Abstract
Immobilized DNA templates, glycerol gradient centrifugation, and native gel analysis were utilized to isolate and compare functional RNA polymerase II (RNAPII) elongation complexes from Saccharomyces cerevisiae and human cell nuclear extracts. Yeast elongation complexes blocked by incorporation of 3′-O-methyl-GTP into the nascent transcript exhibited a sedimentation coefficient of 35S, were less tightly associated to the template than their human counterparts, and displayed no detectable 3′-5′ exonuclease activity on the associated transcript. In contrast, blocked human elongation complexes were more tightly bound to the template, and multiple forms were identified, with the largest exhibiting a sedimentation coefficient of 60S. Analysis of the associated transcripts revealed that a subset of the human elongation complexes exhibited strong 3′-5′ exonuclease activity. Although isolated human preinitiation complexes were competent for efficient transcription, their ability to generate 60S elongation complexes was strikingly impaired. These findings demonstrate functional and size differences between S. cerevisiae and human RNAPII elongation complexes and support the view that the formation of mature elongation complexes involves recruitment of nuclear factors after the initiation of transcription.
mRNA synthesis in eukaryotic cells is a highly complex and regulated process. The transcription cycle on a protein-coding gene consists of at least six mechanistic stages, each of which could potentially be regulated. Numerous experiments have demonstrated that gene-specific activators and repressors can function, at least in part, by modulating the structure of chromatin (1, 33, 44) and the recruitment of the transcription machinery to the promoter to form a preinitiation complex (PIC) (29, 35, 51). Although PIC formation is clearly subject to regulation, a number of eukaryotic genes are known to be regulated by early blocks to elongation (18, 36, 42), and it has been proposed that promoter-proximal pausing of RNA polymerase II (RNAPII) may be a general rate-limiting step for transcription (21). Moreover, numerous accessory factors that alter the elongation efficiency (processivity) of RNAPII in vivo and in vitro have been identified (9, 41), and transcriptional activators can enhance the processivity of elongating RNAPII (2, 4, 5, 32, 46).
In addition to its role in the regulation of mRNA synthesis, elongation has been implicated in such diverse processes as nucleotide excision repair (23, 37, 38, 45), genetic recombination (6), and mRNA processing. mRNA is processed by 5′ capping, 3′ polyadenylation, and splicing, and an interaction between the hyperphosphorylated (elongating) form of the largest subunit of RNAPII and several mRNA processing factors has been demonstrated (3). These include mRNA capping enzyme (7, 19, 24, 31, 39, 47), several components of the mRNA splicing machinery (13, 27, 48), and the CPSF complex, a complex responsible for proper 3′ mRNA processing (11). Consistent with these studies, RNAPII is an essential mRNA polyadenylation factor in vitro (15-17), and sites of high concentrations of splicing factors in vivo coincide with sites of active RNAPII transcription (26, 28). Moreover, cells expressing RNAPII with a truncated carboxy-terminal domain (CTD) exhibit specific defects in mRNA splicing and polyadenylation (25).
Although the evidence that elongation plays an important role in transcription regulation and mRNA processing is convincing, the underlying mechanisms are poorly understood. Several authors have hypothesized the existence of an mRNA-synthesizing and -processing factory, or “transcriptosome” (10, 43). This complex would theoretically comprise the factors necessary for both efficient transcription elongation and mRNA processing. While this hypothesis is attractive, no direct evidence for the existence of this large complex has been discovered. Moreover, just how and when the many elongation-associated factors interact with the complex remains unclear.
Several studies have begun to address the question of the composition of RNAPII elongation complexes. A recent study utilizing a tandem affinity purification scheme demonstrated that several Saccharomyces cerevisiae factors implicated in transcription elongation copurify with RNAPII (20). Earlier biochemical studies involved the use of either purified basal transcription components, PICs purified from extracts, or elongation complexes that were extensively washed (30, 32, 50). Importantly, the composition, properties, and even the molecular mass of functional RNAPII elongation complexes from crude nuclear extracts have yet to be determined. In this paper we report the isolation and partial characterization of functional RNAPII elongation complexes from both yeast and human cell nuclear extracts. Our results demonstrate that yeast and human elongation complexes differ in molecular mass and functional properties, and they provide support for the hypothesis that formation of mature, DNA-bound elongation complexes involves the postinitiation recruitment of nuclear factors.
MATERIALS AND METHODS
Construction of immobilized DNA templates.
DNA fragments to be immobilized were PCR amplified using an upstream 5′-biotinylated primer (bio-SP476) and one of two downstream primers (SP6 and SP2331). For CYC1-Dual and AdML-Dual, a 1.6-kb fragment containing both G-less cassettes and 475 bp of upstream plasmid sequence were amplified using primers bio-SP476 and SP6 and plasmid pCYC1-Dual or pAdML-Dual, respectively. For CYC1-110 and CYC1-210, plasmid pG5CG-D2 was used as a PCR template with bio-SP476 and downstream primer SP6 or SP2331, respectively. For AdML-110 and AdML-210, plasmid pADML-D2 was used as a PCR template with bio-SP476 and downstream primer SP6 or SP2331, respectively. PCR products were resolved on agarose gels, and the desired fragment was excised from the gel. Gel slices were dissolved in 6 M NaI at 55°C for 10 min, and 1 mg of washed (sterile water) M280 streptavadin-coated Dynabeads (Dynal) was added. The mixture was shaken at 37°C for 1 h, and the beads were magnetically harvested, washed five times with 0.5 ml of sterile water, and resuspended in 100 μl of Tris-EDTA (TE). This procedure routinely produced 1 to 2 μg of immobilized DNA per mg of Dynabeads. Lineages of the plasmids used in this study are available upon request.
Nuclear extracts and in vitro transcription. (i) Yeast cells.
Nuclear extracts were prepared from the protease-deficient strain BJ2168 as described previously; they typically contained 20 to 30 mg of protein per ml (34). Transcription reaction mixtures (30 μl) contained 120 μg of nuclear extract, 50 mM HEPES-KOH (pH 7.6), 50 mM potassium acetate, 10% glycerol, 5 mM EGTA, 10 mM magnesium acetate, 2.5 mM dithiothreitol, 40 U of RNasin (Promega Inc.), 30 mM creatine phosphate, 1.5 U of creatine kinase/ml, 400 μM ATP, 400 μM CTP, 1.5 μM UTP, and 10 μCi of [α-32P]UTP. Immobilized templates (100 fmol of DNA; 50 to 100 ng), 3′-O-methyl-GTP, and GTP were added as described in the figure legends. Reaction mixtures were incubated for the indicated times at room temperature, reactions were stopped by addition of 200 μl of stop buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA, and 10 U of RNase T1), and mixtures were incubated for an additional 20 min. Proteinase K (50 μg) and sodium dodecyl sulfate (SDS) (final concentration, 0.5%) were added, and the mixtures were incubated at 37°C for 20 min. RNA transcripts were ethanol precipitated and subjected to electrophoresis on 7% denaturing polyacrylamide gels.
(ii) Human cells.
HeLa cells, harvested at a density of 1 × 109 to 2 × 109 cells per liter, were obtained from the National Cell Culture Center (Minneapolis, Minn.), and nuclear extracts (20 to 30 mg of protein per ml) were prepared as described elsewhere (40). Transcription reaction mixtures (30 μl) contained 60 μg of nuclear extract, 25 mM HEPES-KOH (pH 7.9), 50 mM potassium chloride, 10% glycerol, 200 μM EGTA, 200 μM EDTA, 8.3 mM magnesium chloride, 2 mM dithiothreitol, 150 ng of poly(dI-dC), 4% polyvinyl alcohol, 10 U of RNase T1, 40 U of RNasin (Promega), 30 mM creatine phosphate, 1.5 U of creatine kinase/ml, 400 μM ATP, 400 μM CTP, 1.5 μM UTP, and 10 μCi of [α-32P]UTP. Immobilized templates (100 fmol of DNA; 50 to 100 ng), 3′-O-methyl-GTP, and GTP were added as described in the figure legends. Reaction mixtures were incubated for the indicated times at 30°C, reactions were stopped by addition of 370 μl of HeLa stop buffer (50 mM Tris-HCl [pH 7.5], 1% SDS, and 5 mM EDTA), and reaction products were extracted twice with phenol-chloroform. RNA transcripts were ethanol precipitated and subjected to electrophoresis on 7% denaturing polyacrylamide gels.
Native gel analysis of RNAPII elongation complexes. (i) Yeast complexes.
Transcription reactions were performed as described above except that stop buffer was omitted. Reaction mixtures were incubated for 20 min at room temperature, and immobilized templates were then magnetically harvested. Where indicated in the figure legends, templates were washed with 30 μl of buffer D [10 mM HEPES-KOH (pH 7.6), 50 mM potassium acetate, 10% glycerol, 5 mM EGTA, 10 mM magnesium acetate, 2.5 mM dithiothreitol, 40 U of RNasin, 1.5 μg of poly(dI-dC)] and/or incubated for 30 min at room temperature with 30 μl of buffer D containing 40 U of DraI. Beads were magnetically harvested, and supernatants were analyzed on a 3.5% polyacrylamide (80:1)-0.5% agarose composite native gel. Composite native gels were prepared by taking 65 ml of 0.62% SeaKem (FMC) agarose (melted, cooled to 50°C), adding 700 μl of 10% ammonium persulfate, and then immediately pouring the gel after the addition of 15 ml of solution 2 (8 ml of 0.5 M Tris [pH 8.8]-0.5 M glycine, 7 ml of 40% polyacrylamide [80:1], 70 μl of N,N,N′,N′-tetramethylethylenediamine [TEMED]). Gels were run at 4°C in 25 mM Tris-190 mM glycine (pH 8.3) at 200 V for 4 h. Gels were fixed in 50% methanol-15% acetic acid for 5 min and were dried overnight under a vacuum without heat, and complexes were visualized by autoradiography.
(ii)Human complexes.
Transcription reactions were performed as described above except that stop buffer was omitted. Reaction mixtures were incubated at 30°C for 45 min, and immobilized templates were magnetically harvested and washed once with 30 μl of buffer HD [10 mM HEPES-KOH (pH 7.9), 50 mM potassium chloride, 10% glycerol, 200 μM EGTA, 200 μM EDTA, 8.3 mM magnesium chloride, 2 mM dithiothreitol, 40 U of RNasin, 1.5 μg of poly(dI-dC)]. The washed templates were digested for 20 min at 30°C with 30 μl of buffer HD containing 40 U of DraI, and supernatants were analyzed on native gels as described above.
Slot immunoblotting.
Native gel slices containing RNAPII elongation complexes were subjected to electroelution at 4°C for 2 h at 200 V in 25 mM Tris-190 mM glycine (pH 8.3) by using an Elu-Trap electroeluter (Schleicher and Schuell). Eluants were slot blotted onto an Immobilon-P membrane (Millipore) and were probed with a monoclonal antibody specific for the phosphorylated form of the largest subunit of RNAPII (27) (B3; immunoglobulin M [IgM]; dilution, 1:2,000), followed by goat anti-mouse IgM coupled to horseradish peroxidase (dilution, 1:50,000; Jackson Laboratories). Immune complexes were visualized by addition of Blaze ECL chemiluminescent substrate (Pierce) and autoradiography.
Glycerol gradient sedimentation analysis.
Reactions were performed as described for native gel analysis except that the reactions were scaled up 10-fold and the blocked complexes were washed once with 300 μl of transcription buffer and liberated in 100 μl of transcription buffer containing 100 U of DraI. For analysis of human complexes, gradients contained 10 mM HEPES-KOH (pH 7.9), 50 mM potassium chloride, 200 μM EGTA, 200 μM EDTA, 8.3 mM magnesium chloride, 2 mM dithiothreitol, and a gradient of either 12 to 30% or 15 to 50% glycerol. For analysis of yeast complexes, acetate salts were substituted for the chloride salts. Gradients (5.2 ml) were poured at 0.2 ml/min by using an FPLC P500 pump (Pharmacia), and the entire supernatant of the DraI digestion was layered on top. For analysis of complexes derived from isolated PICs, the supernatant from a 90-μl reaction containing isolated PICs was mixed with an equal volume of transcription buffer lacking polyvinyl alcohol, and 100 μl was layered on the gradient. Gradients were spun for 6 h at 45,000 rpm in an SW55 Ti rotor and were fractionated from the bottom. Fractions (200 μl) were collected by hand, and a portion was analyzed on a native gel or treated with RNase T1, ethanol precipitated, and analyzed on a denaturing polyacrylamide gel. Molecular standards used were thyroglobulin, catalase, bovine serum albumin, and Escherichia coli 30S and 50S ribosomal subunits purified from strain BL21 (49). Gradients containing the standards were spun for each experiment, and the protein concentration of each fraction was determined by Bio-Rad protein assays. To visualize the ribosomal subunits, fractions were extracted with phenol-chloroform, ethanol precipitated, resuspended in 1% SDS-10% glycerol-0.25× Tris-borate-EDTA, and analyzed on 0.8% agarose gels. Amounts of the 23S and 16S RNAs were quantitated by ethidium bromide fluorescence.
RESULTS
Functional yeast RNAPII elongation complexes can be isolated from nuclear extracts and visualized on native gels.
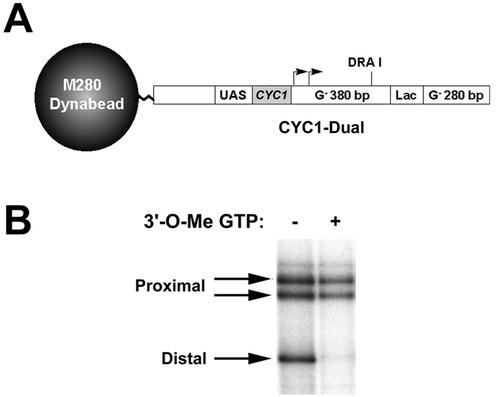
To initiate a characterization of RNAPII elongation complexes, immobilized DNA templates were utilized to isolate blocked elongation complexes from yeast nuclear extracts. Previous studies demonstrated that elongation by mammalian RNAPII could be arrested both in vivo and in vitro by E. coli Lac repressor protein bound to a lac operator (12, 22), whereas elongation by vaccinia virus RNA polymerase could be arrested by incorporation of 3′-O-methyl-GTP into the nascent transcript (14). To potentially use either of these approaches to block yeast RNAPII elongation complexes, a plasmid was constructed (pCYC1-Dual) that contained the yeast CYC1 promoter fused to a 380-bp G-less cassette, followed sequentially by a 100-bp (G-containing) region with two E. coli Lac repressor binding sites and a distal G-less cassette of 280 bp. By using pCYC1-Dual as a template, a 1.6-kbp DNA fragment containing the CYC1 promoter and both G-less cassettes was amplified by PCR using a 5′-biotinylated upstream primer and was immobilized to streptavidin-coated paramagnetic beads (see Materials and Methods) (Fig. 1A). To determine whether incorporation of 3′-O-methyl-GTP into nascent transcripts blocks yeast RNAPII elongation complexes, transcription assays were performed with CYC1-Dual and yeast nuclear extract both in the presence and in the absence of 3′-O-methyl-GTP. In reactions containing GTP and lacking 3′-O-methyl-GTP, two transcripts of approximately 380 nucleotides and one 280-nucleotide transcript were observed (Fig. 1B, lane −). These three transcripts were expected products, since the CYC1 promoter is known to generate two major nascent transcripts (initiating early in the proximal G-less cassette) and the reactions were treated with RNase T1 prior to gel analysis. Because RNase T1 hydrolyzes the 3′-phosphate of guanylate residues in the RNA, transcripts initiating within the proximal G-less cassette and extending through the end of the distal G-less cassette will yield both a 380- and a 280-nucleotide transcript after T1 digestion due to the presence of the intervening G-containing (lac operator) sequence between the two G-less cassettes. In reactions containing 3′-O-methyl-GTP and lacking GTP, transcription elongation through the distal cassette was efficiently blocked, as indicated by the specific loss of the distal transcript (Fig. 1B, lane +). Additional experiments demonstrated that templates lacking the CYC1 promoter did not support significant levels of transcription and that the addition of purified lac repressor also blocked production of the distal transcript (data not shown).
FIG. 1.
Incorporation of 3′-O-methyl-GTP into nascent transcripts blocks yeast RNAPII elongation complexes. (A) Diagram of immobilized template CYC1-Dual. (B) Transcription assays. Reactions were performed with CYC1-Dual in the presence of yeast nuclear extract, ATP, CTP, [α-32P]UTP, and either GTP (lane −) or 3′-O-methyl-GTP (lane +). RNA products were treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis. The positions of the proximal and distal G-less cassette transcripts are indicated.
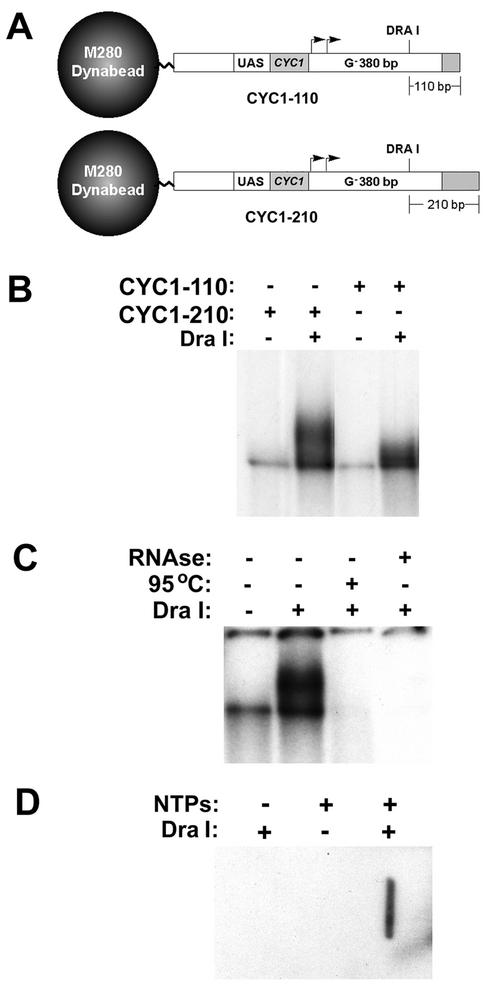
To potentially visualize blocked elongation complexes on native gels, simplified templates were constructed that contained the CYC1 promoter fused to the proximal 380-bp G-less cassette, followed by a minimal amount of G-containing vector sequence (40 or 140 bp) to facilitate blockage of the complexes with 3′-O-methyl-GTP (Fig. 2A). To liberate blocked elongation complexes from the template, a unique DraI restriction site was engineered approximately 70 bp upstream from the end of the G-less cassette. Reactions were performed with immobilized CYC1-110 or CYC1-210 in the presence of nuclear extract, ATP, CTP, 3′-O-methyl-GTP, and [α-32P]UTP. Templates containing blocked elongation complexes were harvested, washed, and then incubated in transcription buffer either in the presence or in the absence of DraI. After incubation, the templates were reharvested and the supernatants were analyzed on composite polyacrylamide-agarose native gels. The results revealed the presence of both DraI-independent and DraI-dependent labeled species in these supernatants (Fig. 2B). The DraI-independent species was variably present and could be removed by modest additional washing (data not shown). The mobility of the DraI-dependent species in the native gel was affected by the length of DNA downstream of the DraI site (Fig. 2B), and the integrity of the species was sensitive to RNase, heat, and proteinase K (Fig. 2C and data not shown). These results indicate that the DraI-dependent species represents ternary complexes containing protein, DNA, and labeled RNA. To determine whether the DraI-liberated complexes contained the hyperphosphorylated (IIO, elongating) form of RNAPII, scaled-up (10×) reactions were performed using unlabeled UTP, and the DraI supernatant was loaded alongside that from a 1× labeled reaction on a native gel. Gel slices corresponding to the positions of the labeled complexes were excised from lanes containing a 10× reaction or, as controls, from 10× reactions that lacked either nucleotides or DraI. The gel slices were subjected to electroelution, and the eluants were analyzed by slot immunoblotting using a monoclonal antibody (B3) specific for the IIO form of RNAPII. The results demonstrated the presence of RNAPIIO in gel slices excised from the experimental, but not from the control, reactions (Fig. 2D). Taken together, these results indicate that the DraI-dependent species are blocked yeast RNAPII elongation complexes.
FIG. 2.
Native gel analysis of radiolabeled yeast RNAPII elongation complexes. (A) Diagram of the immobilized templates used in ternary complex isolation. (B) Native gel analysis. Transcription reactions were performed in the presence of nuclear extract, ATP, CTP, 3′-O-methyl-GTP, and [α-32P]UTP. Immobilized templates containing blocked complexes were harvested, washed, and digested with DraI, and the supernatants were analyzed on composite 0.5% agarose-3.5% polyacrylamide native gels. (C) Elongation complexes were blocked on immobilized templates as described above and then incubated for 5 min at 95°C or in the presence of RNase A (10 μg/ml). The immobilized templates were then treated with DraI, and supernatants were analyzed on composite 0.5% agarose-3.5% polyacrylamide native gels. (D) Slot immunoblotting. Nonradioactive 10× scaled-up reactions (300 μl) were performed with template CYC1-210 as described above for Fig. 2B (rightmost lane) or with the omission of NTPs or the DraI restriction enzyme. Supernatants from the DraI incubation step of these reactions were resolved on a composite native gel alongside labeled complexes as described above. Gel slices corresponding to the positions of the elongation complexes were excised from the native gel and subjected to electroelution. Eluants were slot blotted onto a polyvinylidene difluoride membrane and probed with an antibody specific for the hyperphosphorylated form of RNAPII.
Human RNAPII elongation complexes are larger and more stably bound to the DNA template than their yeast counterparts.
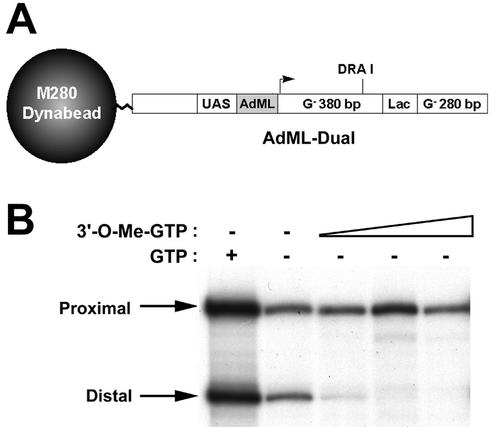
Having established an approach to isolate and visualize yeast RNAPII elongation complexes, we tested whether this method could be used to isolate functional RNAPII elongation complexes from human cell nuclear extracts. An immobilized template containing the adenovirus major late promoter (AdMLP) substituted for the yeast CYC1 promoter was constructed and used in transcription assays with HeLa cell nuclear extracts (Fig. 3A). In the reaction lacking both GTP and 3′-O-methyl-GTP, the proximal 380- and distal 280-nucleotide transcripts were both produced (Fig. 3B), presumably due to the presence of residual GTP in the HeLa extract and/or generation of GTP during the reaction. Nevertheless, as observed with the yeast reactions, the addition of 3′-O-methyl-GTP efficiently blocked production of the distal transcript (Fig. 3B). Additional results demonstrated that templates lacking the promoter did not support significant transcription and that Lac repressor could also block production of the distal transcript (data not shown).
FIG. 3.
Incorporation of 3′-O-methyl-GTP into nascent transcripts blocks human RNAPII elongation complexes. (A) Diagram of the immobilized template AdML-Dual. (B) Transcription assays. Reactions were performed with AdML-Dual in the presence of HeLa nuclear extract, ATP, CTP, GTP, and [α-32P]UTP (leftmost lane) or with the omission of GTP and the addition of increasing concentrations of 3′-O-methyl-GTP (0.33 to 1.0 mM) (three rightmost lanes). RNA products were treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis. The positions of the 380-nucleotide (proximal) and 280-nucleotide (distal) G-less transcripts are indicated.
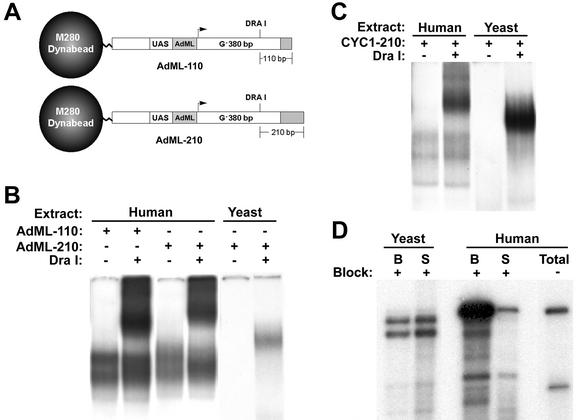
To potentially visualize human RNAPII elongation complexes on native gels and compare them to their yeast counterparts, simplified variants of the AdML-Dual template lacking the distal cassette were constructed as done previously with the yeast CYC1-Dual template (AdML-110 and AdML-210) (Fig. 4A). Immobilized templates containing either the AdML or CYC1 promoter fused to the single G-less cassette were incubated with either yeast or HeLa cell nuclear extracts in the presence of 3′-O-methyl-GTP and [α-32P]UTP. The templates containing blocked elongation complexes were then harvested and incubated in the appropriate transcription buffer either lacking or containing DraI, and the supernatants were subsequently isolated and analyzed on native gels as previously. The results demonstrated that the supernatants from the HeLa reactions contained both a DraI-independent and a larger and more heterogeneous set of DraI-dependent complexes (Fig. 4B). The mobility of the DraI-liberated complexes was dependent upon the size of the DNA fragment liberated, whereas the mobility of the DraI-independent complexes was not. These results suggest that the human DraI-dependent and -independent complexes represent RNAPII elongation complexes that remain bound to the template or have dissociated from the template, respectively. In addition, the relative mobility of the yeast and human ternary complexes suggested that the human complexes were significantly larger than their yeast counterparts (Fig. 4B and C).
FIG. 4.
Comparison of radiolabeled human and yeast RNAPII elongation complexes. (A) Diagram of immobilized templates containing the AdMLP. (B) Native gel analysis of elongation complexes using AdMLP-containing templates. Transcription reactions contained the indicated template in the presence of yeast or HeLa cell nuclear extract, ATP, CTP, 3′-O-methyl-GTP, and [α-32P]UTP. Immobilized templates containing blocked complexes were harvested, washed, and digested with DraI, and supernatants were analyzed on composite native gels. (C) Native gel analysis of elongation complexes using CYC1-containing templates. Blocked yeast or human elongation complexes were formed on immobilized template CYC1-210 and analyzed as described for Fig. 4B. (D) Template association of elongation complexes. Transcription reactions were performed using yeast extract and CYC1-Dual (Yeast) or HeLa extract and AdML-Dual (Human) in the presence of ATP, CTP, 3′-O-methyl-GTP, and [α-32P]UTP (left four lanes) or with 3′-O-methyl-GTP omitted and GTP included (rightmost lane). Before the addition of stop buffer, the immobilized templates were harvested, and the bound fraction (B) and supernatants (S) were treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis.
As noted above, a significant amount of DraI-independent complexes was obtained from the reactions with human nuclear extract. In contrast, very little of these complexes was observed with the corresponding yeast reactions (Fig. 4B and C). These results suggested that either the yeast elongation complexes were more stably associated with the DNA template or a substantial portion of the yeast elongation complexes was less stably bound and had dissociated from the template prior to the DraI (mock) incubation step. To address this issue, transcription reactions were performed with either yeast or human nuclear extracts with the inclusion of 3′-O-methyl-GTP, and the distribution of labeled transcript released into the supernatant or remaining associated with the immobilized template was determined. In reactions with yeast nuclear extract, approximately 40% of the transcript produced remained associated with the immobilized template (Fig. 4D). In contrast, approximately 90% of the transcript synthesized in the reactions with HeLa extract remained associated with the immobilized template (Fig. 4D). These results suggest that blocked human RNAPII elongation complexes possess a greater stability for association with the DNA template and/or the associated transcript than their yeast counterparts.
Human RNAPII elongation complexes that remain associated with DNA exhibit a sedimentation coefficient of approximately 60S.
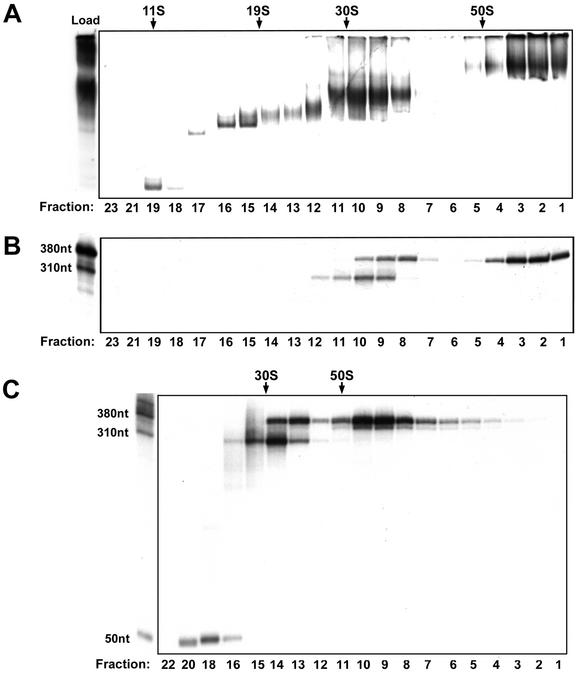
To estimate the molecular mass of human RNAPII elongation complexes, we compared the glycerol gradient sedimentation values of the complexes to those of standards with known Svedberg values (S) and molecular masses that were analyzed in parallel gradients. A scaled-up (10×) reaction was performed, and the supernatant from the DraI digestion was fractionated by centrifugation through a 12 to 30% glycerol gradient. Portions of each fraction were analyzed on a native gel (Fig. 5A) or treated with RNase T1 and analyzed on a denaturing polyacrylamide gel (Fig. 5B). The results demonstrated the existence of multiple forms of the human elongation complex that differed in sedimentation value and/or size of the associated transcript. A series of complexes ranging from approximately 11 to 30S was resolved in the gradient, but their associated transcripts were not detected initially (Fig. 5A and B). To reexamine the sizes of the transcripts associated with these 11-to-30S complexes, the experiment was repeated with the DraI supernatant fractionated on a 15 to 50% glycerol gradient and the denaturing gel electrophoresis conditions were modified in order to retain small transcripts on the gel. The results demonstrated that the gradient fractions containing complexes smaller than 30S (fractions 16 and higher) contained transcripts of approximately 50 nucleotides (Fig. 5C). These results suggest that the 11-to-30S complexes represent elongation complexes that are arrested early in the promoter-proximal region of the G-less cassette.
FIG. 5.
Glycerol gradient sedimentation analysis of human RNAPII elongation complexes. Human elongation complexes were isolated from immobilized template AdML-110 as described in Materials and Methods and were subjected to sedimentation through either a 12 to 30% (A and B) or a 15 to 50% (C) glycerol gradient. Gradients were fractionated, and a portion of each fraction was analyzed on a composite native gel (A) or treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis (B and C). The positions of the 380-, 310-, and 50-nucleotide transcripts are indicated. The migration of molecular weight standards in parallel gradients with known Svedberg (S) values is indicated (catalase, thyroglobulin, and 30S and 50S subunits of purified E. coli ribosomes).
In addition to the 11-to-30S complexes, three major forms of the elongation complex containing discrete RNA species were resolved. The smallest major complex was approximately 33S and was associated with a transcript of approximately 310 nucleotides, whereas the second complex migrated slightly further in the gradient and was associated with the full-length 380-nucleotide transcript. These two complexes (33S and 38S) comigrated with the DraI-independent complex on a native gel. The third and largest complex peaked beyond the 50S subunit (1.6 MDa) of the E. coli ribosome and just prior to the bottom of the gradient. This large human complex, approximately 60S, comigrated with the DraI-dependent ternary complex on a native gel and contained only the full-length 380-nucleotide transcript. To rule out the possibility that the migration of the elongation complexes in the gradient was influenced by the presence of nonspecific DNA-binding proteins on the 110-bp DNA fragment, Southern blot analysis was performed on a portion of each fraction. Since we estimate that only 1 template in 1,000 gives rise to a stable elongation complex, any nonspecific DNA binding proteins contributing to the size of the complexes must be present on virtually all of the templates in the reaction. Southern blot analysis confirmed that the DraI-liberated DNA fragments migrated as a single peak very early in the gradient with an estimated molecular mass equivalent to that of the DNA fragment alone (data not shown). To confirm that the large human complex did not represent precipitated material at the bottom of the gradient, the experiment was repeated with the DraI supernatant fractionated on a 15 to 50% glycerol gradient. The results demonstrated that the large complex sedimented near the middle of the 15 to 50% gradient, again with a sedimentation coefficient greater than that of the 50S standard (Fig. 5C).
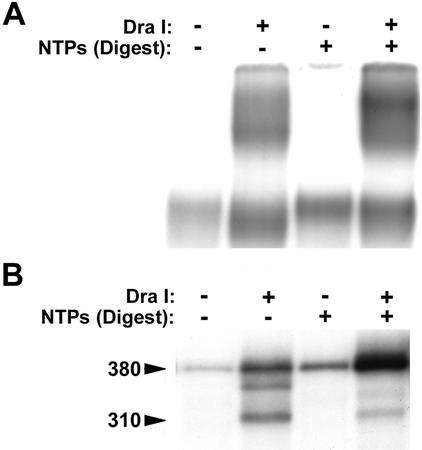
The presence of a 310-nucleotide transcript associated with a subset of the DraI-liberated human complexes was intriguing in light of the fact that the DraI site used to liberate the complexes is located 70 bp upstream from the end of the 380-bp G-less cassette. The 310-nucleotide transcript could result from the 3′-5′ exonuclease activity that is intrinsic to RNAPII and is stimulated by the elongation-associated factor TFIIS (8). Enhanced 3′-5′ exonuclease activity of the polymerase in these complexes could cause the elongation complex to back off of the DraI-liberated DNA fragment after it had excised approximately 70 nucleotides from the 3′ end of the 380-nucleotide transcript. Since the 3′-5′ exonuclease activity of the polymerase can be suppressed by nucleoside triphosphates (NTPs), we tested whether production of the 310-nucleotide transcript could be suppressed by the inclusion of NTPs during the DraI incubation step. Templates containing blocked labeled complexes were harvested, washed, and incubated with DraI either in the absence or in the presence of ATP, CTP, UTP, and 3′-O-methyl-GTP. The supernatants were then either analyzed on a native gel (Fig. 6A) or treated with RNase T1 and analyzed on a denaturing polyacrylamide gel (Fig. 6B). Complexes liberated from the template by DraI digestion in the absence of NTPs displayed heterogeneity in their mobility in the native gel and were associated with transcripts heterogeneous in length. In contrast, complexes liberated by DraI digestion in the presence of NTPs migrated as better-defined species in the native gel and were associated predominantly with full-length transcript. These results indicate that a subset of the human RNAPII elongation complexes exhibits an NTP-reversible 3′-5′ exonuclease activity on the associated transcript.
FIG. 6.
A subset of human elongation complexes contains an NTP-reversible 3′-5′ exonuclease activity. Human elongation complexes were isolated from immobilized template AdML-110 as described for Fig. 4B and were incubated in either the presence or the absence of NTPs (400 μM ATP, 400 μM CTP, 1.5 μM UTP, and 660 μM 3′-O-methyl-GTP) during the DraI liberation step. A portion of each reaction product was analyzed on a composite native gel (A) or treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis (B). The positions of the full-length 380-nucleotide and truncated 310-nucleotide G-less transcripts are indicated.
Native gel analysis of yeast and human complexes indicated that the mobility of the DraI-dependent yeast ternary complexes was similar to that of the DraI-independent human complexes (Fig. 4). Consistent with this result, glycerol gradient analysis of the DraI supernatants from yeast reactions revealed that the yeast ternary complexes migrated as a single species slightly larger than the E. coli 30S ribosome subunit and were associated with full-length 380-nucleotide transcripts (data not shown). Taken together, these results suggest that human RNAPII elongation complexes that remain associated with a 110-bp DNA fragment are approximately 2 MDa, whereas the analogous yeast complexes are approximately 1.2 MDa.
Formation of 60S human RNAPII elongation complexes requires the presence of nuclear factors during elongation.
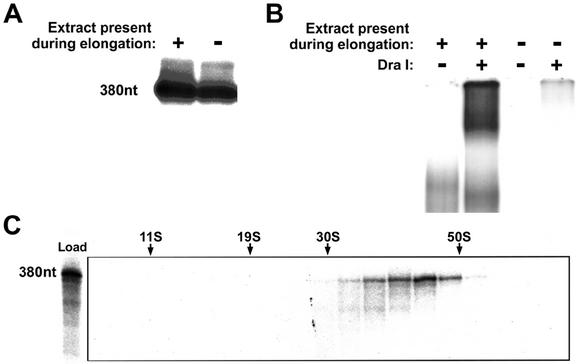
In light of the deduced size of the human elongation complexes, we sought to determine whether these complexes were composed entirely of components present in PICs or were a product of PICs that had initiated transcription and subsequently recruited additional factors from the nuclear extract to the elongation complex. To address this, we compared the sizes of the DraI-liberated complexes from reactions that contained nuclear extract during transcription through the G-less cassette to those from reactions where PICs were preformed on the immobilized template and magnetically purified, and transcription performed in the absence of nuclear extract. Surprisingly, reactions containing the isolated PICs did not yield any significant amount of DraI-liberated ternary complex, despite the fact that these reactions yielded approximately 60% of the transcript synthesized in reactions containing nuclear extract during elongation (Fig. 7A and B). These results suggested that the presence of nuclear extract during the elongation phase was required for the production of elongation complexes that remain associated with the DNA template. Accordingly, we reasoned that the majority of the elongation complexes derived from isolated PICs had dissociated from the template after transcribing through the G-less cassette. To estimate the sizes of these complexes, the supernatant of the transcription reaction containing isolated PICs was fractionated on a glycerol gradient, and each fraction was treated with RNase T1 and analyzed on a denaturing polyacrylamide gel. The results revealed that elongation complexes derived from isolated PICs were present in the reaction supernatant and migrated through the glycerol gradient with a sedimentation coefficient of approximately 40S (Fig. 7C). Combined with the analyses in Fig. 5, these results suggest a postinitiation recruitment of nuclear factors to the human RNAPII elongation complex that confer a dramatic increase in the mass of the complex (to approximately 2 MDa) and an accompanying ability to remain associated with the DNA template.
FIG. 7.
Formation of stable 60S human RNAPII elongation complexes requires the presence of nuclear extract during elongation. (A) Transcription assays. Template AdML-110 was preincubated with HeLa nuclear extract for 45 min. The template was either left in the presence of nuclear extract or magnetically harvested to isolate PICs, and reactions were initiated by the addition of ATP, CTP, 3′-O-methyl-GTP, and [α-32P]UTP. RNA products were treated with RNase T1 and analyzed by denaturing polyacrylamide gel electrophoresis. (B) Native gel analysis. Templates from the reactions with isolated PICs or PICs in the presence of nuclear extract were magnetically harvested, washed, and treated with DraI, and the supernatants were analyzed on native gels. (C) Glycerol gradient sedimentation analysis. Transcription was performed using isolated PICs as described for panel A. Immediately after the reaction incubation, the templates were harvested and the supernatant was isolated and subjected to sedimentation through a 12 to 30% glycerol gradient. Gradient fractions were treated with RNase T1, and the RNA was analyzed by denaturing polyacrylamide gel electrophoresis. The migration of molecular weight standards with known Svedberg (S) values in parallel gradients is indicated (catalase, thyroglobulin, and 30S and 50S subunits of purified E. coli ribosomes).
DISCUSSION
Functional yeast and human RNAPII elongation complexes from nuclear extracts are megadalton-sized complexes that can be visualized on native gels.
A native gel assay was used to visualize both yeast and human RNAPII elongation complexes (Fig. 2 and 4). In this assay, elongation complexes were labeled by incorporation of [32P]UTP into the associated transcripts, were blocked at the end of a G-less cassette by use of 3′-O-methyl-GTP, and were liberated from immobilized DNA templates by DraI restriction enzyme digestion and analyzed on composite acrylamide-agarose native gels. Approximately 90% of the transcript synthesized in the reactions with HeLa extract was found to remain associated with the immobilized template, whereas about 40% of the transcript produced in the yeast reactions remained associated (Fig. 4D). These results suggest that the blocked human RNAPII elongation complexes possessed a greater stability for the DNA template and/or the associated transcript than their yeast counterparts. The human elongation complexes liberated by DraI digestion migrated more slowly through the native gels than their yeast counterparts, and the sizes of the yeast and human complexes were not affected by whether they were formed on templates containing the yeast CYC1 or AdML promoter (Fig. 4). An effect of promoter strength was observed, however, with the AdML promoter giving rise to more complexes in the HeLa nuclear extracts and the CYC1 promoter giving rise to more complexes in the yeast nuclear extracts.
By use of glycerol gradient sedimentation analysis, we confirmed and extended the results from the native gel assays and assigned molecular mass estimates to the elongation complexes from both yeast and human extracts. In reactions containing yeast nuclear extract, a single major form of the RNAPII elongation complex was observed that was associated with the DraI-liberated 110-bp DNA fragment. This complex contained only the full-length 380-nucleotide transcript and exhibited a sedimentation coefficient of approximately 35S, corresponding to an estimated molecular mass of 1.0 to 1.2 MDa. In reactions containing HeLa nuclear extract, a number of elongation complexes were identified. A series of smaller complexes, ranging from approximately 11 to 30S, were resolved and found to be associated with transcripts of approximately 50-nucleotides. These smaller complexes most likely correspond to elongation complexes spontaneously arrested in the promoter-proximal region of the G-less cassette. We observed that addition of exogenous TFIIS stimulated the formation of the proximal transcript, consistent with the presence of spontaneously arrested complexes in this region (data not shown). Since these complexes are visualized by virtue of a small associated transcript, it appears that there are a significant number of these promoter-proximally paused complexes on the immobilized templates.
In addition to the 11-to-30S complexes, three major forms of the human elongation complex were resolved. The smallest complex (complex I) exhibited a sedimentation coefficient of approximately 33S and was associated with a 310-nucleotide transcript. The inclusion of NTPs during the DraI incubation step suppressed the formation of the 310-nucleotide transcript (Fig. 6B), consistent with the view that production of the 310-nucleotide transcript was due to enhanced 3′-5′ exonuclease activity of the polymerase, possibly due to the presence of TFIIS in this complex. We calculate that approximately 1 to 2 fmol of this human complex is liberated by DraI treatment of a 10× (300-μl) reaction. Thus, further scaling up of this procedure is necessary for the direct biochemical determination of the presence of TFIIS in this complex, as well as for identification of additional components of these functional elongation complexes.
The second human complex identified in this study (complex II) was slightly larger than complex I and was associated with the full-length 380-nucleotide transcript. Complex II corresponds to the DraI-independent complex and most likely represents an elongation complex that transcribed the entire G-less cassette and then dissociated from the template. The third major human complex (complex III) remained associated with the 110-bp DraI DNA fragment and exhibited a sedimentation coefficient of approximately 60S, corresponding to an estimated molecular mass of about 2 MDa. It should be noted that the presence of the 110-bp DNA fragment in complex III is not sufficient to explain the large size difference between complex III and complexes I and II. It is unclear whether complexes I and II represent distinct and smaller forms of the elongation complex or are the result of a partial dissociation of the DNA and several factors from complex III. Interestingly, the DraI-dependent human complex III is significantly larger than its yeast counterpart (1.0 to 1.2 MDa). Since the combined molecular mass of RNAPII, the 380-nucleotide transcript, and the 110-bp liberated DNA fragment is approximately 600 to 700 kDa (19S), the estimated mass of both the yeast and human ternary complexes suggests that they contain a substantial number of additional factors.
The discovery that numerous mRNA processing factors interact with the hyperphosphorylated form of the RNAPII CTD has led to the proposal that in vivo mRNA synthesis is carried out by a large mRNA-synthesizing and -processing factory, or transcriptosome (10, 43). The 60S human complex isolated in this study is large enough to accommodate the core ternary complex (RNAPII, RNA, and DNA) plus as much as 1.4 MDa of additional factors. It is likely that, even under our mild isolation conditions, additional associated factors have been lost from this complex. Thus, our size estimate of 2 MDa most likely reflects a lower limit on the size of native human elongation complexes. Since most human type II genes contain introns whereas less than 4% of yeast mRNAs undergo splicing (43), the larger size of the human elongation complexes (about 1 MDa larger than their yeast counterparts) might reflect the differential requirement for splicing factors in these complexes.
Formation of 60S human RNAPII elongation complexes requires the presence of nuclear factors during elongation.
We investigated whether reactions containing isolated human PICs give rise to the same elongation complexes that are produced in reactions containing nuclear extract during both the initiation and elongation phases. Although the overall level of transcription from isolated PICs was comparable to that in reactions containing nuclear extract, the ability of the isolated PICs to generate stable 60S ternary elongation complexes was strikingly impaired (Fig. 7). Rather, elongation complexes derived from isolated PICs were released from the immobilized template after synthesis of the 380-nucleotide transcript and exhibited a sedimentation coefficient of approximately 40S (Fig. 7C). These results strongly suggest postinitiation recruitment of nuclear factors to the human RNAPII elongation complex that results in both a dramatic increase in the size of the complex (to approximately 2 MDa) and an increased association of the complex with the DNA template. Such factors might include those that specifically modulate RNAPII elongation processivity, as well as factors involved in mRNA processing and/or packaging and nuclear transport. It will be of significant interest to extend and modify the biochemical approaches described in this work in order to precisely define the molecular composition of these complexes and to examine the effects of transcriptional regulatory proteins on both their composition and their functional properties.
Acknowledgments
We thank Ron Berezney for the B3 antibody, Judith Jaehning for plasmid pJJ460, the National Cell Culture Center for HeLa cells, and Ken Blumenthal and members of the Ponticelli laboratory for helpful discussions and comments on the manuscript.
This work was supported by a Public Health Service grant (GM51124) from the National Institutes of Health to A.S.P.
Footnotes
This work is dedicated to the memory of Alfred Ponticelli, Sr.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, A., G. Faye, and D. L. Bentley. 1996. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 15:4654-4664. [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347-351. [DOI] [PubMed] [Google Scholar]
- 4.Blau, J., H. Xiao, S. McCracken, P. O'Hare, J. Greenblatt, and D. Bentley. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16:2044-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S. A., A. N. Imbalzano, and R. E. Kingston. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479-1490. [DOI] [PubMed] [Google Scholar]
- 6.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, E., T. Takagi, C. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, K. R., D. E. Awrey, A. M. Edwards, and C. M. Kane. 1994. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37. J. Biol. Chem. 269:936-943. [PubMed] [Google Scholar]
- 9.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 10.Corden, J. L., and M. Patturajan. 1997. A CTD function linking transcription to splicing. Trends Biochem. Sci. 22:413-416. [DOI] [PubMed] [Google Scholar]
- 11.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 12.Deuschle, U., R. A. Hipskind, and H. Bujard. 1990. RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science 248:480-483. [DOI] [PubMed] [Google Scholar]
- 13.Du, L., and S. L. Warren. 1997. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J. Cell Biol. 136:5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagler, J., and S. Shuman. 1992. Structural analysis of ternary complexes of vaccinia RNA polymerase. Proc. Natl. Acad. Sci. USA 89:10099-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 16.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 19.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumm, A., L. B. Hickey, and M. Groudine. 1995. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 9:559-572. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn, A., I. Bartsch, and I. Grummt. 1990. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature 344:559-562. [DOI] [PubMed] [Google Scholar]
- 23.Leadon, S. A., and D. A. Lawrence. 1992. Strand-selective repair of DNA damage in the yeast GAL7 gene requires RNA polymerase II. J. Biol. Chem. 267:23175-23182. [PubMed] [Google Scholar]
- 24.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 26.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 27.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neugebauer, K. M., and M. B. Roth. 1997. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 11:1148-1159. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 30.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 31.Pillutla, R. C., Z. Yue, E. Maldonado, and A. J. Shatkin. 1998. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 273:21443-21446. [DOI] [PubMed] [Google Scholar]
- 32.Ping, Y. H., and T. M. Rana. 1999. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399-7404. [DOI] [PubMed] [Google Scholar]
- 33.Pollard, K. J., and C. L. Peterson. 1998. Chromatin remodeling: a marriage between two families? Bioessays 20:771-780. [DOI] [PubMed] [Google Scholar]
- 34.Ponticelli, A. S., and K. Struhl. 1990. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol. Cell. Biol. 10:2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 36.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. [DOI] [PubMed] [Google Scholar]
- 37.Schaeffer, L., V. Moncollin, R. Roy, A. Staub, M. Mezzina, A. Sarasin, G. Weeda, J. H. Hoeijmakers, and J. M. Egly. 1994. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 13:2388-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeffer, L., R. Roy, S. Humbert, V. Moncollin, W. Vermeulen, J. H. Hoeijmakers, P. Chambon, and J. M. Egly. 1993. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science 260:58-63. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro, D. J., P. A. Sharp, W. W. Wahli, and M. J. Keller. 1988. A high-efficiency HeLa cell nuclear transcription extract. DNA 7:47-55. [DOI] [PubMed] [Google Scholar]
- 41.Shilatifard, A. 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 12:1437-1446. [DOI] [PubMed] [Google Scholar]
- 42.Spencer, C. A., and M. Groudine. 1990. Transcription elongation and eukaryotic gene regulation. Oncogene 5:777-785. [PubMed] [Google Scholar]
- 43.Steinmetz, E. J. 1997. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell 89:491-494. [DOI] [PubMed] [Google Scholar]
- 44.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 45.Tantin, D., A. Kansal, and M. Carey. 1997. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 17:6803-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yankulov, K., J. Blau, T. Purton, S. Roberts, and D. L. Bentley. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749-759. [DOI] [PubMed] [Google Scholar]
- 47.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yusupov, M. M., and A. S. Spirin. 1988. Hot tritium bombardment technique for ribosome surface topography. Methods Enzymol. 164:426-439. [DOI] [PubMed] [Google Scholar]
- 50.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 51.Zawel, L., and D. Reinberg. 1993. Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acid Res. Mol. Biol. 44:67-108. [DOI] [PubMed] [Google Scholar]