Abstract
Transcriptional silencing of the gene coding for amoebapore A (AP-A) was observed when trophozoites of Entamoeba histolytica were transfected with a hybrid plasmid construct containing the ap-a gene flanked by the upstream and downstream segments of the original Ehap-a gene. Transfectants were totally devoid of ap-a transcript and AP-A protein. An identical silencing effect was observed upon transfection with a plasmid that contained only the 5′ upstream region of ap-a. Removal of the selecting antibiotic enabled the isolation of plasmidless clones, which retained in their progeny the silenced phenotype. E. histolytica cells were able to overexpress ap-a when transfected with a plasmid containing the gene flanked by the 5′ and 3′ regions of the EhRP-L21 gene. This plasmid, however, could not express ap-a in the retransfected, cloned trophozoites lacking AP-A. This is the first report of gene silencing in E. histolytica, and the mechanism appears to belong to transcriptional gene silencing and not to posttranscriptional gene silencing. This conclusion is based on the following results: (i) silencing was achieved by transfection of homologous 5′ flanking sequences (470 bp of the Ehap-a gene), (ii) transcription initiation of Ehap-a was found to be blocked, and (iii) short double-stranded RNA fragments of the ap-a coding and noncoding sequences were not detected. Trophozoites lacking AP-A are nonpathogenic and impaired in their bacteriolytic capability.
The amoebapores (AP) are an important virulence factor of Entamoeba histolytica (12, 30). Three isoforms of the small (77 amino acid) AP protein exist as mature and potentially active peptides inside distinct cytoplasmic granules of the trophozoite (30-32). The present view of the mode of action of AP is that following lectin-mediated recognition and intimate adherence between the trophozoite and its target cell, the AP molecules are inserted into the membranes of the latter without depending on the interaction with a specific membrane receptor and that antibodies against AP are thus unable to inhibit its toxic effect. To investigate the specific role of AP-A, the most abundant among the three isoforms (38), in the pathogenicity of the parasite, the levels of AP-A expression were modulated by transfection of trophozoites with different hybrid plasmid constructs. Down-regulation (60%) of expression of AP-A by antisense mRNA caused a drastic reduction in amoeba pathogenicity and clearly demonstrated its importance in the parasite's virulence (12). Interestingly, overexpression (fourfold) of AP-A also caused a dramatic reduction in virulence (13). This result has been attributed to an observed spillover of AP-A from the granules into the cytoplasm and a continued release of AP-A by viable trophozoites into the surrounding medium. In an attempt to overcome the problem of the apparent mislocalization of the overexpressed AP-A, we prepared another hybrid plasmid construct in which the Ehap-a gene was inserted into the vector, flanked by its original 5′ and 3′ regulatory elements. Transfection of trophozoites of E. histolytica virulent strain HM-1:IMSS (HM-1) with this plasmid surprisingly abolished the transcription and translation of both the plasmid and endogenous ap-a genes.
In the present report, we describe the characteristics of this newly discovered silencing phenomenon in Entamoeba. Transgene-induced silencing of gene expression has been reported in almost all eukaryotes, including fungi, Saccharomyces, Drosophila, plants, and mammals (16, 18, 21, 26, 34, 45). In most cases, the molecular mechanisms by which gene silencing is achieved are still poorly understood. Gene silencing has been described as occurring at two main levels: transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS). TGS in transgenic plants has been shown to occur following the insertion of multiple homologous repeats of a transgene promoter region (33, 37). This can cause an inheritable suppression of the endogenous gene that is under the control of the same promoter. TGS in plants has been frequently correlated with condensation of chromatin and DNA methylation (29, 34). In contrast, PTGS in plants, as well as quelling in Neurospora, has been shown to require only the transgene insertion of homologous transcribed sequences (16). A frequently found and notable characteristic of PTGS is the presence of short, nonproductive RNA molecules (26). Our present results suggest that silencing of the Ehap-a gene in Entamoeba belongs to the TGS principle.
MATERIALS AND METHODS
Strain and culture conditions.
Trophozoites of E. histolytica strain HM-1:IMSS were grown at 37°C in TYI-S-33 medium (19). Transfected trophozoites were grown in the presence of the neomycin derivative G418, as previously described (12).
Plasmid construction.
The pEhActNeo shuttle vector, which served as the basic construct, contains the Neo gene that confers resistance to G418 flanked by the 5′ and 3′ untranslated regions (UTRs) of the amoeba actin 1 gene (1, 36) and the E. histolytica autonomous replication sequence, both cloned in pBluescript SK(−). The plasmid psAP-1 was constructed by inserting into the above-described plasmid vector a PCR fragment of the ap-a gene (amplified from genomic DNA of strain HM-1:IMSS) that includes 470 bp of the 5′ flanking region, the open reading frame (ORF), and 331 bp of the 3′ regulatory region. Primers 1 and 2 (Table 1) were prepared according to the sequence information available at Gene Bank accession no. x-70851. Using primers within psAP-1 as indicated (Table 1 and Fig. 1), the other plasmids (psAP-2 to psAP-7) were constructed as described above. Transfections of trophozoites were carried out essentially as previously described (24).
TABLE 1.
Primers used for preparation of plasmid constructs
| Primer | Sequence | S or ASa | Posi- tion | Cloning site |
|---|---|---|---|---|
| 1 | TCCCCGCGGCTTGCTGCACCCTTTG | S | −474 | SacII |
| 2 | TCCCCGCGGCTTCAGGATGGAACAG | AS | +674 | SacII |
| 3 | TCCCCGCGGGATTGTTTGTAAGATATG | AS | −1 | SacII |
| 4 | TCCCCGCGGCGTGCCACCTTCGATC | S | −354 | SacII |
| 5 | TCCCCGCGGCACGGTGATGTGGCTG | AS | −68 | SacII |
| 6 | TCCCCGCGGAAGATCGAAGGTGGCACG | AS | −340 | SacII |
| 7 | TCCCCGCGGCAGCCACATCACCGTG | S | −47 | SacII |
S, sense; AS, anitsense.
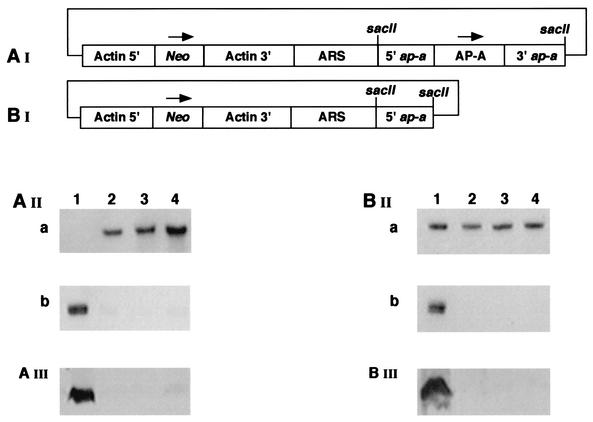
FIG. 1.
(AI) psAP-1 plasmid in which the ap-a gene, including sequences from its upstream and downstream regulatory regions, was inserted into a SacII site of the pEhActNeo shuttle vector (1, 36). ARS, autonomous replication sequence. (AII) Northern blot analysis of amoebic RNA extracted from the following sources: parent strain HM-1:IMSS (lane 1) and psAP-1 transfectants grown in the presence of 6 (lane 2), 12 (lane 3), and 24 (lane 4) μg of G418 ml−1. The DNA probes used were Neo (AIIa) and ap-a (AIIb). (AIII) Western blot of SDS-PAGE reacted with anti AP-A antibodies. Lanes depict results for the same cultures as described above. (BI) psAP-2 plasmid in which only the 470 bp of the 5′ flanking region was cloned in the same vector as described above. (BII) Northern analysis of psAP-2 transfectant. Lane 1, HM-1:IMSS parent strain; lanes 2, 3, and 4, psAP-2 transfectants grown in the presence of 6 (lane 2), 12 (lane 3), and 24 (lane 4) μg of G418 ml−1. The DNA probes used were ribosomal protein (BIIa) and ap-a (BIIb). (BIII) Western blot of SDS-PAGE reaction with anti AP-A. Lanes depict results for the same cultures as described above.
Northern blotting.
Total RNA was prepared using the RNA isolation kit TRI Reagent (Sigma). RNA (5 μg) was size fractionated on a 4% polyacrylamide denaturing gel containing 8 M urea and subsequently blotted to a nylon membrane. Using stringent conditions, hybridization was carried out with different probes (0.1% sodium dodecyl sulfate [SDS], 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]). Probes were randomly labeled using the Rediprime II kit (Amersham Life Science).
Search for small RNA molecules.
A fraction enriched in small RNA molecules was prepared as described previously (15). In brief, extraction from freshly harvested trophozoites was performed with phenol-chloroform and the nucleic acids were precipitated with 3 volumes of absolute ethanol and 1/10 volume of 3 M sodium acetate at −20°C. The washed sediments were resuspended in 2× distilled water, and the solution was incubated on ice (30 min) with polyethylene glycol (molecular weight [MW], 8,000) at a final concentration of 5% and 500 mM NaCl, after which the high-MW nucleic acids were precipitated while the small RNAs remained in the solution. The supernatants were then precipitated with ethanol as described above. Low-MW RNAs (50 μg) were separated by electrophoresis in 0.5× Tris-borate-EDTA buffer through 15% polyacrylamide 7 M urea gels. To control the size and polarity of the low-MW RNAs, sense and antisense oligonucleotides (25 nucleotide) were prepared based on the coding sequence of the ap-a gene and used as size markers. Double-stranded synthetic 25-bp oligonucleotides from the promoter area of the gene were added prior to the extraction to one-half of the freshly disrupted nontransfected trophozoites, which served as an internal control for size and efficiency of small RNA extraction.
Nuclear run-on analyses of gene transcription.
The procedure used was adapted from that used for Trypanosoma and Giardia (43, 49). Freshly grown trophozoites (3 × 107 to 5 × 107) were washed in serumless TYI medium and then twice in buffer A (150 mM sucrose, 20 mM potassium glutamate, 20 mM HEPES [pH 7.5], 3 mM MgCl2, 1 mM dithiothreitol, 10 μg of leupeptin ml−1). Trophozoites were suspended in 3 ml of buffer A and incubated for 5 min on ice, followed by permeabilization with lysolecithin palmitate (Sigma; final concentration, 150 μg ml−1) for 1 min with gentle mixing. The cells were washed by rapid centrifugation (1 min at 1,000 × g) and resuspended in a minimal volume of buffer A. Permeabilization was monitored by staining of an aliquot with propidium iodide (Sigma; 5 μg ml−1), which stains only the nuclei of permeabilized cells. An equal volume of 2× stock solution of transcription buffer (2× stock: 20 mM HEPES [pH 7.5], 180 mM potassium glutamate, 7 mM MgCl2, 1 mM dithiothreitol, 10 μg of leupeptin ml−1, 50 mM phosphocreatine, 1.2 μg of creatine kinase ml−1, 4 mM ATP, 2 mM GTP, 2 mM CTP, 1 mCi of [α-32P]UTP [Amersham; 3,000 Ci/mM]/ml) was added to the cells, and transcription was allowed to proceed for 15 min at 28°C. After transcription, 10-fold (vol/vol) TRI-Reagent (Sigma) was added to each of the incubation mixtures and RNA was extracted according to the manufacturer's directions. The recovered radiolabeled RNA was used immediately to probe dot blots (300 ng of DNA each) containing the ap-a gene as well as other amoebic genes as indicated. Hybridizations were done at 42°C for 48 h, and washing was stringent.
SDS-polyacrylamide gel electrophoresis (PAGE) and Western blots.
Soluble extracts from trophozoites prepared as previously described (12) were subjected to separation on a 20% polyacrylamide gel (20) under nonreducing conditions. Gels were blotted on a nitrocellulose membrane and subjected to immunoreactions with polyclonal antibodies prepared against high-pressure liquid chromatography-purified AP-A kindly supplied by M. Leippe, U. Wuerzburg, Wuerzburg, Germany. The blots were washed and incubated with horseradish peroxidase conjugated to donkey anti-rabbit immunoglobulin whole antibody (Amersham Pharmacia Biotech) and developed by an enhanced chemiluminescence kit (ECL) (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). The procedure was carefully optimized using various protein and antibody concentrations.
Quantitation of plasmid copies in transfectants.
Using the Wizard Genomic DNA purification kit (Promega), genomic DNA was prepared from strain HM-1 and transfectants psAP-1, psAP-2, and psAP-4. DNA from 5 × 105 trophozoites was denatured by 0.3 M NaOH, neutralized by ammonium acetate, and blotted onto a nylon membrane on a slot blotter. A DNA sample of plasmid psAP-1 was treated in the same way and served for calibration of the copy number. The blots were hybridized with a number of labeled probes under stringent conditions as previously described (12).
Southern blots of genomic DNA.
DNA was prepared as mentioned above from trophozoite control cultures of HM-1:IMSS and silenced, plasmidless, cloned (clone G3) trophozoites. DNA samples were digested with different restriction enzymes, separated on 0.8% agarose gels, and subjected to alkaline transfer to positively charged nylon membranes. The membrane was then hybridized with different probes as mentioned above.
Cytopathic activity on cell monolayers.
Baby hamster kidney (BHK) cells were grown to confluency as monolayers in Dulbecco's modified Eagle's medium, supplemented with 10% of fetal calf serum, in 24-well plates. The destruction rate of the BHK cell monolayer by nontransfected or transfected trophozoites was evaluated in triplicate experiments as previously described (11).
Cytotoxic activity.
The cytotoxic activity of viable trophozoites was determined by vital dye exclusion (31). Freshly harvested BHK cells were washed and resuspended in TYI-S-33 medium without serum. The cells were incubated with trophozoites in a ratio of 6:1 at 37°C. Samples were examined microscopically in a hemocytometer chamber at different time points during the incubation. Viability was indicated by exclusion of trypan blue (0.1%). The cytotoxic activity is expressed as the percentage of stained cells in each sample minus the percentage of stained cells in the sample with BHK cells alone for the same time point. Averages of triplicate experiments are given.
Induction of amoebic liver abscesses in hamsters.
Inoculation of trophozoites directly into the liver was performed after laparotomy in Syrian Golden hamsters (females, 6 weeks old; four animals in each group) as previously described (3). Hamsters were sacrificed after 7 days, and the sizes of the liver abscesses were assessed.
Attachment and solubilization of Esherichia coli cells by trophozoites.
Trophozoites (106) were incubated with [14C]glucose-labeled E. coli strain 346 (109), a type I pilated bacterium, for 30 min at 37°C in 1 ml of phosphate-buffered saline as previously described (10). The separation of the bacteria that attached to the trophozoites from the free bacteria was performed by discontinuous density gradient centrifugation with Percoll as previously described (9). The gradient fraction containing the trophozoites with the attached bacteria was recovered, and aliquots were counted to determine the amount of radioactively labeled bacteria that was attached. The integrity of the bacteria that attached to the trophozoites was determined by their susceptibility to solubilization with 0.2% Triton X-100. After exposure to Triton X-100, the cell mixture was sedimented at 6,000 × g, and the amount of radioactivity in the soluble and sedimented fractions was determined by counting aliquots in a Packard Tri-Carb 1500 scintillation counter. The percentage of the soluble count in the total count indicated the degree of disruption caused to the attached bacteria.
Ingestion of GFP-labeled E. coli cells.
E. coli cells (109) containing a plasmid which expresses green fluorescent protein (GFP) (obtained from G. Frankel, Imperial College, London, United Kingdom) were associated in culture medium with different types of trophozoites (106). The trophozoites with bacteria were harvested after 1 h and washed by sedimentation, and a portion (25%) was fixed with formaldehyde (3.7%) and extensively washed. The remaining trophozoites were resuspended in fresh culture medium to which Claforan (Sigma; 50 μg/ml) was added to kill all remaining bacteria that were not ingested by the trophozoites. The trophozoites were harvested after 24 h, washed, and fixed with formaldehyde as described above. The trophozoites and the fluorescent bacteria were examined by confocal microscopy (Olympus Fluoview FV500).
RESULTS
Silencing of the Ehap-a gene following transfection with a hybrid plasmid containing the gene flanked by its 5′ and 3′ regulatory sequences.
In an attempt to overexpress the AP-A gene (Ehap-a) in trophozoites of E. histolytica virulent strain HM-1:IMSS, a hybrid plasmid (psAP-1) based on the pEhActNeo vector (1, 35, 36) was prepared (Fig. 1, panel AI). A cassette with the ap-a gene flanked by segments from its own 5′ upstream (470 bp) and 3′ downstream (331 bp) regulating elements was inserted into the vector. The 5′ and 3′ flanking regions which were used were long enough to include the known regulatory motifs, and their particular lengths were arbitrarily determined according to the convenience of preparing oligonucleotide primers at those sites. Trophozoites were transfected with psAP-1 by electroporation and selected with G418 as previously reported (12). Northern blot analysis of the psAP-1 transfectants grown in the presence of different G418 concentrations revealed, to our surprise, no detectable transcripts of ap-a in comparison to those of nontransfected controls (Fig. 1, panel AIIb). The inhibition of transcription of ap-a in the psAP-1 transfectants from both the chromosomal and the plasmid-coded ap-a genes was already complete at the low G418 concentration of 6 μg ml−1. As previously demonstrated (12), the neo transcript levels correlated well with the G418 concentration present in the culture (Fig. 1, panel AIIa). The AP-A protein content of the same cells, as examined by Western blotting, revealed a similar picture; namely, psAP-1-transfected cells were devoid of any AP-A protein (Fig. 1, panel AIII).
Silencing following transfection with a hybrid plasmid containing only the 470-bp segment of the 5′ flanking region of Ehap-a gene.
To understand which part of the ap-a gene cassette in the psAP-1 construct was responsible for the silencing phenomenon, we prepared a new hybrid plasmid, psAP-2 (Fig. 1, panel BI), in which we introduced only the above-mentioned 470-bp ap-a 5′ flanking segment into the pEhActNeo vector, and then generated with it a new transfectant. The results of Northern and Western blot analysis of this transfectant were identical with those obtained with psAP-1; the trophozoites were already devoid of ap-a transcripts (Fig. 1, panel BIIb) as well as of AP-A protein (Fig. 1, panel BIII) at a G418 concentration of 6 μg of ml−1 in culture. The transcript of ribosomal protein L-21 served as a control for the RNA samples and gave identical results in all cultures (Fig. 1, panel BIIa). The growth rates of transfectant psAP-1 and psAP-2 were very similar to that of the nontransfected parent strain HM-1:IMSS, and SDS-PAGE results for Coomassie-stained trophozoite lysates of the transfectants were practically identical with those of nontransfected cells (data not shown).
Transfections with different sections of the Ehap-a 5′ flanking fragment.
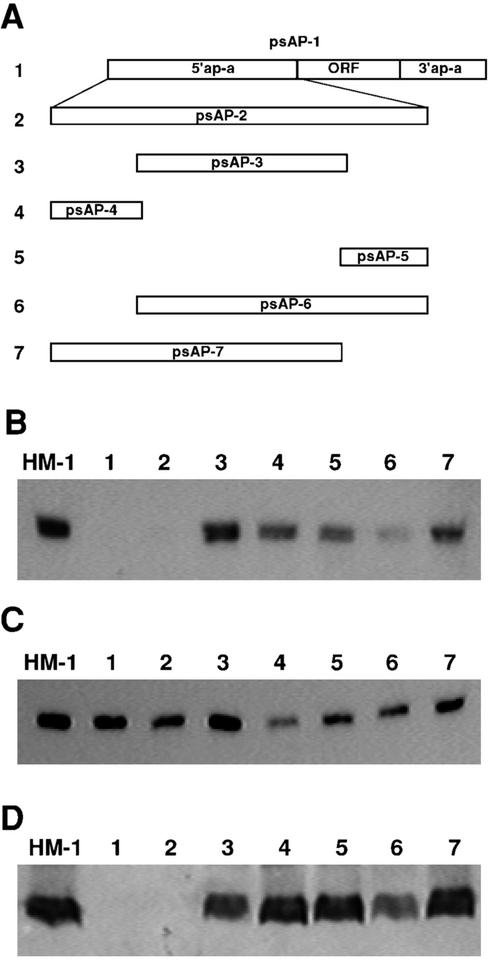
To further identify putative DNA regions responsible for the silencing phenomenon, a series of plasmid constructs containing only certain parts of the 5′ 470-bp flanking element used in psAP-2 were prepared based on sequence and bioinformatic analysis. Plasmid psAP-3 (Fig. 2A) had two deletions; the first was of approximately 120 bp at the distal end of the 470-bp fragment (bp −470 to −352). The second deletion in psAP-3 was of the proximal 50-bp sequence (bp −1 to −50) which is immediately upstream to the ATG start codon of the ap-a gene and includes the short UTR and TATA-like motif. Plasmid psAP-4 contained only the above-mentioned 5′ distal 120-bp segment (bp −352 to −470), and plasmid psAP-5 contained only the 65-bp proximal sequences (bp −1 to −66). In each of the last two plasmids, psAP-6 and psAP-7, either the distal or the proximal part of the 5′ element was omitted. Each of the above-described plasmid constructs was independently transfected into trophozoites, and the resulting steady-state levels of ap-a mRNA and AP-A protein found in the respective transfectants are shown in Fig. 2. The results show that for the full silencing effect of the ap-a gene, the entire stretch of 470 bp of the 5′ flanking region was required. The presence of the ap-a coding sequences was not necessary. When tested, shorter fragments of the 470-bp 5′ flanking region did not induce silencing. However, a partial decrease in ap-a transcription and AP-A protein expression was observed in psAP-6, which lacks only the distal section, but not in psAP-7, from which the proximal region was deleted (Fig. 2B). Transcription levels were normalized on the Northern blots by hybridization with a probe specific for ribosomal protein L21 (Fig. 2C). The AP-A protein levels of the transfectants correlate well with the levels of transcription. The above results indicate that the 5′ UTR, which is present in psAP-5, did not in itself cause any silencing, whereas sequences in the region of −50 to −120, as well as a sizable (>400 bp) fragment, appear to be important for the silencing to occur.
FIG. 2.
(A) Schematic map of the plasmids used to examine the regions of the 5′ upstream 470-bp segment, which may be responsible for silencing. Using the primers described in Table 1, plasmids psAP-2 to psAP7 were derived from plasmid psAP-1. (B) Northern blots probed with ap-a gene. (C) Northern blot probed with the ribosomal protein gene EhRP-L21. (D) Western blot reacted with anti-AP-A antibodies and horseradish peroxidase anti-rabbit antibodies. Lane HM-1, control parent strain; lane 1, psAP-1; lane 2, psAP-2; lane 3, psAP-3; lane 4, psAP-4; lane 5, psAP-5; lane 6, psAP-6; lane 7, psAP-7.
Plasmid copy number in transfectants.
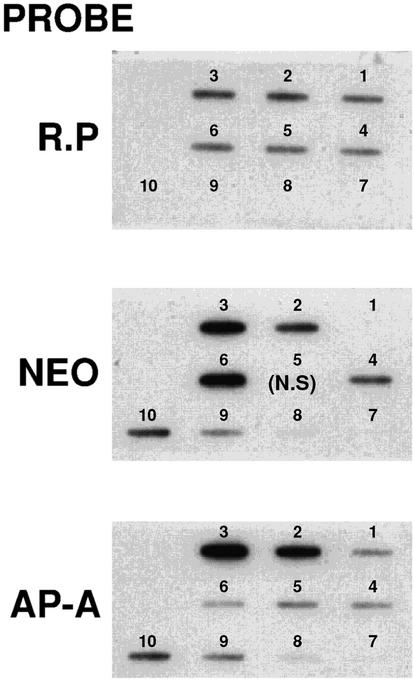
Using different probes, the average number of plasmid copies present in cultures of trophozoites of transfectants psAP-1 and psAP-2, as well as in the control psAP-4 culture grown in the presence of G418 at the indicated concentrations (Fig. 3), was determined by slot blot hybridization of DNA corresponding to 5 × 105 trophozoites. The hybridization with EhRP-L21 served to normalize the DNA amount (Fig. 3), and plasmid DNA of psAP-1 served for calibration and calculation of the plasmid number. Hybridization with Neo (which is absent in the nontransfected HM-1:IMSS trophozoites) revealed the presence of approximately 20 plasmid copies/cell in cultures growing with 6 μg of G418 ml−1 and the presence of 80 copies when 100 μg ml−1 was present in the culture (Fig. 3). Hybridization with the ap-a ORF (Fig. 3) revealed a fivefold increase in ap-a gene signal in transfectant psAP-1 in comparison to that seen with the control transfectants psAP-4 and psAP-2, which have a plasmid devoid of the ap-a ORF, as well as the nontransfected control. These findings indicate that even at low concentrations of G418 (6 μg ml−1), the transfectants have a severalfold excess of homologous sequences of the ap-a gene promoter region.
FIG. 3.
Quantitation of plasmid copies in DNA of 5 × 105 transfected amoeba from the following sources: HM-1 (slot 1); psAP-1 grown with 6 (slot 2) and 100 (slot 3) μg of G418 ml−1; psAP-2 grown with 6 μg of G418 ml−1 (slot 4); and psAP-4 grown with 6 (slot 5) and 100 (slot 6) μg of G418 ml−1. Slots 7, 8, 9, and 10 show samples from plasmid psAP-1 at a multiplicity (compared to the genomic DNA) of 0.8 (slot 7), 2 (slot 8), 8 (slot 9), and 20 (slot 10). Each blot was probed with different labeled probes. (R.P) With ribosomal protein L-21, all samples showed similar levels and the plasmid control showed no signal. (NEO) The nontransfected HM-1 strain (slot 1) showed no signal. Differences were seen between the samples grown at 6 μg of G418 ml−1 (slots 2 and 4) and those grown at 100 μg ml−1 (slots 3 and 6), and psAP-1 plasmid signal levels were relative to the amount spotted. (AP-A) Only samples 2 (slot 2) and 3 (slot 3), containing psAP-1 plasmid, had extra copies of the ap-a gene compared to those of HM-1. The other transfectant showed the same number of copies as the control strain (slot 1). N.S, no sample in the slot.
Stability of the ap-a transcriptional silencing state.
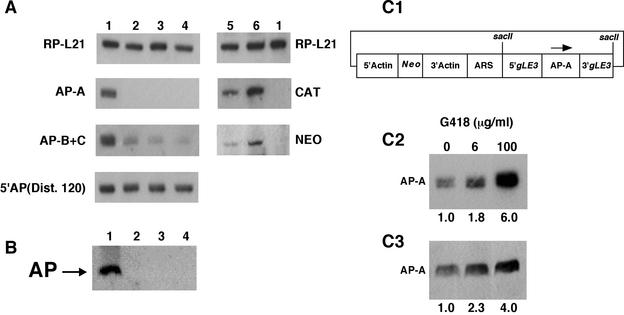
Omission of the selective drug (G418) from the cultures of transfectant psAP-2 for over 90 days drastically reduced the plasmid content of the trophozoites but did not cause any reversal of the ap-a silencing effect. Three independent clones (G1 to G3) were isolated from the cultures growing without G418, and even though all three of them were found to be devoid of plasmid (as evidenced by lack of PCR amplification of the Neo gene), they still showed no AP-A protein expression, indicating that gene silencing remained in force. Clone G3 was chosen for further experiments (Fig. 4A and B, lanes 3). The results obtained were identical to those of the psAP-2 parent culture (Fig. 4A and B, lanes 2). In addition, we attempted to see whether retransfection with plasmid pA-7 (Fig. 4, panel C1), which contains the ap-a gene under the control of regulatory elements of the EhRP-L21gLE3 gene copy (13), of clone G3 trophozoites that lack plasmid and AP-A would restore AP-A expression. As shown in Fig. 4 (panels C2 and C3), plasmid pA7 was able to overexpress AP-A in trophozoites of strain HM1:IMSS (1.8-fold transcript and 2.3-fold protein levels at 6 μg of G418 ml−1) but was incapable of producing any AP-A protein even when the concentration of G418 for the clone G3-retransfected trophozoites was raised to 24 μg ml− (Fig. 4A and B, lanes 4). Furthermore, using primers which amplify 120 bp at the 5′ end of the ap-a mRNA, no Ehap-a transcript was detected by reverse transcription-PCR analysis of RNA extracts from clone G3 trophozoites retransfected with plasmid pA7 (data not shown). On the other hand, retransfection of the same ap-a-silenced clone G3 trophozoites with plasmid pTS-1 (35), which contains the chloramphenicol acetyltransferase (CAT) reporter gene under the control of the same EhRP-L21gLE3 regulatory elements as described for pA-7, was capable of transcribing CAT (Fig. 4A, lane 5). The levels of CAT transcript in strains G3 and HM-1:IMSS transfected with pTS1 correlate with the levels of plasmid, as shown by the Neo transcription results (Fig. 4A, lanes 5 and 6). The expression of CAT in strain G3 indicates that silencing is specific for the ap-a gene sequences. The ribosomal protein probe served as a standard control (Fig. 4A).
FIG. 4.
Characterization of the plasmidless AP-A lacking trophozoite clone G3. (A) Northern blots of various transfectants as well as plasmidless trophozoites probed with a variety of probes. Lanes 1, HM-1:IMSS nontransfected parent strain; lane 2, transfectant psAP-2; lane 3, plasmidless clone G3 isolated from cultures of transfectant psAP-2 grown in the absence of G418; lane 4, clone G3 retransfected with plasmid pA7; lane 5, clone G3 retransfected with plasmid pTS1, which contains the CAT gene (35); lane 6, strain HM-1:IMSS transfected with plasmid pTS1. Probes were as marked. RP-L21 served as loading control. The AP-A probe revealed a complete absence of transcript in lanes 2, 3, and 4. Probe AP-B+C contained in tandem the sequences of genes ap-b and ap-c and was used for their detection. The result revealed a lower level of transcription. Probe 5′AP(Dist. 120), which consists of the distal 120 bp of the 470-bp 5′ flanking element of gene Ehap-a, hybridized to an unidentified transcript. (B) Western blot of SDS-PAGE results for lysates of trophozoites reacted with anti AP-A. Lanes contain the same samples as described above. Lanes 2, 3, and 4 showed no detectable AP-A. (C1) pA7 plasmid in which the ap-a gene is flanked by the 5′ (5′gLE3) and 3′ (3′gLE3) elements of the EhRP-L21 gene (13). ARS is the sequence containing the autonomous replication element. (C2 and C3) Northern and Western blots, respectively, of strain HM-1:IMSS and of HM-1:IMSS trophozoites transfected with plasmid pA7 and grown with G418 at the indicated concentrations.
Does silencing affect the transcription of the other AP genes?
E. histolytica has three AP isoforms (A, B, and C) in the respective ratios of 21:9:1 (38). They all share certain structural similarities and exhibit pore-forming activity on artificial membranes (31). Northern blots of ap-a-silenced trophozoites were hybridized with a probe containing in tandem the coding sequences of both ap-b and ap-c. The specificity of the probe was tested on the respective gene templates. As shown in Fig. 4A, transcription in all of the silenced cultures (lanes 2, 3, and 4) revealed levels of transcription of genes ap-b and ap-c that were still detectable but significantly lower than those of the control. The 120-bp distal segment of the 5′ flanking region of Ehap-a (−470 to −350) hybridized to a transcript which was present at comparable levels in the RNA extracts of all the trophozoites (Fig. 4A). This 120-bp fragment shares sequence homology to several contigs (for example, contig ENTBT19F) in the E. histolytica genome (published by the Institute for Genomic Research), and its sequence appears to include an ORF. Database searches did not yield homology to any known protein.
Sequence and Southern blots of genomic ap-a in silenced trophozoites.
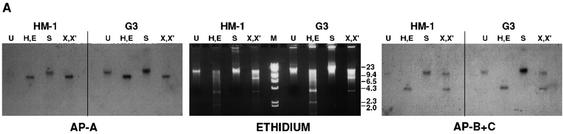
PCR amplification of the genomic ap-a segment from its 5′ upstream region down to the 3′ region (1,100 bp) of the silenced transfectant psAP-2 as well as the nontransfected control revealed a fragment of identical size and DNA sequence, indicating that transfection of plasmids containing the homologous 470-bp promoter region caused no sequence changes. Furthermore, Southern blots of genomic DNA from silenced trophozoites of clone G3 and strain HM-1 (cleaved with various restriction enzymes as indicated) (Fig. 5A) did not reveal any differences in the fragments which appeared either in ethidium staining or after blot hybridization with the ap-a probe (left panel) or with the probe for ap-b and ap-c (right panel), a finding which indicates that no rearrangement took place. Use of McrBC enzyme, which specifically cleaves at methylated cytidine residues, yielded no differences either (data not shown). Both DNA methylation and histone hypoacetylation are modifications which are frequently associated with repression of gene expression (22, 48). Possible connections between these modifications were investigated by treating growing cultures of the transfected psAP-2 trophozoites with trichostatin A (Sigma) (42), a potent inhibitor of histone deacetylase, or with 5′ azacytidine (Sigma), which inhibits methylation of DNA (46). No resumption of ap-a expression was observed after treatment of cultures of psAP-2 transfectants for 24 h with either 5′ azacytidine (1 μM) or with trichostatin A (100 nM) or with both drugs combined (Fig. 5B). Growth of trophozoites with higher concentrations of 5′ azacytidine (20 μM) did not reverse the silencing.
FIG. 5.
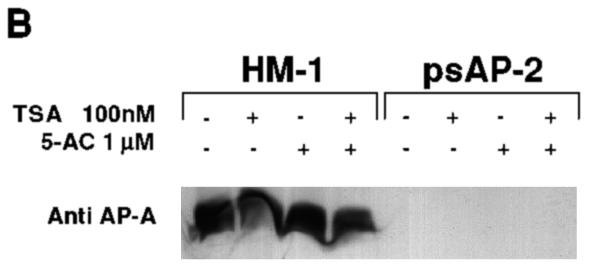
(A) Southern blots of restriction digests of DNA samples from nontransfected parent strain HM-1:IMSS or plasmidless, AP-A-lacking clone G3. Samples were undigested (U) or digested with HindIII and EcoRI (H,E), SalI (S), or XhoI and XbaI (X,X′). (Middle panel) Ethidium-stained agarose gel. (Left and right panels) Blots were probed under stringent conditions with probes specific for ap-a and with a probe containing in tandem the sequences of ap-b and ap-c genes, respectively. (B) Western blot analysis of AP-A with trophozoites grown for 24 h with trichostatin A (TSA) (100 nM), 5′aza-2′deoxycytosine (5-AC) (1 μM), or with both compounds. The amount of AP-A in the lysates was monitored with antibodies against AP-A (Anti AP-A). The trophozoites were from strain HM-1:IMSS (HM-1) and psAP2 (as marked).
Nuclear run-on analysis of initiation of transcription of the ap-a gene.
Nuclear run-on experiments were performed to determine whether silencing is at the level of transcription initiation. Nascent radiolabeled RNA was produced using permeabilized trophozoites (43), and as shown in Fig. 6, no hybridization to the ap-a gene was detected in the silenced, plasmidless G3 trophozoites, whereas positive hybridization as seen with the nontransfected trophozoites was observed with other transcripts. The transcript levels of the two other Ehap genes (ap-b and ap-c), whose natural abundance is significantly lower (38), was indeed very low and was nearly undetectable in the silenced culture.
FIG. 6.
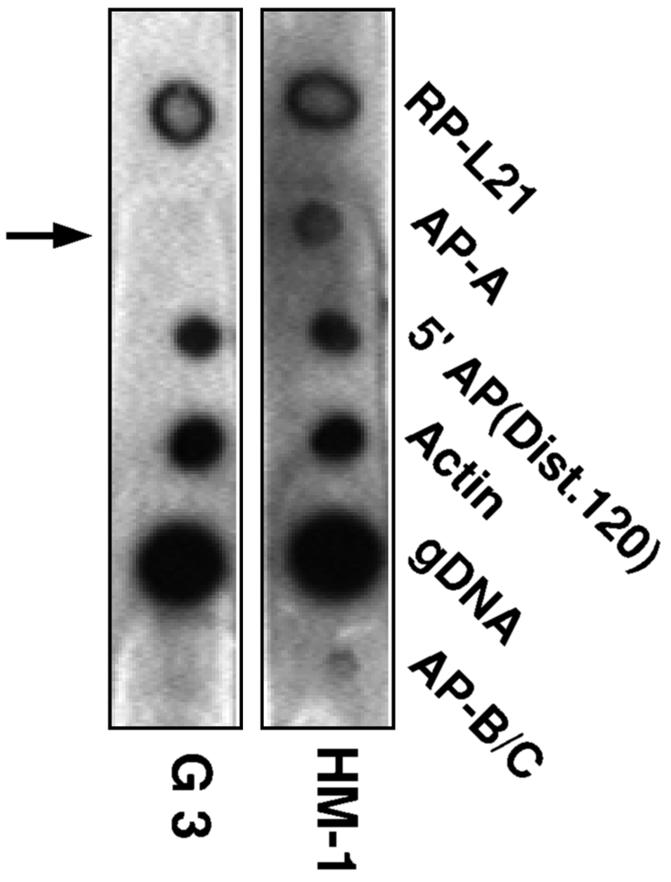
Hybridization of blots with radiolabeled nascent RNA extracted from nuclear run-on experiments of control nontransfected HM-1:IMSS trophozoites and AP-A-lacking trophozoites of clone G3. E. histolytica DNA samples on the blots were prepared by PCR on E. histolytica DNA and included the ribosomal protein L21 (RP-L21), the ap-a gene (AP-A), a 5′ flanking fragment (120 bp) of the ap-a gene (bp −470 to −350) [5′ AP(Dist.120)], actin, genomic DNA (gDNA), and DNA of tandemly linked ap-b and ap-c genes (AP-B/C). 300 ng of each DNA was spotted and denatured prior to hybridization.
Search for siRNA molecules.
The suppression of transcription initiation indicates that silencing of the ap-a gene is due to an TGS mechanism (33, 37) rather than to PTGS (16, 45). To further strengthen this notion, a search for small RNAs, a typical result of PTGS, was performed. These short (22 to 25 nucleotide) RNA fragments of both sense and antisense polarity correspond to the target gene and are produced by the processing of larger double-stranded RNAs (dsRNAs) (25, 50). Extraction and detection of short dsRNAs were done according to the method of Catalanotto et al. (15) from trophozoites of transfectants psAP-1 and psAP-2, clone G3 and G3 trophozoites retransfected with plasmid pA7, and trophozoites from the nontransfected HM-1 control. Hybridizations of Northern blots with a probe for the ap-a coding sequence as well as with a probe for the 5′ flanking region of Ehap-a or of EhRP-L21 failed to detect any small RNA molecules (data not shown). Controls designed to test the efficiency of the small-RNA extraction and detection procedures performed as expected.
Phenotype of ap-a-silenced trophozoites. (i) Virulence determinations.
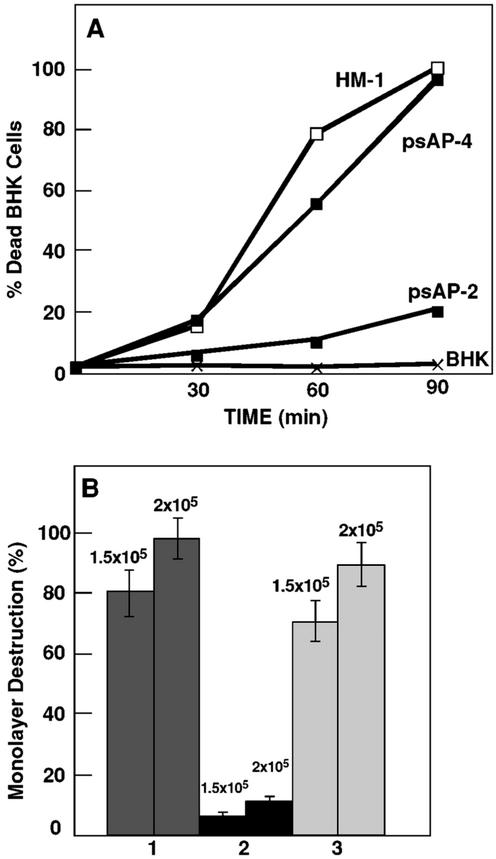
psAP-2 transfectants growing with a low G418 concentration of 6 μg ml−1 were found to be avirulent in both in vitro and in vivo tests (12). Trophozoites were incapable of killing BHK cells in suspension or of destroying monolayers of BHK cells grown in tissue cultures (Fig. 7A and B). As a control for all of these experiments, we used psAP-4 transfectants growing at the same low G418 concentration of 6 μg ml−1. This transfectant has a normal level of production of AP-A (Fig. 2B and D, lanes 4), and its virulence was found to be very similar to that of the nontransfected HM-1:IMSS parent strain. The same avirulent phenotype was obtained for psAP-1 transfectants. Trophozoites of the plasmidless, AP-A-lacking clone G3 were also avirulent, and as determined by their interaction with suspended BHK cells, their cytotoxicity after 90 min was less than 20% of that of the control parent strain trophozoites. Trophozoites of transfectants psAP-1 and psAP-2 and clone G3 did not induce the formation of liver lesions in hamsters, even at inoculations of one million trophozoites/liver (Table 2).
FIG. 7.
(A) Cytotoxic activity. The mortality rates of BHK cells incubated in suspension with freshly harvested trophozoites of strain HM-1:IMSS, psAP-2 transfectants, and psAP-4 transfectants (which served as control) are shown. Both transfectants were grown with 6 μg of G418 ml−1. The number of trypan blue-stained BHK cells was monitored as a function of incubation time. The percentage of BHK cells that incorporated the dye in the absence of trophozoites remained low (<4%) during the period tested. (B) Cytopathic activity of different trophozoite cultures of HM-1:IMSS (lane 1), psAP-2 transfectants (lane 2), and psAP-4 control transfectants (lane 3). Both transfectants were grown with 6 μg of G418 ml−1. Two concentrations (1.5 × 105 and 2 × 105) of trophozoites were added per well for each type of trophozoite; experiments were repeated in triplicate. Bars represent standard errors. Statistical significance was determined by single-tailed t testing (P < 0.05).
TABLE 2.
Induction of liver lesions in hamsters by different trophozoites
| Type of trophozoite (conditions) | No. of tropho- zoites injected/ liver (105) | No. of animals with abscesses/ total no. | Abscess size (mm) |
|---|---|---|---|
| HM-1:IMSS | 2.5 | 4/4 | 20-30 |
| 5.0 | 4/4 | 30-35 | |
| Control psAP-4 | 5.0 | 4/4 | 30-35 |
| (6 μg of G418 ml−1) | - | ||
| Silenced psAP-1 | 5.0 | 0/4 | None |
| (6 μg of G418 ml−1) | |||
| Silenced psAP-2 | 5.0 | 0/4 | None |
| (6 μg of G418 ml−1) | |||
| Silenced clone G3 | 10.0 | 0/2 | None |
(ii) Ingestion of bacterial cells.
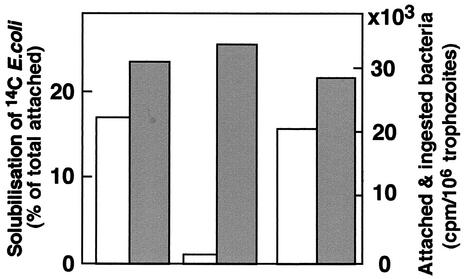
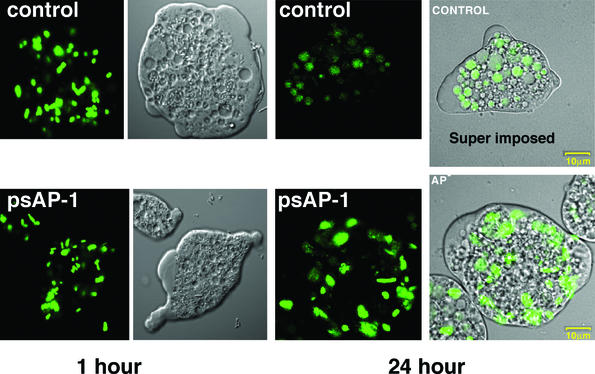
The association of trophozoites with 14C-labeled E. coli cells at a ratio of 1:1,000 was previously shown to cause the rapid ingestion and degradation of the bacterial cells (11). The bacteria which attach and become ingested by the trophozoites have been shown to become sensitive to solubilization by Triton X-100 (0.2%) (11). A very similar amount of 14C-labeled E. coli cells was found to attach to trophozoites of nontransfected strain HM-1 and of transfectants psAP-2 and psAP-4 (Fig. 8). Nevertheless, following treatment with Triton X-100, a much larger portion of the total amount of bacteria which attached and were ingested by control trophozoites or by control transfectant psAP-4 was solubilized in comparison to that which attached and were ingested by psAP-2 trophozoites (Fig. 8), indicating that the AP-A-lacking trophozoites were unable to degrade the ingested bacteria. The drastic difference between the fate of the bacteria ingested by the nontransfected trophozoites and that of the ap-a-silenced trophozoites was also seen upon ingestion of GFP-labeled E. coli cells. As shown in Fig. 9, the differences in the locations and shapes of the ingested fluorescent bacteria in the trophozoite vacuoles seen after 60 min were not large. However, after an additional 23 h of growth of the cultures of trophozoites which had ingested the bacteria, most of the fluorescence in the control trophozoites could be seen (Fig. 9) diffused in the vacuoles, whereas in trophozoites of transfectant psAP-1, the fluorescence remained associated with distinct rod-like structures in many cases. A similar difficulty in digestion was seen when human red blood cells (hRBC) were associated with trophozoites. The amounts of ingested hRBCs during the first hour were similar for the psAP-1 transfectants and the controls, but upon further cultivation of the trophozoites (23 h), many intact hRBCs were still visible in vacuoles of trophozoites of transfectant psAP-1 (in contrast to the control trophozoites, where only remnants could be seen) (data not shown).
FIG. 8.
The total amount of 14C-labeled E. coli cells which attached and became ingested by the trophozoites, as determined after removal of the nonattached bacteria by Percoll gradients (10), is depicted using filled bars. The percentage of radiolabeled bacteria (out of the total attached) which solubilized after treatment of trophozoite samples with Triton x-100 is depicted using empty bars. Left bar pair, strain HM-1:IMSS; center bar pair, transfectant psAP-2, right bar pair, transfectant psAP-4.
FIG. 9.
Confocal fluorescent microscopy of trophozoites that ingested GFP-labeled E. coli cells. Trophozoites were associated with the bacteria for 1 h, after which they were harvested and washed. A portion of the total amount of trophozoites was then fixed in formaldehyde, and the other portion was resuspended in fresh medium for further incubation (23 h) in the presence of antibiotics (Claforan) to prevent the growth of the noningested bacteria. After that period, the trophozoites were harvested, washed, and fixed in formaldehyde. The panel labeled “Super imposed” combines the image obtained by phase contrast (Nomarski optics) with the fluorescent image to show the bacterial location in the vacuoles.
DISCUSSION
The development of plasmid vectors which, after transfection into trophozoites, can up- or down-regulate the expression of certain genes of E. histolytica has in recent years enabled the study of the role of a number of virulence factors, such as those of the cysteine proteases, the Gal/GalNAc-inhibitable lectin, meromyosin, and the AP, in the pathogenesis of E. histolytica (2, 4, 5, 12, 27, 47, 52). During an attempt to overexpress AP-A using a hybrid plasmid (psAP-1) with a cassette containing the ap-a gene flanked by segments of the 5′ upstream and 3′ downstream regions of the endogenous gene, we discovered, to our surprise, that instead of overexpression, a complete suppression of transcription and translation of both the episomal and chromosomal ap-a genes had occurred. The observed silencing was unexpected, because we had previously obtained overexpression of ap-a when transfection was done with a similar hybrid plasmid (pA-7) in which the ORF of the ap-a gene was flanked by the 5′ and 3′ segments of another gene (EhRP-L21gLE-3) (13). In that study, the level of overexpression of AP-A protein in pA7-transfected trophozoites of strain HM-1:IMSS was approximately fourfold higher than that in the nontransfected control (13). Furthermore, some overexpression of AP-A was also obtained (unpublished results) when using the inducible plasmid pEhHYG-TetR-O-ap-a (23).
The remarkable total silencing observed with plasmid psAP-1 implied that something within the regulatory flanking elements of the ap-a gene was responsible for the suppression of transcription. The results obtained with plasmid psAP-2, which was constructed to contain only the 5′ flanking segment (470 bp) without the coding or 3′ flanking sequences, clearly demonstrated that the same silencing of ap-a could be achieved by introducing this 5′ flanking element alone. Preliminary sequence analysis of the 470 bp of the 5′ flanking region indicated that it contains promoter motifs with a TATA-like box similar to those which have been previously found in numerous genes of E. histolytica (14, 39, 40). The 470-bp flanking segment of ap-a also contains at its 5′ distal end a 120-bp region which appears to be present in additional sequences of the E. histolytica genome and which encodes an unidentified transcript. To learn more about the sequences which are important for the silencing effect, a number of plasmid constructs containing partial segments of the 470-bp 5′ fragment were prepared. The transfectants which were produced with these plasmid constructs revealed that only the intact 470-bp fragment can reproduce the complete silencing effect, whereas transfectants containing constructs with only parts of the 5′ fragment were incapable of reproducing it. The only construct which showed a reduction of transcription was psAP-6, which has a truncation of 120 bp in its 5′ end. The requirement for a minimal size of homology of 400 bp is already known for two kinds of TGS mechanisms, the repeat induced point mutation and methylation induced premeiotically (16).
Homology-dependent silencing of gene expression is a well recognized phenomenon in many organisms, including yeasts, plants, and humans (16, 18, 26, 34, 45). Gene silencing through mechanisms that are based on the recognition of nucleic acid sequence homology is achieved via diverse strategies, namely, inactivation at the transcriptional level (TGS) (33, 37) or the posttranscriptional level (PTGS) (16, 41). We demonstrate in this report for the first time that gene silencing also occurs in E. histolytica, and our present results indicate that the suppression of ap-a expression is via a TGS mechanism. The experimental evidence that supports this conclusion is as follows. (i) Silencing of the ap-a gene occurred upon transfection of several (∼20) copies of the upstream promoter region of this gene. Neither the ORF nor the 3′ regulatory sequences were required. (ii) Nuclear run-on experiments indicate that there is no transcription initiation of the ap-a gene in the silenced trophozoites. (iii) Short dsRNA fragments of the ap-a gene, a typical result of silencing via the PTGS mechanism (26), were not detected in RNA extracts of silenced amoeba.
It has been established that the homologous duplicate sequences that are associated with gene silencing can trigger epigenetic changes, such as DNA methylation, histone deacetylation, or methylation, and changes in chromatin structure (29, 34). No DNA rearrangements or changes in the restriction pattern of genomic DNA of the silenced trophozoites were detected using methylation-dependent enzymes or other restriction enzymes. In addition, we investigated the possibility that treatment of silenced trophozoite cultures with inhibitors of DNA methylation, such as 5-azacytidine (46), or of histone deacetylation, such as trichostatin A (42), restores AP-A expression but found that it did not. Although our present data do not imply that DNA methylation or histone acetylation is involved in maintaining the silencing of the ap-a gene, we still cannot rule out the possible involvement of other types of methylations or of trichostatin-resistant histone deacetylases such as members of the Sir2 family (28, 34), nor can we rule out the possible involvement of locus-specific occlusion of histone acetyltransferase accessibility; further investigations are required to clarify these possibilities.
One of the most interesting aspects of the silencing phenomena in E. histolytica is its remarkable stability, which persisted even after removal of the plasmid that contained the homologous sequences. A plasmidless clone isolated from this culture (strain G3) continued to lack AP-A expression even after 6 months in culture. What was even more striking was the fact that transfection of the silenced, plasmidless G3 trophozoites with a plasmid containing the ap-a gene under the control of a different promoter failed to express the AP-A protein. This inability was only restricted to the ap-a gene, as transfection of G3 trophozoites with an analogous plasmid in which the ap-a gene was replaced by the CAT gene transcribed and produced CAT. Silencing appears to be specific for the ap-a gene; other genes, such as those encoding ribosomal protein L-21, actin, lectin, and cysteine proteinase, show no changes in expression due to the silencing. Some effect was, however, observed with the other two isoforms of AP (AP-B and AP-C). A reduction in their transcription was detected, which might be evidence of a feedback inhibition explainable by the fact that AP-A was missing.
The most commonly reported form of gene silencing (or gene quelling, as it is termed in the case of Neurospora) is the posttranscriptional (PTGS) type, which requires the transgene insertion of homologous transcribed sequences (16, 17). Such silencing of gene expression is currently understood to be the consequence of mRNA destruction which appears to be triggered by the presence of dsRNA (50). The small dsRNA fragments (7, 15) are generated by the transfected cells and cause degradation of the cognate mRNA. In the case of E. histolytica, transcription initiation was shown to be blocked and careful and repeated analysis of trophozoite RNA extracts failed to detect any small RNA fragments in either of the AP-A-silenced trophozoites, suggesting that silencing belongs to the TGS principle. TGS has been shown to occur in a number of systems, including those of plants, following the insertion of multiple homologous repeats of a transgene promoter region and in the absence of transcribed sequences of the endogenous gene (33, 37, 48). Such silencing has been shown to be inheritable in progeny, and it can persist even when the silencer sequence was excised (53). In Saccharomyces, an epigenetic formation of a heterochromatin-like structure results in a TGS which is inherited for many mitotic divisions (51). Transformation of the plant pathogen Phytophthora infestans with constructs containing the elicitin inf1 gene resulted in the specific transcriptional silencing of the transgene as well as the endogenous gene, and the silencing status also remained in effect in nontransgenic progeny (44). Silencing was not due to gene disruption, and it was not based on high turnover of inf1 mRNA. A number of possible epigenetic mechanisms have been proposed for locking a gene into a silenced state, and these seem to be unique for each of the different species in which they were discovered. In some cases, the silencing mechanisms involve DNA or protein modifications which trigger the assembly of a repressed chromatin structure. Well-documented modifications include DNA methylation (8, 48), histone deacetylation (22), and histone 3 methylation at specific lysine or even arginine residues (6, 29).
Our present working hypothesis is that in E. histolytica, the initial silencing event, namely, the introduction of multiple copies of the ap-a promoter region, triggers the expression of a diffusible factor which acts in trans and recognizes the ap-a sequence, preventing its transcription. A major goal of our ongoing research is to dissect the mechanism of this promoter sequence homology-dependent TGS phenomenon. We are searching the ORF as well as the 5′ flanking segment of the ap-a gene for DNA modifications as well as for DNA binding proteins and their domains. We are hopeful that these studies will help us to better understand the molecular elements and events which cause the TGS.
The avirulent phenotype found for the AP-A-lacking trophozoites was to be expected, since a significant decrease in amoebic virulence was already previously observed when we succeeded in inhibiting (∼60%) the expression of the ap-a gene by antisense RNA (12). The complete absence of AP-A expression, as shown with transfectants psAP-1 and psAP-2 as well as with the plasmidless clone G3, has enabled us to study additional functions of AP-A, such as its role in disruption of phagocytosed bacteria or hRBCs. Phagocytosis was not impaired, but the disruption of the ingested cells was much slower and many bacteria remained undigested even after 24 h. These observations clearly support the envisioned functions of the AP for both the killing of adhered target cells and the disruption of phagocytosed cells (12, 30-32). Trophozoites of the plasmidless and AP-A-lacking clone G3, which are attenuated in their virulence, nevertheless grow very well in cultures, and they are currently being examined for their potential use as a live vaccine. The retention by the plasmidless clone of the silenced phenotype obviates the need to test its effects using a transfectant with an inducible plasmid, as is often recommended.
Acknowledgments
This investigation was supported by grants from the Center for the Study of Emerging Diseases, Jerusalem, Israel, as well as from Henry J. Meyer, Jr.
We thank M. Leippe, Wuerzburg U., Wuerzburg, Germany, for the anti-AP-A antibodies and E. Tannich from BNI, Hamburg, Germany, for the genomic clone of Ehap-a.
REFERENCES
- 1.Alon, R. N., R. Bracha, and D. Mirelman. 1997. Inhibition of expression of the lysine-rich 30 kDa surface antigen of Entamoeba dispar by the transcription of its antisense RNA. Mol. Biochem. Parasitol. 90:193-201. [DOI] [PubMed] [Google Scholar]
- 2.Ankri, S., F. Padilla-Vaca, T. Stolarsky, L. Koole, U. Katz, and D. Mirelman. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNAc lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol. 33:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Ankri, S., T. Stolarsky, R. Bracha, F. Padilla-Vaca, and D. Mirelman. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases in Entamoeba histolytica does not affect cytopathic or hemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. [DOI] [PubMed] [Google Scholar]
- 5.Arhets, P., J. C. Olivo, P. Gounon, P. Sansonetti, and N. Guillen. 1998. Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. Mol. Biol. Cell 9:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 7.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 9.Bracha, R., D. Kobiler, and D. Mirelman. 1982. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect. Immun. 36:396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracha, R., and D. Mirelman. 1983. Adherence and ingestion of Escherichia coli serotype 055 by trophozoites of Entamoeba histolytica. Infect. Immun. 40:882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracha, R., and D. Mirelman. 1984. Virulence of Entamoeba histolytica trophozoites: effects of bacteria, microaerobic conditions, and metronidazole. J. Exp. Med. 160:353-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bracha, R., Y. Nuchamowitz, M. Leippe, and D. Mirelman. 1999. Antisense inhibition of amoebapore expression in Entamoeba histolytica causes a decrease in amoebic virulence. Mol. Microbiol. 34:463-472. [DOI] [PubMed] [Google Scholar]
- 13.Bracha, R., Y. Nuchamowitz, and D. Mirelman. 2002. Amoebapore is an important virulence factor of Entamoeba histolytica. J. Biosci. (Bangalore) 27:579-587. [Google Scholar]
- 14.Bruchhaus, I., M. Leippe, C. Lioutas, and E. Tannich. 1993. Unusual gene organization in the protozoan parasite Entamoeba histolytica. DNA Cell Biol. 12:925-933. [DOI] [PubMed] [Google Scholar]
- 15.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2002. Involvement of small RNAs and the role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoni, C. 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55:381-406. [DOI] [PubMed] [Google Scholar]
- 17.Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker, and G. Macino. 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15:3153-3163. [PMC free article] [PubMed] [Google Scholar]
- 18.Csink, A. K., A. Bounoutas, M. L. Griffith, J. F. Sabi, and B. T. Sage. 2002. Differential gene silencing by trans-heterochromatin in Drosophila melanogaster. Genetics 160:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 20.Fling, S., and D. S. Gregerson. 1986. Peptide and protein molecular weight determination by electrophoresis using a high-molarity Tris buffer system without urea. Anal. Biochem. 155:83-88. [DOI] [PubMed] [Google Scholar]
- 21.Grishok, A., and C. C. Mello. 2002. RNAi (Nematodes: Caenorhabditis elegans). Adv. Genet. 46:339-360. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 23.Hamann, L., H. Buss, and E. Tannich. 1997. Tetracycline-controlled gene expression in Entamoeba histolytica. Mol. Biochem. Parasitol. 84:83-91. [DOI] [PubMed] [Google Scholar]
- 24.Hamann, L., R. Nickel, and E. Tannich. 1995. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 92:8975-8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton, A. J., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 27.Hellberg, A., R. Nickel, H. Lotter, E. Tannich, and I. Bruchhaus. 2001. Overexpression of cysteine proteinase 2 in Entamoeba histolytica or Entamoeba dispar increases amoeba-induced monolayer destruction in vitro but does not augment amoebic liver abscess formation in gerbils. Cell. Microbiol. 3:13-20. [DOI] [PubMed] [Google Scholar]
- 28.Imai, S.-I., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Gen. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 30.Leippe, M. 1997. Amoebapores. Parasitol. Today 13:178-183. [DOI] [PubMed] [Google Scholar]
- 31.Leippe, M., J. Andrä, and H. J. Müller-Eberhard. 1994. Cytolytic and antibacterial activity of synthetic peptides derived from amoebapore, the pore-forming peptide of Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 91:2602-2606. [DOI] [PMC free article] [PubMed]
- 32.Leippe, M., J. Andra, R. Nickel, E. Tannich, and H. Muller-Eberhard. 1994. Amoebapores, a family of membranolytic peptides from cytoplasmic granules of Entamoeba histolytica: isolation, primary structure and pore formation in bacterial cytoplasmic membranes. Mol. Microbiol. 14:895-904. [DOI] [PubMed] [Google Scholar]
- 33.Matzke, M. A., M. F. Mette, and A. J. M. Matzke. 2000. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43:401-415. [DOI] [PubMed] [Google Scholar]
- 34.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 35.Moshitch-Moshkovitch, S., R. Petter, T. Solarsky, and D. Mirelman. 1997. Regulation of expression of ribosomal protein L21 genes of Entamoeba histolytica and E. dispar is at the posttranscriptional level. Mol. Microbiol. 27:677-685. [DOI] [PubMed] [Google Scholar]
- 36.Moshitch-Moshkovitch, S., T. Stolarsky, D. Mirelman, and R. N. Alon. 1997. Stable episomal transfection and gene expression in Entamoeba dispar. Mol. Biochem. Parasitol. 83:257-261. [DOI] [PubMed] [Google Scholar]
- 37.Muskens, M. W. M., A. P. A. Vissers, J. N. M. Mol, and J. M. Kooter. 2000. Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43:243-260. [DOI] [PubMed] [Google Scholar]
- 38.Nickel, R., C. Ott, T. Dandekar, and M. Leippe. 1999. Pore-forming peptides of Entamoeba dispar: similarity and divergence to amoebapores in structure, expression and activity. Eur. J. Biochem. 265:1002-1007. [DOI] [PubMed] [Google Scholar]
- 39.Perez, D. G., C. Gomez, E. Lopez-Bayghen, E. Tannich, and E. Orozco. 1998. Transcriptional analysis of the EhPgp5 promoter of Entamoeba histolytica multidrug-resistant mutant. J. Biol. Chem. 273:7285-7292. [DOI] [PubMed] [Google Scholar]
- 40.Purdy, J. E., L. T. Pho, B. J. Mann, and W. A. J. Petri. 1996. Upstream regulatory elements controlling expression of the Entamoeba histolytica lectin. Mol. Biochem. Parasitol. 78:91-103. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz, F., L. Vayssié, C. Klotz, L. Sperling, and L. Madeddu. 1998. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell 9:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selker, E. U. 1998. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95:9430-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullu, E., and C. Tschudi. 1990. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 3319-3326. [DOI] [PMC free article] [PubMed]
- 44.van West, P., S. Kamoun, J. W. van't Klooster, and F. Govers. 1999. Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3:339-348. [DOI] [PubMed] [Google Scholar]
- 45.Vaucheret, H., and M. Fagard. 2001. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17:29-35. [DOI] [PubMed] [Google Scholar]
- 46.Venkatasubbarao, K., S. Ammanamanchi, M. G. Brattain, D. Mimari, and J. W. Freeman. 2001. Reversion of transcriptional repression of Sp1 by 5 aza-2-deoxycytidine restores TGF-β type II receptor expression in the pancreatic cancer cell line MIA PaCa-21. Cancer Res. 61:6239-6247. [PubMed] [Google Scholar]
- 47.Vines, R. R., G. Ramakrishnan, J. B. Rogers, L. A. Lockhart, B. J. Mann, and W. A. J. Petri. 1998. Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a β2 integrin motif. Mol. Biol. Cell 9:2069-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolffe, A. P., and M. A. Matzke. 1999. Epigenetics: regulation through repression. Science 286:481-486. [DOI] [PubMed] [Google Scholar]
- 49.Yee, J., M. R. Mowatt, P. P. Dennis, and T. E. Nash. 2000. Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J. Biol. Chem. 275:11432-11439. [DOI] [PubMed] [Google Scholar]
- 50.Zamore, P. D. 2001. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 8:746-750. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Z., K.-I. Shibahara, and B. Stillman. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408:221-225. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Z., L. Wang, K. B. Seydel, E. Li, S. Ankri, D. Mirelman, and S. L. Stanley, Jr. 2000. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol. Microbiol. 37:542-548. [DOI] [PubMed] [Google Scholar]
- 53.Zou, Y. R., M.-J. Sunshine, I. Taniuchi, F. Hatam, N. Killeen, and D. R. Littman. 2001. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 29:332-336. [DOI] [PubMed] [Google Scholar]