Abstract
Activation of the Snf1 kinase requires at least two events, phosphorylation of the activation loop on threonine 210 and an Snf4-dependent process that is not completely defined. Snf4 directly interacts with a region of the regulatory domain of Snf1 that may otherwise act as an autoinhibitory domain. In order to gain insight into the regulation of Snf1 kinase by Snf4, deletions in the regulatory domain of the catalytic subunit were engineered and tested for their effect on Snf1 function in the absence of Snf4. Deletion of residues 381 to 488 from the Snf1 protein resulted in a kinase that was activated by glucose limitation even in the absence of the Snf4 protein. A larger deletion (amino acids 381 to 608) encompassing virtually the entire regulatory domain resulted in complete inactivation of the Snf1 kinase even in the presence of Snf4. A genetic screen for amino acid substitutions that conferred an Snf4-independent phenotype identified four point mutations in the Snf1 catalytic domain. One very conservative mutation, leucine 183 to isoleucine, conferred nearly wild-type levels of Snf1 kinase function in the absence of the Snf4 protein. Purified Snf1 kinase was inactive when isolated from snf4Δ cells, whereas the Snf1-L183I kinase exhibited significant activity in the absence of Snf4. Our data support the idea that Snf1 kinase activity is constrained in cis by an autoinhibitory domain and that the Snf4-mediated activation of Snf1 can be bypassed by subtle conformational changes in the catalytic domain of the Snf1 kinase.
The Snf1 kinase of Saccharomyces cerevisiae is a member of a highly conserved subfamily of serine-threonine protein kinases that includes the AMP-activated protein kinase (AMPK) found in mammalian cells (9). The Snf1 and AMPK enzymes function as heterotrimers, containing a catalytic alpha subunit and noncatalytic beta and gamma subunits (4, 23). Members of this kinase family are distinguished from other serine-threonine kinases by a high degree of sequence identity in the kinase domain and by the presence of a C-terminal regulatory domain that mediates interaction with the beta and gamma subunits (13). In mammalian cells, two genes have been identified for both the alpha and beta subunits (2, 26), while three gamma subunit genes have been found (2). The presence of multiple genes for each subunit combined with differences in tissue-specific expression results in the presence of numerous, distinct AMPK enzyme complexes that differ in subunit composition. Defining the exact role of these complexes remains a challenge.
In yeast, the subunit composition is less complex, with a single alpha subunit gene, SNF1, and a single gamma subunit gene, SNF4. However, yeast does carry three beta subunit genes, SIP1, SIP2, and GAL83, and therefore has the potential to express three distinct Snf1 enzyme complexes. The specific role of each complex is beginning to be understood. We have shown that the presence of a beta subunit is required for kinase function (19) and that yeast strains expressing a single beta subunit have distinct growth phenotypes as well as differing abilities to phosphorylate the Sip4 protein (19). Work by Vincent et al. has shown that the beta subunits confer different subcellular localizations to the enzyme complex (25). Thus, the three forms of the Snf1 kinase present in yeast cells are likely to have specialized roles determined by different localization patterns and substrate specificities.
The focus of this study is on the regulatory role played by the gamma subunit of the Snf1 kinase complex. The gamma subunit, encoded by SNF4, is essential for the full activation of Snf1 kinase (1). Previous studies have found that the Snf1 and Snf4 proteins are held in the kinase complex through constitutive binding to the beta subunit (13). In addition, Snf4 makes direct contact with the regulatory domain of the catalytic alpha subunit, and this interaction is regulated by the availability of glucose (12). How binding of Snf4 to the regulatory domain controls the activity of the alpha subunit is not fully understood. One model for this regulation proposes the existence of an autoinhibitory domain present in the alpha subunit (3, 12). The gamma subunit and the kinase domain compete for binding to the autoinhibitory domain. Under conditions of glucose excess, the kinase domain binds the autoinhibitory domain, thereby forming an inactive complex. When glucose is limiting, the gamma subunit binds the autoinhibitory domain, displacing and thereby relieving the inhibition of the kinase domain. A second event, phosphorylation of a conserved threonine residue in the activation loop, is also required for the full activation of the Snf1 and AMPK enzymes (10, 14). However, these two events are not dependent on one another, since phosphorylation of Snf1 threonine 210 occurs normally in cells lacking the Snf4 protein (14).
In order to better understand the Snf4-mediated regulation of the Snf1 kinase complex, we used a combination of protein engineering and random mutagenesis to isolate variants of the catalytic subunit that were no longer dependent on Snf4 for activation. Our results support the idea that the Snf4 subunit counteracts the effect of the autoinhibitory domain. Surprisingly, we also found that subtle changes in the catalytic domain are able to bypass the need for the gamma subunit.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
The S. cerevisiae strains utilized in this study were FY1193 (MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 snf1Δ10) and MSY563 (MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 snf1Δ10 snf4Δ1). For carbon sources, glucose was present at 2% (grams/100 ml), while the glycerol-ethanol mixture was present at 3% (vol/vol) glycerol and 2% (vol/vol) ethanol. Raffinose medium contained 2% raffinose and 0.05% (grams/100 ml) glucose as the carbon source and antimycin A at 1 μg/ml. Plasmid transformations of yeast strains was by the lithium acetate procedure (7).
Site-directed mutagenesis.
Deletions and point mutations in the SNF1 gene were constructed by site-directed mutagenesis (5) with the oligonucleotides listed in Table 1. The integrity of all deletion junctions and point mutations was confirmed by DNA sequencing.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequencea | Restriction change |
|---|---|---|
| SNF1-T390 | GGTTATAGAGTACGCCGGGAACG | NAb |
| SNF1-B1922 | CATTCTTTTACGTTCCACCATC | NA |
| SNF1Δ381-415 | gaagacactcctgcattcaacgaaattaggACTTTTCAACAACAAAGCAAATCCC | Size |
| SNF1Δ381-488 | gacactcctgcattcaacgaaattaggGAAGCTTCTAAAATATCTCCTCTTGTAAC | Size |
| SNF1Δ381-608 | gaagacactcctgcattcaacgaaattaggAGTACTTTTTCAGCCTACCCATTTTTAC | Size |
| SNF1-L1831 | CATAGAGATCTGAAGCCTGAAAACaTtCTACTAGATGAGCATCTGAATG | XmnI |
| SNF1-K192R | CATCTGAATGTAcgtATTGCCGATTTTGGTTTGTCAAAC | BsaAI |
| SNF1-1241N | GTGTGGTCATGTGGGGTTAaCCTTTATGTTATGCTTTGTCGT | HpaI |
Changes between upper- and lowercase signify boundaries between wild-type and altered sequences.
NA, not applicable.
Random mutagenesis and genetic selection.
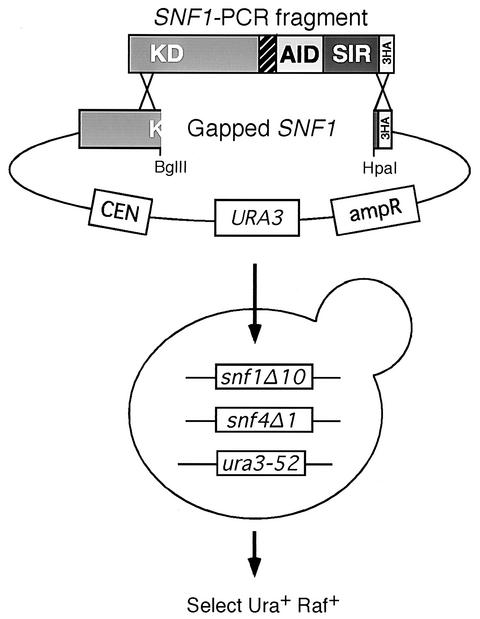
Random mutations in the SNF1 gene were introduced by PCR amplification of a DNA fragment encompassing the SNF1 open reading frame from amino acid 130 through end of the frame, using primers Snf1-T390 and Snf1-B1922 (Table1). Taq polymerase was used in reaction mixtures containing 2 mM MgCl2 and a 0.2 mM concentration of each deoxynucleoside triphosphate. The amplified fragment was then cotransformed with the centromeric plasmid pRS314 (21) containing an epitope tagged SNF1 gene that had been gapped at codons 175 and 628 by treatment with BglII and HpaI (see Fig. 3). Transformants of the yeast strain MSY563 (snf1Δ10 snf4Δ1) were selected on SC medium (17) lacking uracil and replica plated to medium containing raffinose and antimycin A. Cells that were able to grow on raffinose medium (Snf+) were retained for further study. Plasmids isolated from Snf+ colonies were amplified in Escherichia coli and sequenced.
FIG. 3.
PCR Mutagenesis strategy. Schematic representation of the pSNF1-316 plasmid gapped by digestion with BglII-HpaI and the PCR fragment used for gap repair are shown. The recipient strain (MSY563 [snf1Δ10 snf4Δ1]) is represented along with the selection strategy. Abbreviations are as described for Fig. 1.
Western blots.
Hemagglutinin (HA)-tagged Snf1 proteins were detected by Western blotting (20) with a mouse monoclonal antibody directed against the HA epitope (Santa Cruz Biotechnology).
Enzyme assays.
Quantitative invertase assays were performed as previously described (20). Specific activity was defined in terms of milliunits of invertase activity (with 1 U being equal to the activity required to release 1 mmol of glucose per min) per unit of optical density at 600 nm of cells assayed. Snf1 kinase activity was assayed in reaction mixtures containing kinase buffer (20 mM HEPES [pH 7.0], 0.5 mM EDTA, 0.5 mM dithiothreitol, and 5 mM Mg-acetate), 0.2 mM [γ-32P]ATP (1,000 cpm/pmol), 10 μg of glutathione S-transferase (GST)-Mig1 protein per ml, and approximately 2.5 ng of Snf1 kinase per ml. Reaction mixtures were incubated at 30°C for 20 min, and reactions were stopped by addition of 10 volumes of ice-cold 10% trichloroacetic acid. Samples were precipitated, washed in acetone, and resolved on a sodium dodecyl sulfate (SDS)-polyacrylamide gel. Dried gels were subjected to autoradiography.
Snf1 purification.
The Snf1 protein was tagged and purified by the tandem affinity purification method (16) followed by an additional chromatography step on a 1-ml MonoQ column as described elsewhere (14a).
Structural model of the Snf1 kinase domain.
A structural model of the Snf1 kinase domain was prepared by ProMod II as part of the Swiss-Model Automated Protein Modelling Server (8, 15). The Snf1 model used the Protein Databank coordinates of the related kinases CHK1 and PKA (PDB files 1IA8, 1FOT, 1YDS, 1YDR, and 1YDT) and included Snf1 residues 48 to 317. Coordinates for the Snf1 structural model are available on request.
RESULTS
Mutants with deletions in the Snf1 regulatory domain.
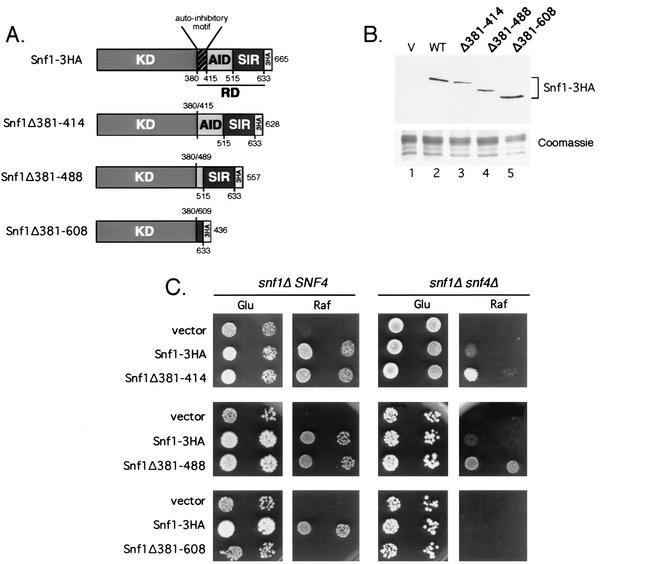
Many proteins contain autoinhibitory domains that regulate enzyme activity in cis. Deletions in the regulatory domain of AMPK indicate that such an autoinhibitory mechanism plays a role in its regulation (3). Current models of the regulation of Snf1 kinase invoke a similar regulatory role for a region immediately C terminal of the Snf1 kinase domain (12, 13). Sequence alignments between the AMPK alpha subunits from yeast, Drosophila, and various mammalian species identified a conserved amino acid motif (amino acids 379 to 415 in Snf1) located directly after the kinase domain that was suggested to function as an autoinhibitory domain (3). To test this model directly, we engineered specific internal deletions in the regulatory domain of the Snf1 kinase alpha subunit by site-directed mutagenesis. The catalytic kinase domain, amino acids 1 to 380, was left intact. Three progressively larger deletion mutations were made starting at amino acid 381 and including amino acid 414, 488, or 608 (Fig. 1). The Δ381-414 deletion specifically removes the putative autoinhibitory motif identified by Crute et al. (3). The Δ381-488 deletion removes the majority of the regulatory domain that interacts with gamma subunit, Snf4 (12, 13). The largest deletion, Δ381-608, removes all but the final 30 amino acids of the C-terminal regulatory domain, including the entire Snf4-interacting region and most of the beta subunit-interacting region (13). The wild-type protein and all deletion constructs contained three copies of the HA epitope at the C terminus, allowing detection by Western blotting. The internal deletions did not reduce the accumulation of the Snf1 protein, since the wild type and all deletion mutants were detected at comparable levels (Fig. 1B). Indeed, the mutant with largest internal deletion, which removed amino acids 381 to 608, appeared to be more abundant than the wild-type protein when normalized to the loading control. An even greater increase in abundance was observed in mammalian cells when the regulatory domain was removed from the AMPK alpha subunit (3).
FIG. 1.
Deletion of an internal inhibitory domain confers Snf4 independence. (A) Schematic representation of the Snf1 protein. Wild-type Snf1 and three internal deletion mutants are shown. Shown are the kinase domain (KD) (residues 1 to 380), the autoinhibitory domain (residues 380 to 415), the extended inhibitory domain (AID) (residues 380 to 515), the SIP-interacting region (SIR) (residues 515 to 633), the C-terminal epitope tag (3HA), and the regulatory domain (RD). (B) Western blot of Snf1 and internal deletion mutants. Equivalent aliquots of yeast whole-cell extracts (20 μg of protein) were resolved by SDS-polyacrylamide gel electrophoresis and either transferred to a nylon membrane and probed with antibodies against the HA epitope or stained in Coomassie blue. Cells were transformed with vector (V) (lane 1), wild-type Snf1-3HA (WT) (lane 2), or internal deletion mutants (lanes 3 to 5). (C) Growth phenotypes of cells expressing no Snf1, wild-type Snf1, or internal deletion mutants. Cells (snf1Δ10 SNF4 or snf1Δ10 snf4Δ1) transformed with the indicated plasmids were normalized to an optical density at 600 nm of 0.2, and 10-fold serial dilutions were spotted onto SC medium lacking uracil and containing either glucose (Glu) or raffinose (Raf) as the carbon source.
The ability of the internal deletion mutants to provide Snf1 kinase function in vivo was assayed by testing the ability of these constructs to complement an snf1Δ10 mutant for growth on raffinose medium. Since the Snf4 subunit is thought to counteract the effect of the autoinhibitory domain, these constructs were tested in the presence and absence of the SNF4 gene (Fig. 1C). In the presence of Snf4 protein, the SNF1Δ381-414 and SNF1Δ381-488 alleles were functional and indistinguishable from wild-type SNF1, indicating that these residues are not required for Snf1 function in this assay. In contrast, the SNF1Δ381-608 allele was completely nonfunctional even though the protein is expressed at least as well as the wild type. In the absence of Snf4 protein, Snf1 kinase function is compromised and cells grow poorly on raffinose. Deletion of amino acids 381 to 414 and 381 to 488 from the Snf1 protein produces a significant increase in the ability of the snf4Δ1 cells to grow on raffinose relative to the cells expressing wild-type Snf1. The Δ381-488 deletion consistently provides a greater degree of Snf4-independent function to the Snf1 kinase than the Δ381-415 deletion.
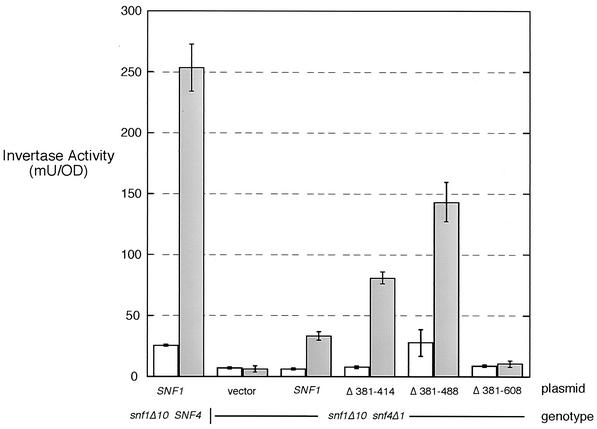
Invertase activity assays provide an additional measure of Snf1 kinase function. To assess the activity of the internal deletion mutants in the absence of SNF4, plasmids carrying wild-type SNF1, the internal deletion mutations, or no insert (vector) were transformed into cells lacking chromosomal copies of SNF1 and SNF4. Cells grown under glucose-repressing and -derepressing conditions were collected and assayed for their invertase activity levels (Fig. 2). The SNF1Δ381-608 allele was not functional in this assay, since invertase expression was not induced more than with the vector control. Furthermore, the SNF1Δ381-608 allele did not induce invertase in the presence of SNF4 (not shown). In contrast, the SNF1Δ381-415 and SNF1Δ381-488 alleles both caused significant induction of invertase compared to the wild-type SNF1 plasmid and vector control. The larger deletion (Δ381-488) was consistently found to provide greater levels of Snf4-independent activity. Also, the Δ381-488 deletion appeared to be weakly active under repressing conditions, although the effect is not large and variability in the measurement tempers this conclusion. Taken together, these data indicate that the autoinhibitory domain extends beyond the conserved motif present within amino acids 381 to 415. Second, amino acids 381 to 488 are not required for Snf1 function, and their absence confers Snf4-independent function to the Snf1 kinase. Third, amino acids 488 to 608 are required for Snf1 function even in the absence of the autoinhibitory domain.
FIG. 2.
Invertase activity of Snf1 deletion mutants. Centromeric plasmids expressing the indicated Snf1 protein were transformed into the indicated strains (snf1Δ10 SNF4 or snf1Δ10 snf4Δ1). The mean invertase activity and the standard error from three independent transformants that were grown in 2% glucose (repressed) (open bars) or 0.05% glucose (derepressed) (shaded bars) are plotted. OD, optical density unit.
Genetic selection for Snf4-independent alleles of SNF1.
Additional Snf4-independent alleles of SNF1 were identified in a genetic screen using PCR-mediated, random mutagenesis within the SNF1 gene (Fig. 3). A centromeric plasmid carrying wild-type SNF1 driven by its endogenous promoter was digested by BglII and HpaI, thus removing codons 175 through 628. The gapped plasmid and an overlapping PCR fragment amplified with Taq polymerase were used to transform an snf1Δ10 snf4Δ1 strain to Ura+. Uracil prototrophs were recovered and screened for the ability to grow on raffinose medium. Plasmids conferring Snf4-independent growth following retransformation into naive snf1Δ10 snf4Δ1 cells were screened by Western blotting for the ability to produce full-length Snf1 protein (data not shown). Nine clones that satisfied these criteria were sequenced. Each clone contained a unique collection of mutations that produced four to seven amino acid changes (Table 2) as well as several silent third-position codon changes (not shown). Two amino acid changes, L183I and K192R, were found in more than one independent clone, suggesting that these changes might confer the observed Snf4-independent phenotype. These mutations were introduced into SNF1 by site-directed mutagenesis and were found to confer Snf4 independence comparable to that of the original isolates (data not shown). A combination of subcloning and site-directed mutagenesis was used to identify two additional point mutations, Y167H and I241N, that were able to confer Snf4 independence. Two alleles of SNF1 contained multiple amino acid changes, none of which was able to confer an Snf4-independent phenotype on its own. These alleles, SNF1-203 and SNF1-214, were not studied further.
TABLE 2.
Mutations conferring Snf4 independence
| Clone | Amino acid changesa |
|---|---|
| SNF1-201 | L1831, R248H, P374S, Q476L, V639A |
| SNF1-202 | E187K, 1241N, M244L, E311K, K3191 |
| SNF1-203 | D304N, 1553F, F573Y, E607K |
| SNF1-205 | L1831, D186E, E326K, N3471, A434T, L474V |
| SNF1-207 | K192R, D304N, S353A, K4001, V4021, Q412L, M519L |
| SNF1-208 | K192R, L227P, A382P, D407Y, Q453R, S513T, M624L |
| SNF1-211 | Y167H, 1193V, V468D, M6241 |
| SNF1-213 | L1831, E365K, N3991, S513L |
| SNF1-214 | M2861, L314P, L319I, P339S, S370L, T4461 |
Mutations that by themselves confer Snf4 independence are boldfaced.
Characterization of Snf4-independent point mutants.
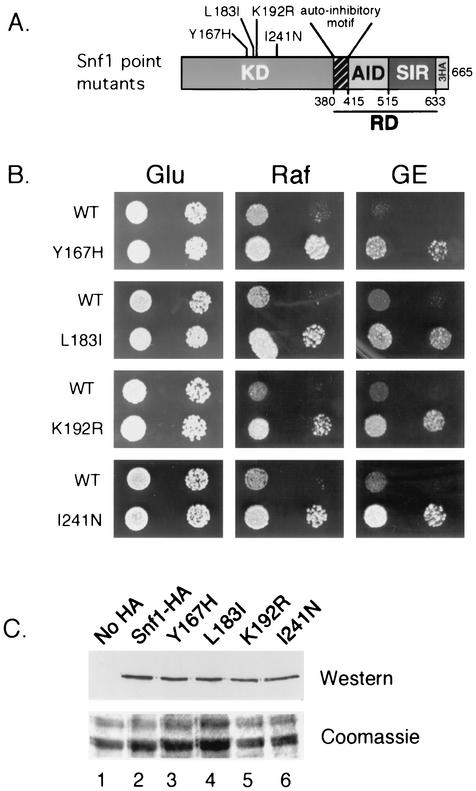
Random mutagenesis identified four point mutations in the kinase domain of the SNF1 gene that each conferred some level of Snf4 independence (Fig. 4A). These alleles of SNF1 were characterized further for their growth phenotypes on two carbon sources that require Snf1 kinase activity. An snf1Δ10 snf4Δ1 strain was transformed with a centromeric plasmid expressing either wild-type Snf1 or one of the four point mutants, and serial dilutions of liquid cultures were spotted onto agar plates (Fig. 4B). All four point mutants confer enhanced growth compared to wild-type SNF1 on both raffinose and glycerol-ethanol media. All four point mutants are expressed at levels equivalent to that observed for wild-type protein when assayed by Western blot (Fig. 4C).
FIG. 4.
Point mutations in SNF1 that confer Snf4 independence. (A) The locations of four point mutations in the SNF1 gene are drawn to scale. Abbreviations are as described for Fig. 1. (B) Growth phenotypes of wild-type Snf1 (WT) and point mutants on glucose (Glu), raffinose (Raf), and glycerol-ethanol (GE) media. All transformants were in the snf1Δ10 snf4Δ1 strain. (C) Steady-state protein levels were analyzed by Western blotting (upper panel). Aliquots of protein extracts (20 μg) from MSY563 cells transformed with control vector (no HA) or Snf1-expressing plasmids as indicated were probed with monoclonal antibodies directed against the HA epitope. As a loading control, equivalent aliquots were analyzed in parallel by Coomassie blue staining (lower panel).
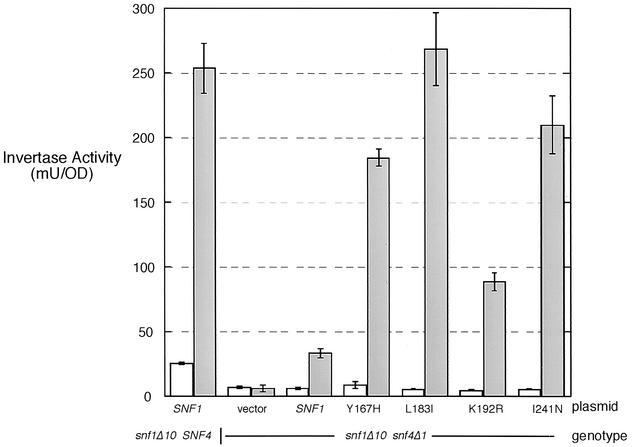
Invertase expression was also measured in cells expressing the four Snf1 point mutants in both SNF4+ and snf4Δ1 backgrounds. In the presence of SNF4, all four point mutations were indistinguishable from wild-type SNF1 (not shown). In the absence of SNF4, the four point mutations all confer levels of invertase expression significantly higher than that observed for wild-type SNF1 (Fig. 5). The K192R allele is the weakest of the four Snf4-independent alleles. The L183I allele confers levels of invertase expression that are comparable to that observed in a wild-type SNF4 background. None of the four point mutations are constitutively activating alleles of SNF1, since they do not cause invertase induction under high-glucose conditions. The regulation of invertase in response to glucose was normal; however, the need for SNF4 was bypassed by these single amino acid changes.
FIG. 5.
Invertase activities of Snf1 point mutants. Centromeric plasmids expressing the indicated Snf1 protein were transformed into the indicated strains (snf1Δ10 SNF4 or snf1Δ10 snf4Δ1). The mean invertase activity and the standard error from three independent transformants that were grown in 2% glucose (repressed) (open bars) or 0.05% glucose (derepressed) (shaded bars) are plotted. OD, optical density unit.
In vitro kinase activity in the absence of Snf4 protein.
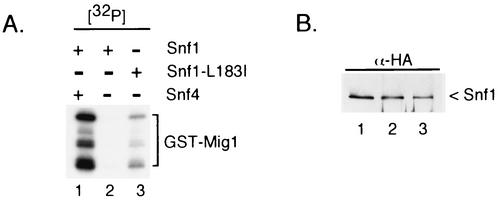
Snf1 kinase complexes were purified by using a modification (14a) of the tandem affinity purification (TAP) protocol devised by Rigaut et al. (16). Wild-type SNF1 and SNF1-L183I were TAP tagged at their C termini, and kinase complexes were purified from SNF4 and snf4Δ1 cells. Kinase activity was assayed in vitro for the ability to phosphorylate a recombinant GST fusion protein containing amino acids 202 to 414 from the yeast Mig1 protein, a known substrate of the Snf1 kinase (22, 24). Kinase complexes purified from cells expressing wild-type Snf1 and Snf4 proteins efficiently phosphorylated GST-Mig1, as well as a set of proteolytic breakdown products (Fig. 6A, lane 1). In the absence of the Snf4 protein, the purified Snf1 kinase exhibited greatly reduced activity (lane 2). When the L183I amino acid substitution was present in the Snf1 subunit, substantial kinase activity was restored to the complex lacking the Snf4 subunit (lane 3). In all reactions, equivalent levels of the catalytic subunit were used as judged by Western blotting (Fig. 6B). We conclude that the single amino acid substitution L183I is able to restore in vitro kinase activity to complexes lacking the Snf4 subunit.
FIG. 6.
In vitro kinase activity of Snf1-L183I. (A) Purified Snf1 kinase was assayed in reaction mixtures containing GST-Mig1 (amino acids 202 to 414) and [γ-32P]ATP. TAP-tagged wild-type Snf1 (lanes 1 and 2) and Snf1-L183I (lane 3) were purified from cells that lacked genomic copies of either SNF1 (lane 1) or SNF1 and SNF4 (lane 2 and lane 3). Phosphorylated GST-Mig1 protein was resolved on an SDS-polyacrylamide gel and detected by autoradiography. (B) Levels of the catalytic Snf1 subunit were normalized by Western blotting with a monoclonal antibody against the HA epitope. Enzyme preparations were the same as in panel A.
DISCUSSION
The activities of the Snf1 kinase and its mammalian homologue, AMPK, are tightly regulated in vivo. Current models of the regulation of Snf1 kinase activity propose at least two steps (14, 18). One step is the phosphorylation of the activation loop threonine 210 by one or more upstream kinases. The second step is mediated by the gamma subunit encoded by the SNF4 gene in S. cerevisiae. Several lines of evidence suggest that the main function of the Snf4 protein is to block the effects of an autoinhibitory domain present in the catalytic subunit (3, 12). The gamma subunit directly interacts with the proposed autoinhibitory domain of the catalytic subunit, and this interaction is regulated by nutrient stress (12). Deletion of the proposed autoinhibitory domain from the catalytic subunit of rat AMPK resulted in higher levels of activity when purified enzyme was assayed with a peptide substrate (3). In this study, we have explored the role of the gamma subunit in the activation of the Snf1 enzyme in two ways. First, specific deletions were engineered in the catalytic subunit. Second, a genetic screen for point mutations that conferred gamma subunit independence was conducted. Snf1 enzyme activity was assessed by using yeast growth assays, invertase induction assays, and, in one case, activity assays of purified Snf1 kinase.
Studies by Crute et al. proposed that a short conserved motif in the catalytic subunit of rat AMPK (amino acids 315 to 349) acted as an autoinhibitory domain (3). Evidence in support of this idea included the observation that AMPK catalytic subunits purified as GST fusions were more active when this region was removed. However, the truncated form of the alpha subunit (α1-312) did not associate with or require the beta and gamma subunits for in vitro activity. Earlier studies had shown that the C-terminal region of the catalytic subunit was required for association with the gamma and beta subunits (3, 26) and that the presence of the beta and gamma subunits was essential for the reconstitution of enzyme activity (6). Therefore, the in vitro activity of the truncated catalytic subunit purified as a GST fusion protein might not accurately reflect the regulation of enzyme activity in vivo. In this study with the Snf1 enzyme, deletion of the region that includes the putative autoinhibitory motif (Snf1Δ381-414) confers partial independence of the gamma subunit as measured by growth on alternative carbon sources (Fig. 1) and derepression of invertase (Fig. 2). However, this small motif does not comprise the entire autoinhibitory domain, since a higher level of Snf4-independent activity was observed in the larger deletion construct, Snf1Δ381-488 (Fig. 1 and 2). Earlier studies using the two-hybrid assay to map protein-protein interaction domains found that the region of Snf1 bound by Snf4 protein was present in residues 392 to 495 (12). Thus, the idea that the main role of the Snf4 subunit is to block the effect of an autoinhibitory domain is supported by the observation that deletion of this region leads to an enzyme that no longer requires the Snf4 subunit for activity. Note, however, that the Snf1Δ381-488 enzyme is not constitutively active. Invertase expression is still repressed by high glucose concentrations (Fig. 2). We conclude that relief from autoinhibition either by the action of Snf4 protein or by the deletion of the autoinhibitory domain is required but not sufficient for Snf1 activation.
The largest deletion construct examined in this study, Snf1Δ381-608, removes all but the final 30 amino acids of the C-terminal domain of Snf1 (full-length Snf1 contains 638 residues), including the region thought to interact with the beta subunit (3, 13). The Snf1Δ381-608 enzyme is completely inactive, even though it accumulates to levels as high or higher than do the other deletion constructs (Fig. 1 and 2). This enzyme lacks the autoinhibitory domain yet is still inactive. This result suggests very strongly that the association with the beta subunits is required for functions in addition to the recruitment of the gamma subunit to the heterotrimeric enzyme. Indeed, we have directly tested for beta subunit requirement and have found that deletion of all three beta subunit genes inactivates the Snf1Δ381-488 enzyme (data not shown). Earlier studies have shown that the beta subunits are important for determining enzyme localization (25) and substrate specificity (19). The data presented in this study support the idea that the beta subunits are important for more than the association of the alpha and gamma subunits.
Our finding that Snf1Δ381-608 is completely inactive is not consistent with the conclusions of an earlier study reported by Jiang and Carlson (12). Jiang and Carlson reported that the Snf1 regulatory domain is not required for activity, since the kinase domain (residues 1 to 392), expressed as a fusion with the Gal4 activation domain, was functional in vivo in the absence of Snf4 protein (12). Their study used invertase derepression as a measure of Snf1 kinase activity. We find that the Snf1Δ381-608 enzyme accumulates but is not active when measured in growth assays or invertase assays (Fig. 1 and 2). We considered the possibility that the Gal4 activation domain and simian virus 40 nuclear localization signal present in the two-hybrid construct might contribute to the ability of the Snf1 kinase domain to derepress invertase. However, we have also tested the Snf1 kinase domain (residues 1 to 361 as well as residues 1 to 392) expressed alone or as a fusion to the Gal4 activation domain and simian virus 40 nuclear localization signal and have been unable to find any evidence for activity using a truncated Snf1 kinase domain (data not shown). It is not clear to us why our results differ from those previously reported by Jiang and Carlson. Our data indicate that the catalytic domain of Snf1 by itself is not functional.
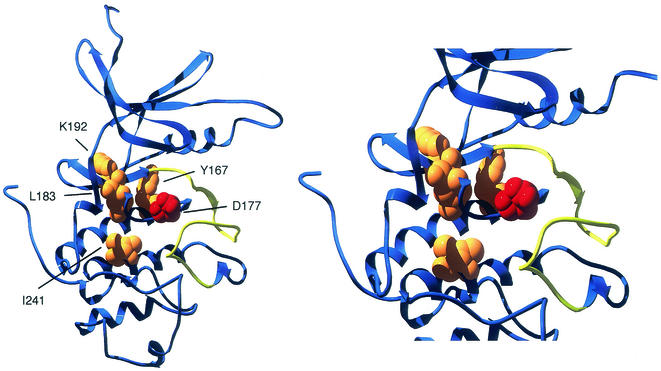
A second approach to examine the role of the gamma subunit was to screen for mutations in the SNF1 gene that conferred Snf4 independence. The source of mutations was the PCR amplification of a large region of the SNF1 gene by using Taq polymerase. Our expectation was that we would recover mutations in the autoinhibitory domain of the Snf1 subunit. Much to our surprise, the four point mutations that by themselves confer some degree of Snf4 independence were found in the catalytic domain and not in the autoinhibitory domain. We should note that our screen was not completely unbiased. The gapped plasmid used to generate our library of SNF1 mutations encompassed codons 175 to 628. Our intended target was the Snf4 interaction domain (residues 392 to 495) mapped by Jiang and Carlson (12), which is entirely within the gap, while our unintended target, the catalytic domain (residues 1 to 360), was only partly covered by the gap. All four point mutations identified in this screen lie in a highly conserved region of the catalytic domain (Fig. 7). Three of the residues identified (Y167, K192, and I241) are invariant in orthologous enzymes from organisms as divergent as yeast, human, fly, nematode, and plant. The fourth residue identified in this screen, L183, is also highly conserved in its hydrophobic nature. Yeast, fly, nematode, and plant all have a leucine at this position, while the human enzyme contains a valine residue. The L183I mutation is particularly intriguing, since it involves the shift of a single methyl group by no more than a few angstroms yet is sufficient to confer Snf4-independent activity to the Snf1 kinase. All four point mutations are located very close in primary sequence and in three-dimensional space to the catalytic aspartate residue (D177) in Snf1 kinase (Fig. 8).
FIG. 7.
Sequence alignment of Snf1 protein and selected orthologues. The amino acid sequence of Snf1 (residues 158 to 247) is shown in a multiple-sequence alignment of the corresponding residues from orthologous proteins. The species used were as follows: Sc, S. cerevisiae; Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana. The positions of the four residues giving rise to Snf4-independent alleles are indicated.
FIG. 8.
Structural model of the Snf1 kinase domain. A ribbon diagram of the Snf1 kinase domain (residues 48 to 317) is shown in blue, and the activation loop (residues 202 to 219) is shown in yellow. The catalytic aspartate residue (D177) is represented in red. The four residues whose change can confer Snf4 independence are shown in orange.
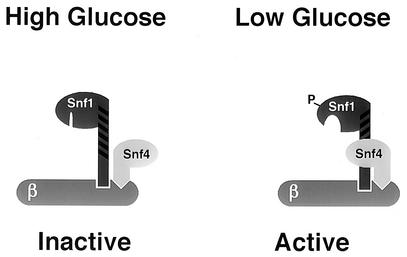
How does the Snf1 autoinhibitory domain control the activity of the catalytic domain? Jiang and Carlson have proposed a competitive binding model in which the autoinhibitory domain is bound to the catalytic domain under high-glucose conditions and thereby blocks kinase activity directly (12). Under low-glucose conditions, the Snf4 subunit binds to the autoinhibitory domain, displacing and freeing the catalytic domain. It is possible that the four point mutations isolated in this study weaken the interaction between the kinase domain and the autoinhibitory domain, thereby bypassing the need for the Snf4 protein. We have attempted to measure the interaction between wild-type and mutant Snf1 kinase domains and the Snf1 autoinhibitory domain by using the two-hybrid system. However, we have been unable to detect this interaction even with the wild-type Snf1 kinase domain (data not shown). Consistent with earlier studies, we do detect strong interaction between the autoinhibitory domain and the Snf4 protein, and this interaction is strongly affected by carbon source. In the absence of a detectable interaction between the kinase domain and the autoinhibitory domain, we propose a model for the regulation of Snf1 kinase activity in which the catalytic activity of the Snf1 kinase is controlled by subtle conformational changes transmitted to the kinase domain by the autoinhibitory domain (Fig. 9). All four residues which confer Snf4-independent activity are located close to the active site of the kinase domain in both primary sequence (Fig. 7) and three-dimensional space, as predicted by a structural model of the Snf1 kinase catalytic domain (Fig. 8). Subtle changes in the packing of the central hydrophobic core of the catalytic domain may be sufficient to change the orientation of the catalytic residues, thereby controlling the activity of the kinase domain. The regulation of protein kinase activity by controlling the orientation of active-site residues is a common regulatory mechanism. For instance, the interactions between the Src kinase SH2 and SH3 domains with their respective ligands control the orientation of residues in the active site (11). We propose that the binding of the Snf4 protein to the autoinhibitory domain of the Snf1 protein promotes a favorable orientation of the active-site residues.
FIG. 9.
Conformational regulation of Snf1 kinase. A model for the regulation of Snf1 kinase is proposed. Under high-glucose conditions, the Snf1 kinase autoinhibitory domain is unbound, but it holds the active site in a closed and inactive conformation. Under low-glucose conditions, the autoinhibitory domain is bound by Snf4 protein, which promotes an open and active conformation of the active site.
Acknowledgments
This work was supported by grant GM46443 from the National Institutes of Health.
We thank Tom Smithgall for assistance with computer graphics.
REFERENCES
- 1.Celenza, J. L., F. J. Eng, and M. Carlson. 1989. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol. Cell. Biol. 9:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung, P. C., I. P. Salt, S. P. Davies, D. G. Hardie, and D. Carling. 2000. Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem. J. 346 Pt 3:659-669. [PMC free article] [PubMed] [Google Scholar]
- 3.Crute, B. E., K. Seefeld, J. Gamble, B. E. Kemp, and L. A. Witters. 1998. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 273:35347-35354. [DOI] [PubMed] [Google Scholar]
- 4.Davies, S. P., S. A. Hawley, A. Woods, D. Carling, T. A. Haystead, and D. G. Hardie. 1994. Purification of the AMP-activated protein kinase on ATP-gamma-Sepharose and analysis of its subunit structure. Eur. J. Biochem. 223:351-357. [DOI] [PubMed] [Google Scholar]
- 5.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 6.Gao, G., C. S. Fernandez, D. Stapleton, A. S. Auster, J. Widmer, J. R. Dyck, B. E. Kemp, and L. A. Witters. 1996. Non-catalytic beta- and gamma-subunit isoforms of the 5′-AMP-activated protein kinase. J. Biol. Chem. 271:8675-8681. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 8.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 9.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 10.Hawley, S. A., M. Davison, A. Woods, S. P. Davies, R. K. Beri, D. Carling, and D. G. Hardie. 1996. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271:27879-27887. [DOI] [PubMed] [Google Scholar]
- 11.Huse, M., and J. Kuriyan. 2002. The conformational plasticity of protein kinases. Cell 109:275-282. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105-3115. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 14a.Nath, N., R. R. McCartney, and M. C. Schmidt. 2002. Purification and characterization of Snf1 kinase complexes containing a defined beta subunit composition. J. Biol. Chem. 277:50403-50408. [DOI] [PubMed] [Google Scholar]
- 15.Peitsch, M. C., T. Schwede, and N. Guex. 2000. Automated protein modelling—the proteome in 3D. Pharmacogenomics 1:257-266. [DOI] [PubMed] [Google Scholar]
- 16.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 17.Rose, M. D., F. Winston, and P. Hoeter (ed.). 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Sanz, P., G. R. Alms, T. A. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt, M. C., and R. R. McCartney. 2000. Beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wolfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, F. C., S. P. Davies, W. A. Wilson, D. Carling, and D. G. Hardie. 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453:219-223. [DOI] [PubMed] [Google Scholar]
- 23.Stapleton, D., G. Gao, B. J. Michell, J. Widmer, K. Mitchelhill, T. Teh, C. M. House, L. A. Witters, and B. E. Kemp. 1994. Mammalian 5′-AMP-activated protein kinase non-catalytic subunits are homologs of proteins that interact with yeast Snf1 protein kinase. J. Biol. Chem. 269:29343-29346. [PubMed] [Google Scholar]
- 24.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods, A., P. C. Cheung, F. C. Smith, M. D. Davison, J. Scott, R. K. Beri, and D. Carling. 1996. Characterization of AMP-activated protein kinase beta and gamma subunits. Assembly of the heterotrimeric complex in vitro. J. Biol. Chem. 271:10282-10290. [DOI] [PubMed] [Google Scholar]