Abstract
The kinetoplastid protozoan spliced leader (SL) RNA is the common substrate pre-mRNA utilized in all trans-splicing reactions. Here we show by fluorescence in situ hybridization that the SL RNA is present in the cytoplasm of Leishmania tarentolae and Trypanosoma brucei. Treatment with the karyopherin-specific inhibitor leptomycin B was toxic to T. brucei and eliminated the cytoplasmic SL RNA, suggesting that cytoplasmic SL RNA was dependent on the nuclear exporter exportin 1 (XPO1). Ectopic expression of xpo1 with a C506S mutation in T. brucei conferred resistance to leptomycin B. A reduction in SL RNA 3′ extension removal and 5′ methylation of nucleotide U4 was observed in wild-type T. brucei treated with leptomycin B, suggesting that the cytoplasmic stage is necessary for SL RNA biogenesis. This study demonstrates spatial and mechanistic similarities between the posttranscriptional trafficking of the kinetoplastid protozoan SL RNA and the metazoan cis-spliceosomal small nuclear RNAs.
The kinetoplastid protozoa use the trans-splicing reaction to resolve individual mRNAs from polycistronically transcribed pre-mRNAs. Transcription initiation for pre-mRNA in these early branching organisms is regulated minimally, if at all, and steady-state mRNA levels are determined posttranscriptionally at the level of trans splicing (6) or transcript stability (11). In trans splicing a 39-nt exon, the spliced leader (SL) is transferred from the SL RNA to the 5′ end of each protein-coding pre-mRNA (2). Mechanistically, cis and trans splicing are related in that they share common machinery, such as U2, U4, and U6 small nuclear RNAs (snRNAs) (46) and associated core proteins (17, 39).
SL RNA is the best-characterized molecule unique to the machinery of trans splicing. All kinetoplastid and nematode SL RNAs studied to date fold into a conserved, three-stem-loop structure (7). An unusual feature of the kinetoplastid SL RNA is the 5′ cap 4 structure that has been characterized for Trypanosoma brucei and Crithidia fasciculata (3, 15, 40). Kinetoplastid cap 4 is comprised of 2′-O-methylation of the sugar groups for the first four nucleotides (AACU), with additional methylations on the bases of first (m6,6A) and fourth (m3U) nucleotides.
A posttranscriptional processing model for trypanosomatid SL RNA maturation was proposed based on mutagenesis studies with Leishmania tarentolae. SL RNA genes are transcribed from individual promoters by RNA polymerase II (8). Transcription termination occurs in a T-tract-producing 3′-extended SL RNA. The Sm protein binding site and downstream stem-loop III structure are required for cytoplasmic 3′-nucleolytic processing of the primary SL RNA transcripts (45). Formation of the mature 3′ end is linked to cap 4 methylation; however, a mature 3′ end is not the sole determinant of cap 4 formation, since some correctly 3′-processed SL RNA mutants do not receive cap 4 (43).
While SL RNA exon content is largely tolerant to mutagenesis in L. tarentolae, intron sequence and structural changes reduce or eliminate trans splicing (43). Similar conclusions regarding exon and intron function were drawn from studies of trans splicing in nematodes (13, 33) and in the kinetoplastid Leptomonas collosoma (32). In contrast, studies with Leptomonas seymouri indicate an essential role for the exon and dispensability of intron structures and sequences (28).
A cotranscriptional model for cap 4 methylation has been presented (31), consistent with the proposal that the SL RNA substrate molecule remains exclusively in the nuclear compartment prior to participation in trans splicing (49-51). The proposed limitation of the SL RNA to the nucleus was based on SL RNA half-life determinations in the range of 3 to 6 min (25, 26, 50) that were considered insufficient to include cytoplasmic trafficking.
Several lines of evidence indicate that SL RNA has a cytoplasmic stage. First, aqueous fractionation shows a significant SL RNA presence in the cytoplasmic fraction (20, 42). Second, SL RNA generated by in vitro transcription with L. tarentolae nuclear extract is in the 3′-extended form, implying the lack of 3′-processing activity in the nucleus; these transcripts also lack cap 4 methylations (45). A similar size difference and lack of modification is observed in T. brucei in vitro extracts (18). Finally, the Sm-binding site dependence of SL RNA maturation (32, 43, 45) suggests that SL RNA travels to the cytoplasm, where it can be bound by Sm protein orthologs. Common Sm proteins have been detected in the cytoplasm of T. brucei (38).
Exportin 1 (XPO1/CRM1) is a pivotal component in the nuclear export machinery responsible for the transport of RNA polymerase II-transcribed U-rich snRNAs, along with particular classes of proteins, in metazoans (23). XPO1 is inactivated specifically and irreversibly by the cytotoxin leptomycin B (LMB) via covalent modification of a conserved binding site (22). In the trafficking of U-rich snRNAs of metazoans, m7G-capped transcripts in the cytoplasm are bound by the Sm protein complex and then hypermethylated to yield the m3G cap structure. Cytoplasmic modification of the associated proteins involves symmetrical dimethylation of arginine in Sm protein by the methylosome and SMN complex (34). Sm proteins and the m3G cap comprise the bipartite signal for import of snRNAs back into the nucleus (35), where final modifications to the RNA, such as RNA-guided 2′-O-methylations and pseudouridinylation, may occur in Cajal bodies (12). Since SL RNA fits the criterion of an RNA polymerase II-derived snRNA, XPO1 could be involved in its nuclear export.
In this study we addressed several issues in the maturation of kinetoplastid SL RNA. We examined cap 4 methylation of transcripts being actively transcribed from L. tarentolae and T. brucei and SL RNA subcellular localization visualized by fluorescence in situ hybridization (FISH). Nuclear export of SL RNA was assayed relative to the activity of exportin 1, which is specifically inhibited by LMB. LMB-induced defects in SL RNA nuclear export resulted in reduced mature 3′ end formation and reduced methylation in the SL RNA cap 4 structure. Our results are consistent with a posttranscriptional model for SL RNA processing that includes a cytoplasmic stage.
MATERIALS AND METHODS
Strains, cell culture, and plasmids.
L. tarentolae (UC strain) was grown at 28°C in brain heart infusion medium supplemented with 10 μg of hemin/ml. All T. brucei strains were derived from transfection strain YTAT KH4A. T. brucei was grown at 28°C in SM medium containing 10% fetal calf serum. T. brucei strains pHD-XPO1 and pHDxpo1-C506S were grown in the presence of 50 μg of hygromycin/ml. pHD-XPO1 and pHDxpo1-C506S were created by transfection (21) with pHD496-derived, ribosomal DNA-spacer targeting plasmids (4) linearized at the NotI site and containing either the T. brucei XPO1 open reading frame (ORF) or the T. brucei xpo1-C506S ORF using a Bio-Rad Gene Pulser II electroporator. pHD-XPO1 and pHDxpo1-C506S were created by replacing the pHD496 HindIII-GFP-BamHI cassette with Tb XPO1 or xpo1-C506S.
Aqueous fractionation.
The fractionation procedure was carried out at 4°C. Mid-log-phase L. tarentolae (109) or T. brucei (5 × 108) was washed twice in buffer A (150 mM sucrose, 20 mM KCl, 3 mM MgCl2, 20 mM HEPES-KOH [pH 7.9], 1 mM dithiothreitol, and 40 U of RNAguard [Pharmacia]/ml). After resuspension in 1.0 ml of buffer A, 0.5 ml was removed and total cell RNA was extracted with phenol-chloroform (1:1) followed by ethanol precipitation. To the remaining 0.5 ml of cells, Nonidet P-40 was added to 0.2% (vol/vol), and the cells were passed through a 26-gauge syringe needle three times to generate cell lysate. This cell lysate was fractionated by centrifugation at maximum speed for 10 min. The supernatant (cytoplasmic fraction) was decanted and spun twice to minimize any particulate material carried over by pipetting, and RNA was extracted as described above. The pellet (nuclear fraction) was passed through a 26-gauge syringe needle 15 times and refractionated by centrifugation at maximum speed for 10 min. The supernatant was discarded, and the pellet was rinsed without resuspension once in 0.5 ml of buffer A, resuspended in 0.5 ml of buffer A, and extracted for RNA as described above.
RNA analysis.
RNA from L. tarentolae and T. brucei was purified with the TriZOL reagent (Invitrogen) using ethanol rather than isopropanol precipitation. RNA blotting, probing, and analyses were performed as described previously (45). Oligonucleotides used for hybridization were the following: TbSL stem-loop I, 5′-CTACTGGGAGCTTCTCATCA (9); and U4, 5′-AACAATCACCTGAGGTTCTTGCGT (52). Primer extension analysis was performed as described previously (45). The primer used on L. tarentolae RNA was LtSL intron, 5′-GTTCCGGAAGTTTCGCATAC (14); for T. brucei, primers used were TbSL stem-loop I, TbSL stem-loop II, 5′-ATGCGTCTGTTGGCCCAGCTG; and TbSL stem-loop III, 5′-CCGACCCCACCTTCC (9). Quantitation of bands was performed using a PhosphorImager (ImageQuant; Molecular Dymamics). The L. tarentolae and T. brucei SL RNA sequences were generated by sequencing pLME12 or pUC-TbSL plasmids with LtSL intron or TbSL stem-loop I oligonucleotides, respectively.
Fluorescence in situ hybridization and FISH oligonucleotide synthesis.
T. brucei and L. tarentolae were grown as indicated, mid-log-phase cells were pelleted, and the supernatant was discarded. Cells were washed once in 1.7 ml of phosphate-buffered saline and resuspended in 1 ml of fixation buffer (4% EM-grade formaldehyde [Polysciences, Inc., Warrington, Pa.], 5% acetic acid in 1× phosphate-buffered saline). Cells were fixed for 20 min at room temperature and pelleted, and the fixation supernatant was discarded. The fixed cells were washed three times with 70% ethanol to remove any traces of formaldehyde and resuspended in 1 ml of 70% ethanol. Ten microliters of this cell suspension was spotted on a microscope slide and dried at room temperature for 10 min, followed by a 10-min incubation at 80°C in a vacuum oven. Slides were pretreated by washing them twice with 30 ml of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min each, followed by two more washes in 40% formamide-2× SSC for 5 min each. Ten nanograms of 3× fluorescein isothiocyanate (FITC)-labeled oligonucleotide probe in 20 μl of hybridization mix (40% formamide, 2× SSC, 2.5 mg of tRNA/ml, 50 U of RNasin, 0.02% bovine serum albumin) was added to the immobilized cell spot. Target RNA and probe were simultaneously denatured on a 75°C heat block for 3 min and slow-cooled to 37°C. Probe was hybridized overnight in the dark at 37°C in a humid chamber. Unbound probe was removed by washing it twice in 30 ml of 40% formamide-2× SSC at 37°C for 15 min each, next twice in 2× SSC, and once in 1× SSC for 15 min each at room temperature. Coverslips were mounted with ProLong Antifade kit (Molecular Probes) spiked with 100 ng of 4′,6′-diamidino-2-phenylindole DAPI/μl. Cells were visualized with the 63× objective of a Zeiss Axiocam fluorescence microscope, and images were captured with a Zeiss digital camera and Zeiss Axiovision software. All directly compared images were captured at the same fluorescence lamp intensity and shutter speed; quantitation of fluorescence (n = 5) was performed using Zeiss Axiovision 3.0 software. L. tarentolae FISH was controlled for background hybridization and hybridization to DNA sequences by comparison with an LtSL DNA control oligonucleotide, a 3× FITC-labeled oligonucleotide complementary to the LtSL intron FISH oligonucleotide. Repeated experiments with LtSL DNA control produced FITC background levels identical to those for unhybridized L. tarentolae samples. Oligonucleotide FISH probes were synthesized with an Expedite 8909 nucleic acid synthesis system (Applied Biosystems). All synthesis reagents were purchased from Glen Research. Oligonucleotide sequences are the following: U3 FISH, 5′-TAAGAGGTTGTACTCATAAAACGATTCTGT (37); TbSL intron FISH, 5′-TGGCCAGCTGCTACTGGGAGCTTCTCATAC (9); LtSL intron FISH, 5′-AGGTTCCGGAAGTTTCGCATACCAATAAAGT (14); LtSL mRNA control, 5′-TCCAAGGCCTTCAAAGCGTATGCAATAAAGT; LtSL DNA control, 5′-ACTTTATTGGTATGCGAAACTTCCGGAACCT (14); and tRNAAla-FISH, 5′-AAGTTGGGTATCGATCCCAATACCTACCGCA (GenBank accession no.AY007788; internal FITC-dT residues are underlined). With the exception of tRNAAla-FISH, all primers have a 5′ FITC cap.
XPO1 cloning.
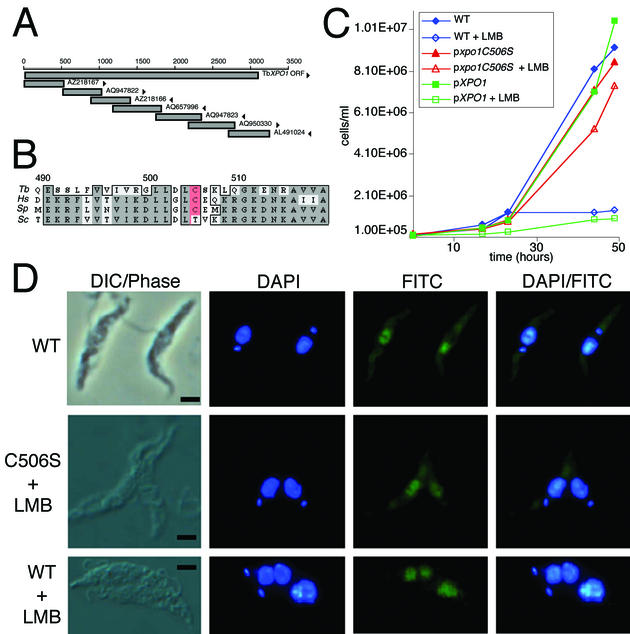
T. brucei XPO1 was identified by TBLASTN searching of the EMBL-EBI T. brucei genome BLAST server (http://www.ebi.ac.uk/blast2/parasites.html) with the protein sequence of S. pombe XPO1. The full-length T. brucei XPO1 sequence was assembled from overlapping contigs (for specifications, see Fig. 3A). The complete T. brucei XPO1 gene was PCR amplified from YTAT genomic DNA using T. brucei XPO1 primers and Pfu polymerase (Pfu Turbo; Stratagene), and the DNA sequence was confirmed (GenBank accession no. AY135380). The xpo1-C506S ORF was generated with the QuickChange XL site-directed mutagenesis kit (Stratagene) using the primers C506Sfwd (5′-GATCGTGGCCTGCTAGACCTTAGTAGCAAGCTACAAGG) and C506Srev (5′-CCTTGTAGCTTGCTACTAAGGTCTAGCAGGCCACGTAC).
FIG. 3.
Mutated XPO1 allows export of SL RNA in the presence of LMB. (A) Schematic map of the contigs assembled to derive the complete sequence for XPO1 from T. brucei. Arrowheads indicate orientation of the sequences. (B) Conservation of XPO1 in the region interacting with the inhibitor LMB. Multiple sequence alignment of the XPO1 conserved core region fragment from T. brucei (Tb), Homo sapiens (Hs) (GenBank no. NP003391), S. pombe (Sp) (GenBank NP593928), and Saccharomyces cerevisiae (Sc) (GenBank# P30822). The cysteine conferring sensitivity to LMB is highlighted in red. (C) Mutation C506S of XPO1 confers resistance to LMB. Growth curves in the absence or presence of 100 ng of LMB/ml were performed on wild-type T. brucei and on T. brucei harboring a ribosomal DNA spacer-integrated copy of either wild-type XPO1 or xpo1-C506S. (D) Wild-type T. brucei and pxpo1-C506S transfectants grown in the presence of 1 μg of LMB/ml for 48 h were used to detect SL RNA by FISH. Untreated T. brucei are shown as a control (WT). Nuclei and kinetoplast DNA are counterstained with DAPI. Bar, 2.5 μm.
RESULTS
SL RNA is cap 4 methylated posttranscriptionally.
T. brucei cap 4 acquisition has been examined on transcripts prematurely terminated by the RNA elongation inhibitor 3′-O-methyl-GTP (31). Transcripts terminated prior to the Sm-binding site were incompletely methylated, while approximately 60% of transcripts terminated downstream of the Sm-binding site had mature cap 4 structures, leading to the conclusion that cap 4 methylation is cotranscriptional. This conclusion contrasts with the posttranscriptional cap 4 modification data from genetic studies with L. tarentolae (43), which indicated that mature cap 4 formation required 3′ processing and was lacking for mutants in the Sm-binding site and stem-loop III regions of the intron. Reduction in cap 4 formation in these intron mutants consisted of the absence of methylation at positions +2, +3, and +4 as measured by primer extension analysis.
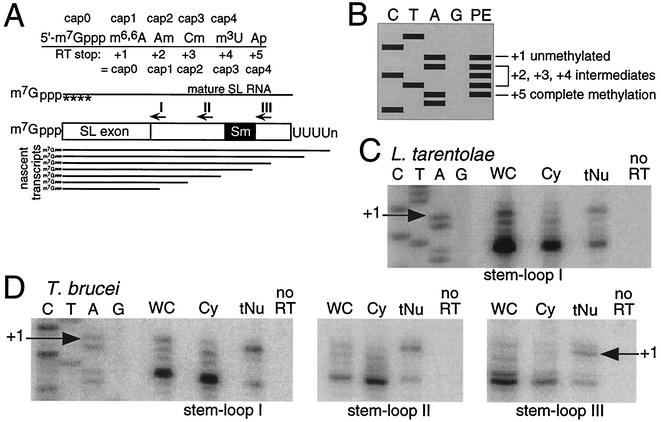
To address the cap 4 conundrum, nascent RNA from L. tarentolae and T. brucei was assayed for cap 4 formation using a different approach. Nascent RNA was enriched by isolating nuclei in an aqueous fractionation (42). Since aqueous fractionation is accompanied by nucleoplasmic leakage (41), we performed extensive washing of the nuclei such that nascent SL RNA tethered to its RNA polymerase would be retained in the nuclear fraction. SL RNA released from the RNA polymerase (i.e., nucleoplasmic SL RNA) would be depleted from the RNA polymerase-tethered nuclear fraction but present in the cytoplasmic fraction. SL RNA engaged in a trans-splicing complex would also comprise part of the nuclear RNA pool. SL RNA is a viable target for nascent transcript enrichment since it is a high-copy-number gene that accounts for a significant percentage of total cellular transcription (5). The 5′-methylation profile of SL RNA was assayed by primer extension (Fig. 1A), a method used for the detection of 2′-O-methyl groups and other RNA modifications (29, 30). Primer extension is the most widely employed assay for monitoring overall SL RNA cap 4 status from a heterogeneous population of SL RNA containing the complete spectrum of methylated species (28, 32, 36, 44). A schematic representation for the interpretation of the five primer extension stops is provided in Fig. 1B.
FIG. 1.
Nascent SL RNA transcripts lack 5′ methylations. (A) Schematic of the 5′-end structure of SL RNA and intermediates cap 0 through cap 4. Primer extension using reverse transcriptase (RT) is terminated adjacent to methylated nucleotides and serves as an assay for partial cap 4 formation as indicated. Nascent-enriched transcripts were enriched from purified nuclei and are extended using SL RNA intron primers specific for stem-loop I, II, or III. (B) Schematic representation of the five possible primer extension (PE) products and their identities. (C) Primer extension analysis of nascent-enriched RNA from L. tarentolae using a stem-loop I-directed primer. A sequence ladder of SL RNA using the same primer is shown, with the transcription start point (+1) indicated. Primer extension reactions on whole-cell (WC), cytoplasmic (Cy), and tethered-nuclear (tNu) RNA fractions are indicated, including a lane lacking reverse transcriptase (no RT) on whole-cell RNA. In all extension reactions, between 1 and 5 μg of RNA was used. (D) Primer extension analysis of tethered-nuclear RNA from T. brucei using three primers at staggered locations through the intron region. A sequence ladder of SL RNA using the stem-loop I primer is shown, with the transcription start point (+1) indicated. The sources of RNA are as indicated in panel B.
Following aqueous fractionation of L. tarentolae or T. brucei cells, we performed primer extensions on SL RNA with oligonucleotides specific for stem-loops I, II, or III in the nascent-enriched nuclear fraction, the cytoplasmic fraction, and whole-cell RNA (Fig. 1C and D and data not shown). Since the aqueous fractionation conditions selected for nascent SL RNA tethered to RNA polymerase, the stem-loop I, II, and III oligonucleotides assayed nascent SL RNA frozen in different stages of transcription. The stem-loop I oligonucleotide extends any nascent SL RNA transcribed beyond nucleotide positions 50 to 55 but does not extend shorter SL RNAs in an earlier stage of transcription. In contrast, the stem-loop III oligonucleotide only extends nascent SL RNAs of more than 127 to 131 nucleotides (nt), which all contain a complete Sm-binding site.
With all three oligonucleotides, primer extension products from SL RNA isolated from purified nuclei displayed two predominant bands of approximately equal intensity at +1 and +5 (Fig. 1C and D, lane tNU). Products derived from the cytoplasmic population of SL RNA were reduced for +1 and contained predominantly +5, with an additional background of +2 and +3 intermediates (Fig. 1C and D, lane Cy). RNA from whole cells showed the relative abundance of all forms in the SL RNA cap 4 methylation process; the +5 species comprised the majority product, while the minor +1, +2, and +3 products were roughly equal (Fig. 1C and D, lane WC). A deoxynucleoside triphosphate titration was performed (200 nM to 2 mM range) to ensure that the experimental concentration (200 μM) did not lead to read-through of methylations (data not shown).
Based on these data, there is no difference in the timing of SL RNA cap 4 acquisition between T. brucei and L. tarentolae. Detection of +5 in the tethered-nuclear RNA preparation is consistent with the presence of mature SL RNA in the nucleus, due either to sequestration within trans-splicing complexes and/or in other structures involved in additional SL RNA modification. The reduction of +1, the appearance of +2 and +3, and the relative increase in +5 primer extension products from the cytoplasmic fraction indicates that SL RNA cap 4 modification takes place posttranscriptionally. Most notably, the cap 4 intermediates predicted by the cotranscriptional model are not detected in the nuclear fraction. Furthermore, cytosolic RNA sampled using a digitonin titration procedure (1) minimally disruptive to the nuclear contents produced +1 primer extension products in addition to +2, +3, and +5 products for SL RNA (data not shown), supporting the cytosolic acquisition of cap 4 methylations on SL RNA.
In situ analysis demonstrates the cytoplasmic presence of SL RNA in L. tarentolae.
Since aqueous fractionation is an imperfect cytoplasmic sampling method due to the potential for nuclear leakage, we adapted FISH to assay convincingly the intracellular localization of SL RNA. FISH oligonucleotide probes (10) were ideal for assessing the subcellular localization of SL RNA. Substrate SL RNA is 96 nt in L. tarentolae and 140 nt in T. brucei (15). A probe targeting the 9-nt exon/22-nt intron junction was chosen based on accessibility to oligonucleotide binding in permeabilized T. brucei cells (48). Low- and medium-resolution RNA blotting with probes hybridizing to this region of the SL RNA did not hybridize to other cellular RNAs (data not shown).
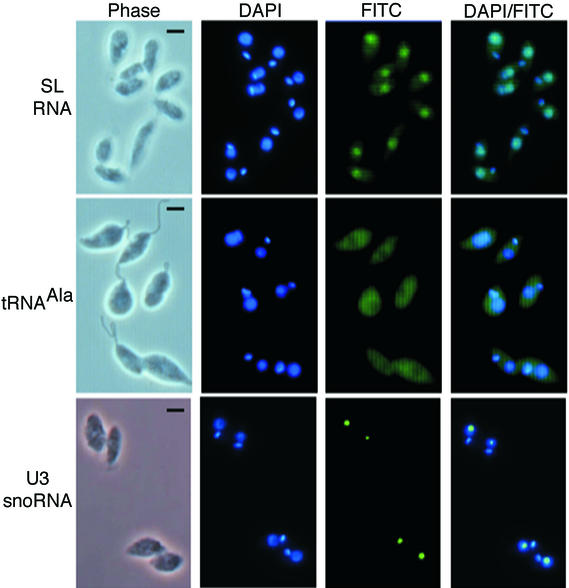
Oligonucleotide probes internally labeled three times with fluorescein isothiocyanate (FITC) were used (10). FISH for L. tarentolae SL RNA (Fig. 2) showed diffuse cell body staining punctuated by an area of more intense staining, identified as the cytoplasm and nucleus, respectively. Nuclear and kinetoplast DNAs were visualized by blue counterstaining with DAPI. As a specificity control for unspliced SL RNA, we created the LtSL mRNA control probe. The LtSL mRNA control hybridizes to 9 nt of SL RNA exon immediately upstream of the exon/intron boundary and contains 22 nt of noncomplementary intron sense strand immediately downstream of the exon/intron boundary. FISH with LtSL mRNA control confirmed that mRNA cross-hybridization with the SL RNA substrate probe was negligible (data not shown). tRNAAla and U3 small nucleolar RNA (snoRNA) showed, respectively, diffuse cytoplasmic staining or a single compact spot of hybridization marking the nucleolus. To control for background due to nonspecific interaction of the L. tarentolae SL RNA probe, hybridization to DNA, or autofluorescence, the LtSL DNA control oligonucleotide probe that is not predicted to hybridize to SL RNA transcripts was assayed along with mock-treated cells. FITC signal from mock-treated and negative controls showed identical background levels that were subtracted from the FITC signal of the indicated probes (data not shown).
FIG. 2.
Direct visualization of SL RNA in the cytoplasm of L. tarentolae. Mid-log-phase L. tarentolae cells were prepared for FISH analysis. Hybridization with specific FITC-labeled oligonucleotides was used to detect SL RNA, tRNAAla, or U3 snoRNA. Nuclei and kinetoplast DNA were counterstained with DAPI. Images were captured using the indicated fluorescence filters. Bar, 2.5 μm.
Substrate SL RNA is present in the cytoplasm and nucleus of L. tarentolae. Similar results were seen when examining SL RNA subcellular localization in T. brucei (see below), indicating that intracellular trafficking of SL RNA is not species specific in the trypanosomatids.
T. brucei is sensitive to LMB treatment.
The exportin 1 gene had not been reported previously in T. brucei. TBLASTN searches identified fragments of potential homology to Schizosaccharomyces pombe XPO1. These fragments were used to reconstruct the gene (Fig. 3A) from the T. brucei nucleotide database, allowing direct manipulation of XPO1. In metazoans, XPO1 has a role in nuclear export and can be inactivated specifically by LMB. A cysteine at position 506 of TbXPO1 suggests sensitivity to the drug in T. brucei (Fig. 3B). LMB was assayed for effects on kinetoplastids and on SL RNA subcellular localization in particular.
T. brucei was sensitive to LMB treatment (≤1 μg/ml), displaying growth (Fig. 3C) and morphological phenotypes, including enlarged nuclei and multiple kinetoplasts (Fig. 3D); L. tarentolae was insensitive to LMB as demonstrated by growth curves and unaltered morphology (data not shown). SL RNA nuclear-cytoplasmic trafficking studies were therefore continued using T. brucei.
The sensitivity of T. brucei to LMB treatment implied the involvement of XPO1 in SL RNA nuclear export. We next sought to confirm the participation of XPO1 in SL RNA maturation by genetically altering TbXPO1 to LMB resistance.
LMB-resistant xpo1 in T. brucei.
The complete XPO1 gene was cloned from T. brucei. Alignment of the predicted protein based on the sequenced T. brucei gene with XPO1 from other eukaryotes showed that T. brucei XPO1 contained a cysteine at position 506, corresponding to C526 in S. pombe, that is critical for LMB binding (see Fig. 3B). To investigate the role of XPO1 in SL RNA transport, we made a single amino acid substitution in the cysteine (xpo1 C506S) associated with LMB resistance in S. pombe (22). Ectopic expression of xpo1-C506S in T. brucei abolished growth inhibition by LMB (Fig. 3C). T. brucei cells expressing wild-type XPO1 ectopically and wild-type cells were equally sensitive to LMB.
One hundred nanograms of LMB/ml was added to the medium of a T. brucei culture, and FISH for SL RNA was performed on cells fixed 48 h post-LMB addition. We performed 100-ng/ml LMB time course experiments, analyzing LMB-treated T. brucei by FISH for SL RNA. Although the cells demonstrated a 35% increase in nuclear SL RNA signal at 16 h relative to mock-treated cells (data not shown), the cytoplasmic SL RNA signal remained constant until 48 h, when it was reduced to 36% of wild-type signal and not visible to the naked eye. FISH analysis of the mutated xpo1-C506S transfectants detected SL RNA in the cytoplasm of LMB-treated cells (Fig. 3D) at levels comparable to those in untreated wild-type cells. In contrast, SL RNA in LMB-treated T. brucei was confined to the enlarged nucleus with no cytoplasmic staining. In all T. brucei FISH experiments, FITC images were captured at the same shutter speed and were not background corrected.
These data suggest that substrate SL RNA is exported from the nucleus via the XPO1 pathway in T. brucei. XPO1 is the only cellular target of LMB in S. pombe; thus, the mechanism of XPO1 inactivation by LMB is evolutionarily conserved. Given the short half-life of SL RNA in T. brucei, we had anticipated a more rapid effect of LMB on T. brucei. However, the effects of LMB were not observed until 48 h. Several variables may be responsible for this observation: (i) kinetics of LMB uptake, (ii) kinetics of XPO1 inhibition, (iii) pleiotropic effects of XPO1 inhibition, since it is responsible for many other cargoes, and (iv) disruption of the RAN gradient, which drives both nuclear import and export, potentially creating an artificial accumulation of SL RNA in the cytoplasm. The most parsimonious interpretation of these results is that SL RNA is present in the cytoplasm as a cargo of XPO1. Under nuclear export inhibition conditions, the molecular phenotype of SL RNA would be revealing for both posttranscriptional processing and subcellular location of processing events.
LMB-treated T. brucei cells accumulate unprocessed SL RNA.
The posttranscriptional processing model of SL RNA biogenesis predicts that a defect in SL RNA nuclear export due to XPO1 inactivation would preclude 3′ processing and impair cap 4 modification. By contrast, in the cotranscriptional cap 4 model all SL RNA processing events should occur under LMB-treatment conditions. To differentiate between these two possibilities, the status of 3′ and 5′ processing was examined on SL RNA extracted from LMB-treated cells.
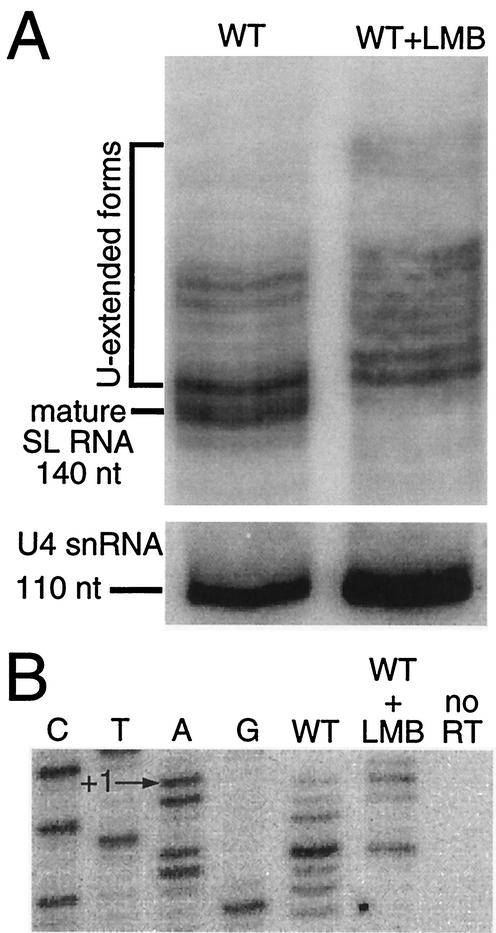
Total cell RNA was harvested from T. brucei treated with 1 μg of LMB/ml for 16 h. We performed both short-term (30 s to 10 min) and long-term (1 to 48 h) time courses of LMB treatment and analyzed SL RNA by RNA blotting and primer extension (data not shown). A molecular phenotype was not visible until 16 h, at which time SL RNA was still visible in the cytoplasm by FISH (data not shown). High-resolution RNA blots probed for SL RNA showed a loss of mature SL RNA and a general shift towards 3′-extended precursor SL RNA in LMB-treated cultures compared to nontreated cultures (Fig. 4A). Control hybridization of the blot with a U4 snRNA probe demonstrated that there were no migration rate artifacts (Fig. 4A). SL RNA cap 4 formation was assayed by primer extension with an intron-specific oligonucleotide, TbSL stem-loop I. Whereas a typical distribution of 20:80 was observed for +1:+5 in nontreated total cell RNA, LMB treatment caused a primer extension termination shift to 50:50, +1:+5, with a minor +2 band (Fig. 4B). FISH analysis of LMB-treated cells revealed normal cytoplasmic levels of SL RNA, with an increase in nuclear SL RNA abundance at 16 h relative to levels for mock-treated T. brucei (data not shown).
FIG. 4.
LMB treatment correlates with an accumulation of cap 0 and 3′-extended SL RNA forms in T. brucei. (A) LMB treatment results in accumulation of 3′-extended SL RNA. Total RNA from wild-type (WT) and LMB-treated cells (WT+LMB) at 16 h was resolved in an 8 M urea-10% polyacrylamide sequencing gel and hybridized with an SL RNA intron-specific or U4 snRNA-specific probe. (B) LMB treatment correlates with an increase in SL RNA cap 0. Total RNA from LMB-treated T. brucei was subjected to primer extension analysis with an SL RNA intron-specific oligonucleotide. A sequence ladder of SL RNA shows the position of the transcription start point.
Under the LMB conditions of this experiment, the total SL RNA pool examined represents two major classes of SL RNA: cytoplasmic SL RNA that was exported from the nucleus prior to LMB treatment and primary SL RNA transcripts that were retained in the nucleus after LMB treatment. The relative increase of a primer extension stop at +1 in total RNA from LMB-treated cells reveals the unmethylated status of these primary SL RNA transcripts. The phenotypic shift of SL RNA in LMB-treated cells supports cytoplasmic trafficking in the posttranscriptional cap 4 modification model.
DISCUSSION
In this study we have confirmed key aspects of a posttranscriptional processing model for the maturation of the SL RNA in the trypanosomatids. At issue are two major points: (i) export of the SL RNA into the cytoplasm and (ii) the posttranscriptional acquisition of methylations that constitute the mature cap 4 structure. Presented here are two approaches supporting the presence of substrate SL RNA in the cytoplasm. Examination of nuclear SL RNA enriched for nascent transcripts demonstrated that cap 4 modification is a posttranscriptional event in T. brucei and L. tarentolae. While the precise kinetics of cap 4 methylation are beyond the scope of this study, indications from the data will be discussed. The presence of SL RNA in the cytoplasm was shown using aqueous fractionation and FISH analysis. A spectrum of cap 4 methylation intermediates was visible in the cytoplasmic SL RNA population, suggesting that at least some methylations in cap 4 formation are occurring in the cytoplasm. The mechanism of SL RNA export was identified as the exportin 1 pathway. Specific inhibition of XPO1 resulted in a loss of SL RNA in the cytoplasm, as well as an accumulation of 3′-extended and incompletely methylated SL RNAs, supporting the contention that SL RNA processing is a posttranscriptional process.
Here we propose an alternative model for the maturation of SL RNA based on the data from this and other studies and by analogy with the processing pathways for metazoan snRNAs (35). The genesis of SL RNA is initiated with transcription by RNA polymerase II (8, 16). During transcription, nascent SL RNA receives an m7G cap structure, or cap 0 (31), but none of the cap 4 methylations, as demonstrated by the absence of +2, +3, or +4 cap 4, intermediates in nascent-enriched nuclear RNA. After newly synthesized, 3′-extended SL RNA transcript is released from RNA polymerase II, some or all of the three cap 1 methylations, any of which results in +2 termination of primer extension reactions (24, 30), are acquired. At least one of these events is Sm independent and could be commencing in the nucleoplasm prior to export, in the cytoplasm, or post-import. The nascent nucleoplasmic SL RNA is bound by an ortholog of the cap-binding complex and delivered to a nuclear pore for export into the cytoplasm via the XPO1 pathway. The nuclear egress of SL RNA primary transcripts is rapid, as supported by the observation that 3′-extended SL RNA precursors are underrepresented in nuclear versus cytoplasmic fractions (data not shown). Cytoplasmic SL RNA is likely to associate with Sm proteins. The disruption of the Sm-binding site results in a loss of 3′ processing (43, 45) and of further cap 4 modifications beyond cap 1 (32, 43, 45), suggesting that they are dependent on the presence of an intact Sm-binding site. The cytoplasmic +5 primer extension products may not be indicative of complete cap 4 methylation, since, similar to the methylations at position 1, primer extension termination products at position 4 could arise from either base or ribose methylations. A combination of 5′-end methylation and snRNP formation may provide the signals for nuclear import. Some processing of SL RNA may occur in the nucleolus, since SLA1 RNA, a snoRNA guiding exon pseudouridinylation, has been localized to that subnuclear compartment; however, SL RNA has not been detected in the nucleolus (27, 42). The precise acquisition of SL RNA cap 4 methylations is an area of continued interest. The exact subcellular localization of cap 4 acquisition will be resolved by the purification and characterization of the methylase(s) responsible for cap 4 formation.
Consistent with the posttranscriptional cap 4 formation scenario are methylation studies using whole-cell, nuclear or cytoplasmic lysates to affect maturation of undermethylated SL RNA isolated under methylase inhibition conditions in which only snRNP-bound SL RNA, and not deproteinized SL RNA, could be cap 4 methylated (49). Although the conclusion that common protein binding is a prerequisite for cap 4 formation is inconsistent with the cotranscriptional model (31, 47), the data and conclusion that SL RNA must be in a protein complex to acquire cap 4 modification are consistent with two key aspects of the model supported here: (ii) mature cap 4 acquisition is posttranscriptional, and (ii) cytoplasmic Sm protein binding is necessary for mature cap 4 formation.
The model presented in this paper contrasts with a cotranscriptional model for cap 4 formation derived from a study using premature SL RNA termination created by inclusion of 3′-O-methyl GTP in the nucleotide mixture (31). When incorporated into RNA in place of GTP, this modified nucleotide can stall transcription without release of the nascent RNA (19), thus allowing the assessment of cap 4 status on SL RNA of truncated size. Application of this terminator to T. brucei cells permeabilized by the detergent lysolecithin showed that truncated SL RNA lacking the Sm-binding site (shorter than 117 nt) were partially methylated and that truncated, Sm-binding-site-containing SL RNA (position 117+) showed variable cap 4 methylation, including the mature cap 4 structure (31). These results can be reconciled with our posttranscriptional model if some truncated molecules were released from the transcription complex and entered the proposed posttranscriptional SL RNA maturation pathway. The cotranscriptional model does not account for the absence of mature cap 4 formation in intron-mutated SL RNA (32, 43), nor for reduced cap 4 methylation in SL RNA transcription termination mutants (43, 45). In this study, the intermediate cap 1 or cap 2 forms predicted by cotranscriptional processing in nascent-enriched SL RNA analyses were not detected.
Cytoplasmic trafficking is a key step in the maturation of SL RNA. The involvement of the exportin 1 pathway is implicated by LMB treatment of T. brucei. We have identified other potential components of the exportin 1 nuclear export complex, including PHAX (23) in L. major and CBP 20 and CBP 80 (23) in T. brucei. These genes, in addition to XPO1, constitute our springboard for further elucidation of the SL RNA biogenesis pathway.
Acknowledgments
This work was supported by NIH grant AI34536 to D.A.C. G.M.Z. is a predoctoral trainee recipient of Microbial Pathogenesis Training Grant 2-T32-AI-07323.
The individual sequences for the T. brucei XPO1 were retrieved from the EMBL-EBI T. brucei Genome BLAST server. We thank Minoru Yoshida (RIKEN, Saitama, Japan) for the generous gift of leptomycin B; Kent Hill for providing the T. brucei YTAT cells and use of the Zeiss Axiocam fluorescence microscope; Jason Underwood and Doug Black for assistance with FISH oligonucleotide synthesis; and Robert Hitchcock, Sean Thomas, and Scott Westenberger for critical reading of the manuscript.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agabian, N. 1990. Trans splicing of nuclear pre-mRNAs. Cell 61:1157-1160. [DOI] [PubMed] [Google Scholar]
- 3.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 4.Biebinger, S., S. Rettenmaier, J. Flaspohler, C. Hartmann, J. Pena-Diaz, L. E. Wirtz, H. R. Hotz, J. D. Barry, and C. Clayton. 1996. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 24:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boothroyd, J. C., D. A. Campbell, and R. E. Sutton. 1985. Expression of surface antigen genes in Trypanosoma brucei involves a novel system of discontinuous transcription, p. 61-66. In R. A. Lerner, R. M. Channock, and F. Brown (ed.), Vaccines 85: modern approaches to vaccines. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 6.Brooks, D. R., H. Denise, G. D. Westrop, G. H. Coombs, and J. C. Mottram. 2001. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J. Biol. Chem. 276:47061-47069. [DOI] [PubMed] [Google Scholar]
- 7.Bruzik, J. P., K. Van Doren, D. Hirsh, and J. A. Steitz. 1988. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature 335:559-562. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, D. A., N. R. Sturm, and M. C. Yu. 2000. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today 16:78-82. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, D. A., D. A. Thornton, and J. C. Boothroyd. 1984. Apparent discontinuous transcription of Trypanosoma brucei variant surface antigen genes. Nature 311:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrand, P., E. Bertrand, R. H. Singer, and R. M. Long. 2000. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 318:493-506. [DOI] [PubMed] [Google Scholar]
- 11.Clayton, C. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq, X., B. E. Jády, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridinylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denker, J. A., P. A. Maroney, Y. T. Yu, R. A. Kanost, and T. W. Nilsen. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2:746-755. [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, J., and D. A. Campbell. 1994. Expression of the Leishmania tarentolae ubiquitin-encoding and mini-exon genes. Gene 144:45-51. [DOI] [PubMed] [Google Scholar]
- 15.Freistadt, M. S., G. A. M. Cross, A. D. Branch, and H. D. Robertson. 1987. Direct analysis of the mini-exon donor RNA of Trypanosoma brucei: detection of a novel cap structure also present in messenger RNA. Nucleic Acids Res. 15:9861-9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncharov, I., Z. Palfi, A. Bindereif, and S. Michaeli. 1999. Purification of the spliced leader ribonucleoprotein particle from Leptomonas collosoma revealed the existence of an Sm protein in trypanosomes. J. Biol. Chem. 274:12217-12221. [DOI] [PubMed] [Google Scholar]
- 18.Günzl, A., E. Ullu, M. Dorner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wunsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 19.Hagler, J., and S. Shuman. 1992. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science 255:983-986. [DOI] [PubMed] [Google Scholar]
- 20.Hartshorne, T., and N. Agabian. 1993. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol. Cell. Biol. 13:144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, K. L., N. R. Hutchings, D. G. Russell, and J. E. Donelson. 1999. A novel protein targeting domain directs proteins to the anterior cytoplasmic face of the flagellar pocket in African trypanosomes. J. Cell Sci. 112:3091-3101. [DOI] [PubMed] [Google Scholar]
- 22.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Sorinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuersten, S., M. Ohno, and I. W. Mattaj. 2001. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11:497-503. [DOI] [PubMed] [Google Scholar]
- 24.Lafontaine, D. L. J., T. Preiss, and D. Tollervey. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 18:2360-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laird, P. W., A. L. ten Asbroek, and P. Borst. 1987. Controlled turnover and 3′ trimming of the trans splicing precursor of Trypanosoma brucei. Nucleic Acids Res. 15:10087-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laird, P. W., J. C. Zomerdijk, D. de Korte, and P. Borst. 1987. In vivo labelling of intermediates in the discontinuous synthesis of mRNAs in Trypanosoma brucei. EMBO J. 6:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, X.-h., Y.-X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lücke, S., G. L. Xu, Z. Palfi, M. Cross, V. Bellofatto, and A. Bindereif. 1996. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans splicing and cap 4 formation. EMBO J. 15:4380-4391. [PMC free article] [PubMed] [Google Scholar]
- 29.Maden, B. E., M. E. Corbett, P. A. Heeney, K. Pugh, and P. M. Ajuh. 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochemie 77:22-29. [DOI] [PubMed] [Google Scholar]
- 30.Maden, B. E. H. 2001. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 25:374-382. [DOI] [PubMed] [Google Scholar]
- 31.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994-28999. [DOI] [PubMed] [Google Scholar]
- 32.Mandelboim, M., C. L. Estraño, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 33.Maroney, P. A., G. J. Hannon, J. D. Shambaugh, and T. W. Nilsen. 1991. Intramolecular base pairing between the nematode spliced leader and its 5′ splice site is not essential for trans-splicing in vitro. EMBO J. 10:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massenet, S., L. Pellizzoni, S. Paushkin, I. W. Mattaj, and G. Dreyfuss. 2002. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 22:6533-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattaj, I. W., and L. Engelmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 36.McNally, K. P., and N. Agabian. 1992. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol. 12:4844-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mottram, J., K. L. Perry, P. M. Lizardi, R. Lührmann, N. Agabian, and R. G. Nelson. 1989. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 9:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palfi, Z., A. Günzl, M. Cross, and A. Bindereif. 1991. Affinity purification of Trypanosoma brucei small nuclear ribonucleoproteins reveals common and specific protein components. Proc. Natl. Acad. Sci. USA 88:9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palfi, Z., S. Lücke, H.-W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Séraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 97:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry, K. L., K. P. Watkins, and N. Agabian. 1987. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. USA 84:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinke, J., and J. A. Steitz. 1985. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 13:2617-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts, T. G., N. R. Sturm, B. K. Yee, M. C. Yu, T. Hartshorne, N. Agabian, and D. A. Campbell. 1998. Three small nucleolar RNAs identified from the spliced leader-associated RNA locus in kinetoplastid protozoans. Mol. Cell. Biol. 18:4409-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturm, N. R., and D. A. Campbell. 1999. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 274:19361-19367. [DOI] [PubMed] [Google Scholar]
- 44.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 273:18689-18692. [DOI] [PubMed] [Google Scholar]
- 45.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tschudi, C., and E. Ullu. 1990. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell 61:459-466. [DOI] [PubMed] [Google Scholar]
- 47.Tschudi, C., and E. Ullu. 2002. Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids. Gene Expr. 10:3-16. [PMC free article] [PubMed] [Google Scholar]
- 48.Ullu, E., and C. Tschudi. 1993. 2′-O-methyl RNA oligonucleotides identify two functional elements in the trypanosome spliced leader ribonucleoprotein particle. J. Biol. Chem. 268:13068-13073. [PubMed] [Google Scholar]
- 49.Ullu, E., and C. Tschudi. 1995. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 270:20365-20369. [DOI] [PubMed] [Google Scholar]
- 50.Ullu, E., and C. Tschudi. 1991. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 88:10074-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ullu, E., C. Tschudi, and A. Günzl. 1996. Trans-splicing in trypanosomatid protozoa, p. 115-133. In D. F. Smith and M. Parsons (ed.), Molecular biology of parasitic protozoa. IRL Press, Oxford, United Kingdom.
- 52.Yu, M. C., T. C. Orlando, N. R. Sturm, L. Zhou, R. M. Saito, L. M. Floeter-Winter, and D. A. Campbell. 2002. Two distinct functional spliced leader RNA gene arrays in Leishmania tarentolae are found in several lizard Leishmania species. Int. J. Parasitol. 32:1411-1422. [DOI] [PubMed] [Google Scholar]