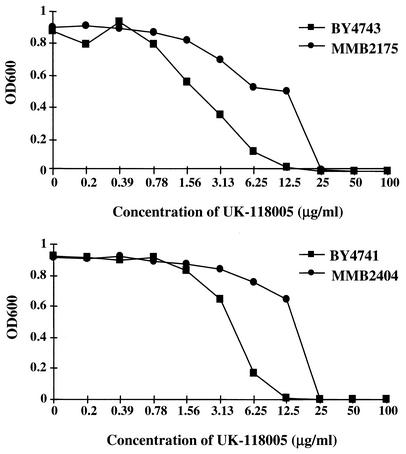
Previous studies on the mechanism of action of antifungal compounds demonstrate the utility of resistant mutants in identifying the target. Of particular value are dominant mutations in the target protein that lead to decreased binding of the inhibitor relative to the substrate (15, 21). This strategy was adopted to identify the molecular target of UK-118005. Diploid BY4743 cells were mutagenized with EMS and plated on medium containing UK-118005. Strain MMB2175 (Table 1) was obtained that has a twofold increase in MIC and a fourfold increase in IC50 (Fig. 2). Tetrad analysis of the diploid mutant was performed and, after two back-crossings, resistance to UK-118005 segregated 2:2 (resistant:sensitive), indicating that a single locus is responsible for the resistance. To verify that the mutation is dominant to the wild-type allele, the resistant haploid strain MMB2177 was crossed to the wild-type haploid MF37. The resulting diploid, MMB2154 (Table 1), also displayed a fourfold increase in IC50 compared to the wild-type parental diploid strain, indicating that the resistance conferred by the mutant gene is dominant to its wild-type allele.
FIG. 2.
Growth inhibition of S. cerevisiae with UK-118005 treatment. Growth inhibition by UK-118005 was assayed as described in Materials and Methods. The two mutant strains (•) are more resistant than the wild-type cells (▪), as indicated by at least a fourfold increase in IC50 after 48 h of treatment with UK-118005. (Top panel) diploid strains; (bottom panel) haploid strains. The IC50 is the concentration of compound that inhibits growth by 50%.
A mutation in RPO31 confers resistance to UK-118005.
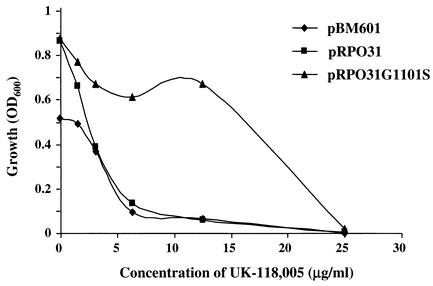
To identify the mutated gene, a genomic DNA library was constructed from the resistant mutant MMB2404 (Table 1). The library was transformed into BY4741 and screened for colonies resistant to UK-118005. Six resistant colonies were isolated that had a twofold increase in MIC and a fourfold increase in IC50 compared to the control strain containing only the vector. Retransformation of the plasmids rescued from these six clones into the MMB1487 strain (Table 1) yielded resistant transformants, indicating that the resistance was due to the plasmid rather than a spontaneous chromosomal mutation. Sequencing of the plasmids showed that RPO31 was the only complete gene common to all six plasmids. RPO31, encoding a 1,460-amino-acid protein, is the largest subunit of the 17-subunit yeast RNA Pol III complex and is a component of the catalytic core of the enzyme (2, 3, 10). Since a 2-micron plasmid was used for cloning, it was possible that the resistance was due to overexpression of wild-type Rpo31p. To eliminate this possibility, the mutant and the wild-type RPO31 genes were cloned into the 2-micron pBM601 vector and transformed into strain BY4741 (Table 1). MIC testing of the transformants showed that only the strain expressing the mutant RPO31 (Fig. 3) was resistant to UK-118005. This demonstrates that a mutation in RPO31, rather than the overexpression of the wild-type protein, confers resistance.
FIG. 3.
Cloned mutant allele of RPO31 confers resistance to UK-118005. MIC determination for the wild-type haploid strain BY4741 containing either pBM601 (vector, no insert [♦]), pRPO31-G1101S (▴), or pRPO31 (▪) after 48 h of treatment with serial dilutions of UK-118005.
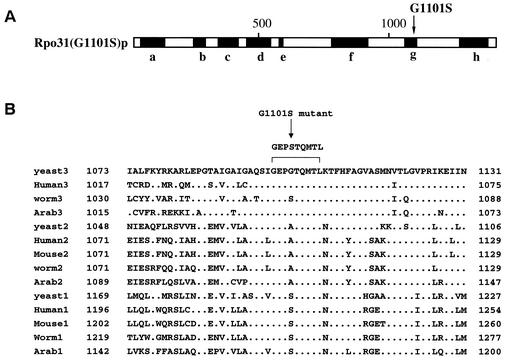
The Rpo31p protein contains eight major conserved regions (a to h) (2, 3, 10, 13, 23). Sequencing the mutant and wild-type RPO31 genes identified a single nucleotide change, G to A, at nucleotide position 3301, changing the amino acid at position 1101 from glycine to serine in region g, which is conserved among RNA Pol I, Pol II, and Pol III from different eukaryotes (Fig. 4), as well as Pols from prokaryotes. Although the function of region g is unclear, our data show that the G1101S mutation leads to resistance of S. cerevisiae cells to UK-118005.
FIG. 4.
(A) Site of mutation in Rpo31-G1101S mutant protein. A schematic diagram showing the eight conserved domains of Rpo31 protein and the single amino acid change from glycine (G) to serine (S) at amino acid residue 1101 in the g domain of the Rpo31 protein is shown. (B) Comparisons of homologous amino acid sequence motif g of analyzed RNA Pol large subunits from S. cerevisiae (yeast), human, mouse, Caenorhabditis elegans (worm), and Arabidopsis thaliana. The Pols from different organisms are indicated by the names of the organisms followed by 1 (Pol I), 2 (Pol II), and 3 (Pol III). Amino acid positions are given at the beginning and end of each sequence. Identical residues with the RPO31p g motif sequence (RNA Pol III) are indicated by points. The G1101S mutation site is indicated by an arrow, and its conserved surrounding amino acid residues (discussed in the text) are overlined and displayed above the overline.
Inhibition of Pol III by UK-118005 correlates with growth inhibition in yeast.
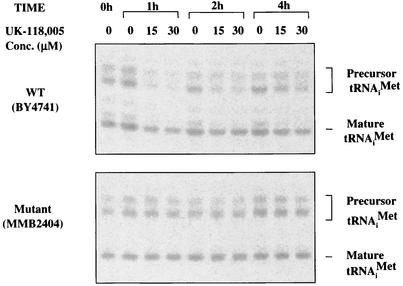
RNA Pol III transcribes 5S rRNA, tRNAs, and various small nuclear RNAs (14, 33). Because Rpo31p is essential for the catalytic activity of RNA Pol III (10, 13, 23), a Northern hybridization experiment was performed to examine whether UK-118005 impairs the activity of RNA Pol III in vivo (Fig. 5). Cells were treated with UK-118005 at doses that partially inhibited growth, and RNA was extracted after 1, 2, and 4 h of treatment. 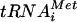 was detected by using a probe that hybridizes to both precursor and mature
was detected by using a probe that hybridizes to both precursor and mature 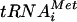 . The compound treatment of wild-type cells led to significant inhibition of the RNA Pol III activity, as evidenced by a rapid decrease in the level of newly synthesized
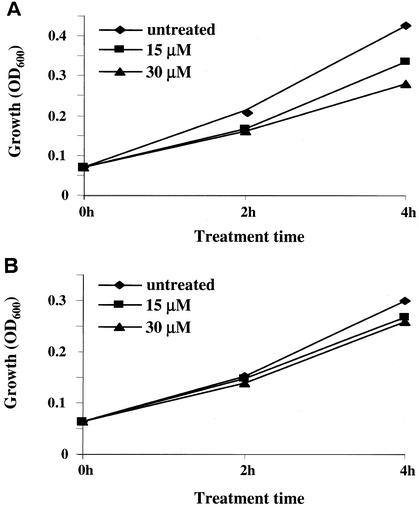
. The compound treatment of wild-type cells led to significant inhibition of the RNA Pol III activity, as evidenced by a rapid decrease in the level of newly synthesized 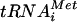 precursors within 1 h of addition of the compound (Fig. 5, top panel). The decrease of in vivo tRNA synthesis was accompanied by a comparable decrease in cell growth rate in a dose dependent manner (Fig. 6). Furthermore, protein synthesis was inhibited, as expected for an inhibitor of tRNA synthesis (data not shown). In contrast, UK-118005 treatment of the resistant mutant MMB2177 had little or no effect on in vivo tRNA synthesis (Fig. 5, bottom panel) or cell growth (Fig. 6). Quantitation of precursor tRNA after 1 h of treatment demonstrates 60 to 63% inhibition for the wild type and 2 to 13% for the RPO31 mutant strain (15 to 30 μM, respectively). The wild-type strain treated with UK-118005 demonstrates some recovery of tRNA synthesis after 4 h since it was treated with sublethal concentrations and is still able to grow (Fig. 6). In summary, we have shown that the inhibition of growth and cellular tRNA synthesis by UK-118005 are tightly correlated. Because both of these effects are mitigated by a dominant mutation in RNA Pol III, it is likely that this enzyme is the target of UK-118005.
precursors within 1 h of addition of the compound (Fig. 5, top panel). The decrease of in vivo tRNA synthesis was accompanied by a comparable decrease in cell growth rate in a dose dependent manner (Fig. 6). Furthermore, protein synthesis was inhibited, as expected for an inhibitor of tRNA synthesis (data not shown). In contrast, UK-118005 treatment of the resistant mutant MMB2177 had little or no effect on in vivo tRNA synthesis (Fig. 5, bottom panel) or cell growth (Fig. 6). Quantitation of precursor tRNA after 1 h of treatment demonstrates 60 to 63% inhibition for the wild type and 2 to 13% for the RPO31 mutant strain (15 to 30 μM, respectively). The wild-type strain treated with UK-118005 demonstrates some recovery of tRNA synthesis after 4 h since it was treated with sublethal concentrations and is still able to grow (Fig. 6). In summary, we have shown that the inhibition of growth and cellular tRNA synthesis by UK-118005 are tightly correlated. Because both of these effects are mitigated by a dominant mutation in RNA Pol III, it is likely that this enzyme is the target of UK-118005.
FIG. 5.
Inhibition of tRNA transcription in vivo by UK-118005. A Northern blot shows a time course of treatment of wild-type BY4741 cells (top panel) and rpo31 mutant MMB2404 cells (bottom panel) either untreated (lanes 0) or treated with 15 μM or 30 μM UK-118005 as indicated (see Materials and Methods for details). The migration of the precursor and mature 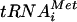 is indicated.
is indicated.
FIG. 6.
Comparison of growth inhibition by UK-118005. Growth inhibition by UK-115005 in wild-type BY4741 (A) and the resistant mutant MMB2404 (B) was measured by taking OD600 readings of the same cultures as analyzed by Northern in Fig. 5 at the times indicated. Strains were treated with 15 μM (▪) or 30 μM (▴) UK-118005 or no UK-118005, i.e., the untreated control (♦).
UK-118005 inhibits Pol III activity in vitro.
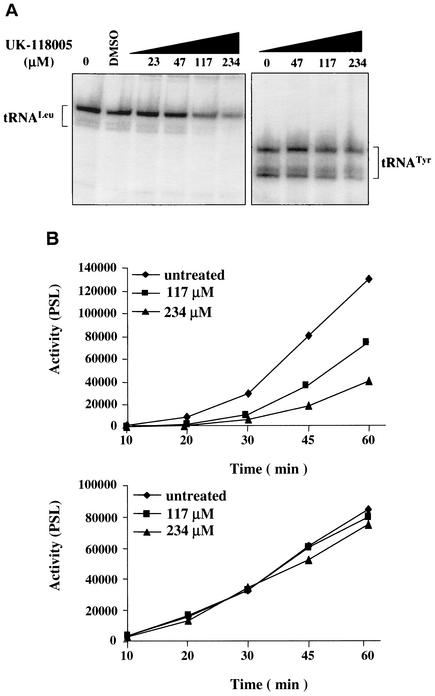
We next examined whether UK-118005 inhibits Pol III activity in cell extracts. Nuclear extracts were prepared from exponentially growing cells of both the parental wild-type and the resistant mutant strains. A portion of each crude nuclear extract was also passed through a Bio-Rex 70 column and eluted with 500 mM KCl to prepare a fraction (BRα) enriched for RNA Pol III and its transcription factors, TFIIIC and TFIIIB (22). In vitro transcription assays were carried out with either the yeast SUP4 gene (encoding tRNATyr) or the SUP53 gene (encoding tRNALeu) as the template, and the tRNA transcripts were quantified after polyacrylamide gel separation. Robust transcription of both templates was observed with the crude subcellular extract (Fig. 7A). This level of transcription was not affected by the addition of DMSO (used to dissolve the inhibitor). However, the addition of UK-118005 markedly reduced the transcription of both templates (up to 70%) in the wild-type crude extract in a dose-dependent manner (Fig. 7A). Similar results were obtained with assays utilizing the BRα fraction (data not shown). From these data, we determined the IC50 for UK-118005 to be 200 μM (Table 2).
FIG. 7.
Inhibition of tRNA transcription in vitro by UK-118005. (A) In vitro transcription of SUP53 (left panel) and SUP4 (right panel) in the presence or absence of UK-118005 for 1 h; (B) quantitation of dose-dependent inhibition of SUP4 in vitro transcription by UK-118005. Quantitation of radiolabeled tRNA is expressed in photostimulated luminescence (PSL) units. (Top panel) transcription with wild-type RNA Pol III nuclear extract; (bottom panel) transcription with RPO31-G1101S mutant RNA Pol III nuclear extract.
A time course of in vitro transcription was carried out in the presence or absence of different concentrations of the compound. After the usual lag phase (∼30 min) for complex assembly, transcription proceeded linearly at a rate that decreased with increasing concentrations of inhibitor (Fig. 7B, top panel). In contrast to the wild-type extract, assays with extracts from the resistant strain showed only a minor effect on RNA Pol III transcription at 200 μM (Fig. 7B, bottom panel). The assays were done with crude nuclear preparations, and there was some assay variability, which may account for some of the differences in kinetics of the two assays. However, this assay was run numerous times, and the same level of inhibition was detected each time. UK-118005 also inhibited transcription with a nonspecific template under assay conditions in which transcription factors are not required for Pol III transcription. These data demonstrate a direct inhibition of RNA Pol III activity by UK-118005. Therefore, both in vivo and in vitro, UK-118005 inhibits yeast cell growth by targeting and impairing the activity of RNA Pol III.
Characterization of structural analogs of UK-118005.
Twenty-nine analogs of UK-118005 were identified and characterized further. To determine the activity against Pol III, each was tested in the in vitro transcription assay. Ten of the twenty-nine compounds inhibited yeast RNA Pol III. One of the more potent analogs is ML-60218. This compound was found to have an IC50 of 32 μM (Table 2). To address whether UK-118005 and ML-60218 exhibit broad-spectrum activity, a subcellular extract was prepared from another yeast, C. albicans, by the same protocol as that used for S. cerevisiae. This extract was active in RNA Pol III gene transcription and, together with an extract from the human cell line HEK293, was used to test the compounds for their inhibitory activity. Results from these assays that were run in triplicate show that both compounds inhibited the in vitro transcription activity of S. cerevisiae, C. albicans, and human RNA Pol III and that ML-60218 is more potent than UK-118005 in all three systems (Table 2). Thus, these compounds represent novel inhibitors with activity against RNA Pol III enzymes from yeast to humans.
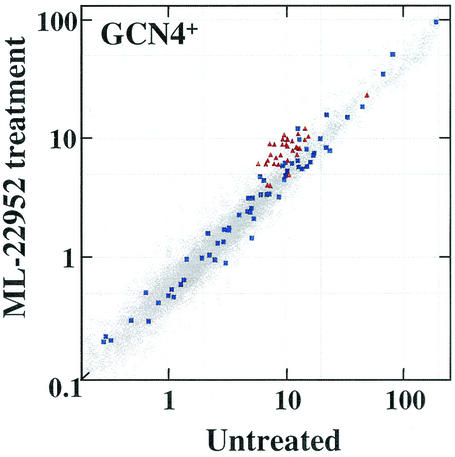
Among the structural analogs of UK-118005 that did not inhibit RNA Pol III activity, one (ML-22952) had been previously identified in a screen for antifungal compounds and had been analyzed by transcript profiling. These profiling results clearly indicated that ML-22952 does not induce amino acid biosynthetic genes but rather induced genes involved in regulated protein degradation (Fig. 8). Thus, the profiling results are consistent with the in vitro Pol III transcription data and indicate that the antifungal activity of ML-22952 is mechanistically distinct from UK-118005. These results demonstrate that similar chemical entities can have very different effects on the cell and that transcript profiling is a valuable tool for classifying compounds at an early stage of the drug discovery process.
FIG. 8.
Transcript profiling reveals that ML-22952 treatment induces proteasome subunit genes. Transcript profiling of ML-22952-treated cells. Scatter plot data are presented as in Fig. 1, as are the genes encoding the subunits of the 26S proteasome (red triangles) and a set of known Gcn4p target genes (blue squares). Although the upregulation of proteasome subunits is not high, this increase is physiologically relevant (12).
DISCUSSION
This report demonstrates a successful effort of using yeast to characterize the mechanism of action of a small bioactive compound, which has led to the identification of novel inhibitors that can prevent cell growth by targeting RNA Pol III.
RNA Pol III is a large and complex enzyme of multiple subunits. The composition of Pol III subunits has been well characterized, especially in yeast cells, by both mutational analysis and two-hybrid screening (7, 13). However, the functions of these proteins and their interactions are not fully understood. Of the few inhibitors of RNA Pol III that have been described to date, those in common use include two natural products: α-amanitin purified from fungi and tagetitoxin from bacteria. The octapeptide α-amanitin is only a weak inhibitor of S. cerevisiae RNA Pol III (compared to the mammalian enzyme) and is significantly more potent against yeast RNA Pol I and Pol II (32, 33, 36). Tagetitoxin, on the other hand, is considered a selective inhibitor of RNA Pol III (34, 35). Neither of these compounds are growth inhibitory for yeast. Through both genetic screening and template-specific in vitro RNA Pol III transcription assays, we identified novel inhibitors of Pol III with significant potency both in vivo and in vitro. These inhibitors represent the first group of small synthetic compounds that target RNA Pol III, suggesting that the RNA Pol III is tractable to inhibition by small molecules.
We have not yet tested the inhibitors against purified RNA Pol I and Pol II and therefore do not know whether they are selective for RNA Pol III. However, several observations suggest that UK-118005 is not a general RNA Pol inhibitor: (i) in vitro transcription by T7 RNA Pol was unaffected by 100 μM UK-118005; (ii) transcript profiling with UK-118005 did not reveal any significant negative affects on RNA Pol II and the transcription of some Pol II genes was induced (Fig. 2); and (iii) a phylogenetic analysis (described below) of the mutation site in conserved region g of the largest subunit of nuclear RNA Pol suggests that UK-118005 may be a specific inhibitor of RNA Pol III. The sequence of domain g around the mutation site (GEPXTQMTL, where X is the mutation site) is conserved in nuclear RNA Pols from yeast to humans (Fig. 4B). In RNA Pol III, a glycine at the mutation site is changed to serine in the resistant mutant. Interestingly, the corresponding position in RNA Pol I is invariantly serine, suggesting that this enzyme may be naturally resistant to UK-118005. RNA Pol II has a conserved alanine residue at this position. Together, the above observations suggest that inhibition by UK-118005 may require a glycine residue (i.e., no side chain) at the site of the resistance mutation in domain g. Presumably, other residues at this site would diminish inhibitor binding due to steric effects.
We present evidence that UK-118005 inhibits transcription of tRNA both in vivo and in vitro. UK-118005 significantly inhibits the wild-type RNA Pol III transcription activity, with only a minor effect on the G1101S mutant Pol III enzyme. Although the IC50 for UK-118005 from the in vitro assay is higher than might be expected compared to the concentration needed to inhibit growth, we have found that these values cannot be directly compared. The in vitro inhibition can vary depending on the assay conditions, such as DNA or nuclear extract concentration. Similarly, the potency of tagetitoxin can vary by as much as fourfold depending on conditions such as the template and nucleotide concentration (27, 34). However, this variability does not exclude the possibility that UK-118005 has two targets within the cell. A putative second target would account for the residual sensitivity of the RPO31 mutant strain to UK-118005, whereas the RPO31 mutant RNA Pol is nearly completely resistant in the in vitro assay.
Transcript profiling has been successfully applied to mechanism of action studies (12, 20, 26). In the case of UK-118005, however, this approach did not lead directly to the targeted pathway. Our transcript profiling of UK-118005-treated cells clearly showed the induction of the GCN4 response, suggesting that this compound inhibits amino acid biosynthesis. Although this is clearly not the case, how inhibition of RNA Pol III induces the expression of GCN4-regulated genes is unclear. Preliminary studies indicate that induction of the GCN4 response by UK-118005 is GCN2 independent (V. Thoroddsen, unpublished results) and does not increase the translation or stability of the Gcn4 protein (Gcn4p; D. Kornitzer, unpublished results). Although there are several reports of GCN2-independent induction of the GCN4 response, in most cases stimulation of Gcn4p translation is still observed, in contrast to our results with UK-118005 (unpublished data). One exception is mutation of the WD protein CPC2. Deletion of CPC2 increases the transcription of GCN4-regulated genes and bypasses the amino acid analogue (e.g., 3-aminotriazole) sensitivity of gcn2Δ mutants (18). Although these effects require GCN4 function, the level and stability of Gcn4p are unchanged in cpc2Δ mutants. Thus, CPC2 is a negative regulator of general amino acid control, and one possible explanation for our results with UK-118005 is that RNA Pol III plays a role in maintaining, directly or indirectly, this negative regulatory network. Perhaps transcript profiling of the rpo31 mutant strain treated with UK-118005 would reveal more subtle or indirect effects of the compound.
We also used transcript profiling to characterize an analog of UK-118005, ML-22952. The results show that ML-22952 clearly has a different mechanism of action, inducing genes involved in protein degradation rather than the GCN4 response. Interestingly, rpn4 mutants, which are defective in the expression of proteasome subunits (12), are hypersensitive to ML-22952 (unpublished data), a finding consistent with the profiling results. Thus, the transcriptional response of yeast cells to subtle changes in structure of the molecules can provide valuable mechanistic information and can guide structure-activity relationship studies in the absence of specific biochemical assays.
Acknowledgments
We thank Xiaoping Wu for assistance with the human Pol III assays.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 2.Allison, L. A., M. Moyle, M. Shales, and C. J. Ingles. 1985. Extensive homology among the largest subunits of eukaryotic RNA polymerases. Cell 42:599-610. [DOI] [PubMed] [Google Scholar]
- 3.Archambault, J., and J. Friesen. 1993. Genetics of eukaryotic RNA polymerase I, II, and III. Microbiol. Rev. 57:703-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, M. W., and J. S. Butler. 1996. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics 143:1149-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo, O., R. Cuesta, J. Anderson, N. Gutierrez, M. T. Garcia-Barrio, A. G. Hinnebusch, and M. Tamame. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4167-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chedin, S., M. L. Ferri, G. Peyroche, J. C. Andrau, S. Jourdain, O. Lefebvre, M. Werner, C. Carles, and A. Sentenac. 1998. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symposia on Quantitative Biology, vol. LXIII. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [DOI] [PubMed]
- 8.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 18:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutler, N. S., J. Heitman, and M. E. Cardenas. 1999. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol. Cell. Endocrinol. 155:135-142. [DOI] [PubMed] [Google Scholar]
- 10.Dieci, G., S. H.-L. Denmat, E. Lukhtanov, P. Thuriaux, M. Werner, and A. Sentenac. 1995. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 14:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, C. F., and D. R. Engelke. 1988. Yeast extracts for transfer RNA gene transcription and processing. Methods Enzymol. 181:439-450. [DOI] [PubMed] [Google Scholar]
- 12.Fleming, J. A., E. S. Lightcao, S. Sadis, V. Thoroddsen, C. E. Bulawa, and R. K. Blackman. 2002. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA 99:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, A., J.-F. Briand, O. Gadal, J.-C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Aced. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiduschek, E. P., and G. P. Tocchini-Valentini. 1988. Transcription by RNA polymerase III. Annu. Rev. Biochem. 57:873-914. [DOI] [PubMed] [Google Scholar]
- 15.Gustafson, G., G. Davis, C. Waldron, A. Smith, and M. Henry. 1996. Identification of a new antifungal target site through a dual biochemical and molecular-genetics approach. Curr. Genet. 30:159-165. [DOI] [PubMed] [Google Scholar]
- 16.Guthrie, C., and G. R. Fink. 1991. Method in enzymology: guide to yeast genetics and molecular biology. Academic Press, Inc., New York, N.Y. [PubMed]
- 17.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. II. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 18.Hoffmann, B., H.-U. Mosch, E. Sattlegger, I. B. Barthelmess, A. Hinnebusch, and G. H. Braus. 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 31:807-822. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, J. R., E. P. Dawson, K. L. Rushing, C. H. Jackson, D. L. Lockshon, D. Conover, C. Lanciault, J. R. Harris, S. J. Simmons, R. Rothstein, and S. Fields. 1997. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 7:1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, T. R., C. J. Roberts, H. Dai, A. R. Jones, M. R. Meyer, D. Slade, J. Burchard, S. Dow, T. R. Ward, M. J. Kidd, S. H. Friend, and M. J. Marton. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25:333-337. [DOI] [PubMed] [Google Scholar]
- 21.Inokoshi, J., H. Tomoda, H. Hashimoto, A. Watanabe, H. Takeshima, and S. Omura. 1994. Cerulenin-resistant mutants of Saccharomyces cerevisiae with an altered fatty acid synthase gene. Mol. Gen. Genet. 244:90-96. [DOI] [PubMed] [Google Scholar]
- 22.Kassavetis, G. A., D. L. Riggs, R. Nigri, L. H. Nguyen, and E. P. Geiduscheck. 1989. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 9:2551-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kock, J., R. Evers, and A. W. C. A. Cornelissen. 1988. Structure and sequence of the gene for the largest subunit of trypanosomal RNA polymerase III. Nucleic Acids Res. 16:8753-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunz, J., and M. N. Hall. 1993. Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem. Sci. 19:334-338. [DOI] [PubMed] [Google Scholar]
- 25.Li, J. J., and T. J. Kelly. 1984. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 81:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marton, M. J., J. L. DeRisi, H. A. Bennett, V. R. Iyer, M. R. Meyer, C. J. Roberts, R. Stoughton, J. Burchard, D. Slade, H. Dai, D. E. Bassett, Jr., L. H. Hartwell, P. O. Brown, and S. H. Friend. 1998. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat. Med. 4:1293-1301. [DOI] [PubMed]
- 27.Mathews, D. E., and R. D. Durbin. 1994. Mechanistic aspects of tagetitoxin inhibition of RNA polymerase from Escherichia coli. Biochemistry 33:11987-11992. [DOI] [PubMed] [Google Scholar]
- 28.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols, M., I. Willis, and D. Soll. 1988. Yeast suppressor mutations and transfer RNA processing. Methods Enzymol. 181:377-394. [DOI] [PubMed]
- 30.Pulido, J. C., and G. M. Duyk. 1994. Construction of small-insert libraries enriched for short-tandem-repeat sequences by marker selection. Greene Publishing Associates/John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 31.Rex, J. H., M. A. Pfaller, M. G. Rinaldi, A. Polak, and J. N. Galgiani. 1993. Antifungal susceptibility testing. Clin. Microbiol. Rev. 6:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz, L. D., and M. N. Hall. 1976. Transcription in yeast: α-amanitin sensitivity and other properties which distinguish between RNA polymerase I and III. Proc. Natl. Acad. Sci. USA 73:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sentenac, A. 1985. Eukaryotic RNA polymerases. CRC Crit. Rev. Biochem. 18:31-90. [DOI] [PubMed]
- 34.Steinberg, T. H., and R. R. Burgess. 1992. Targetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template dependent. J. Biol. Chem. 267:20204-20211. [PubMed] [Google Scholar]
- 35.Steinberg, T. H., D. E. Mathews, R. D. Durbin, and R. R. Burgess. 1990. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 265:499-505. [PubMed] [Google Scholar]
- 36.Valenzuela, P., G. L. Harger, F. Veinberg, and W. Rutter. 1976. Molecular structure of yeast RNA polymerase III: demonstration of the tripartite transcriptive system in lower eukaryotes. Proc. Natl. Acad. Sci. USA 73:1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]