Abstract
The opportunistic pathogen Aspergillus fumigatus is the most frequent cause of deadly airborne fungal infections in developed countries. In order to identify novel antifungal-drug targets, we investigated the genome of A. fumigatus for genes that are necessary for efficient fungal growth. An artificial A. fumigatus diploid strain with one copy of an engineered impala160 transposon from Fusarium oxysporum integrated into its genome was used to generate a library of diploid strains by random in vivo transposon mutagenesis. Among 2,386 heterozygous diploid strains screened by parasexual genetics, 1.2% had a copy of the transposable element integrated into a locus essential for A. fumigatus growth. Comparison of genomic sequences flanking impala160 in these mutants with that of the genome of A. fumigatus allowed the characterization of 20 previously uncharacterized A. fumigatus genes. Among these, homologues of genes essential for Saccharomyces cerevisiae growth have been identified, as well as genes that do not have homologues in other fungal species. These results confirm that heterologous transposition using the transposable element impala is a powerful tool for functional genomics in ascomycota, and they pave the way for defining the complete set of essential genes in A. fumigatus, the first step toward target-based development of new antifungal drugs.
Systemic infections by opportunistic pathogenic fungi have become a clinical problem as the number of immunodeficient patients and the development of severely immunosuppressive therapies are increasing (51, 53). The two main causative agents of opportunistic fungal infections are the commensal polymorphic yeast Candida albicans and the saprophytic filamentous fungus Aspergillus fumigatus (38, 51). Currently available antifungal drugs belong to three main classes: polyenes (e.g., amphotericin B) and azoles (e.g., fluconazole), both of which target fungal membranes (19, 22), and the new echinocandin family (e.g., caspofungin), which targets the enzyme responsible for cell wall β(1,3)-glucan biosynthesis (2, 23). However, systemic fungal infections are still associated with a high mortality, mainly due to the relative toxicity and side effects of antifungal drugs, in addition to often-late diagnosis and the emergence of resistance (15, 19, 49).
A rational approach to increasing our antifungal arsenal relies on the identification of novel targets involved in various aspects of fungal biology (21, 29). Although gene products necessary for virulence are seen as candidate targets (44), no genuine virulence factor in opportunistic fungal pathogens has yet been identified (34, 42). Attractive alternative antifungal targets are to be found among gene products that are essential for fungal growth both in vivo and ex vivo (18, 26). Compendia of essential genes have been obtained for the model eukaryotic microorganism Saccharomyces cerevisiae through various approaches (31), including systematic gene inactivation or random insertional mutagenesis in a diploid background followed by analysis of meiotic progenies (20, 47). A set of genes critical for growth of the yeast C. albicans has also been defined by using inducible expression of antisense RNA molecules (10). Of the 86 C. albicans genes identified, 38% do not have homologues in available databases (10). Differences in essential biological processes between the yeasts S. cerevisiae and C. albicans highlight the need to study the larger and more complex filamentous fungal genomes in order to reveal species-specific and filamentous fungus-specific targets.
A. fumigatus has become the most prevalent airborne filamentous fungal pathogen (38). Dissemination of A. fumigatus occurs by release of asexual spores (conidia) into air (33). They are inhaled daily without major consequences for human health. However, in immunocompromised hosts, A. fumigatus can cause a usually fatal infection, termed invasive pulmonary aspergillosis (13, 33, 36). A. fumigatus is haploid and devoid of a sexual cycle (33), preventing the application of strategies that use classical genetics to define essential genes. Nevertheless, it has previously been shown that the parasexual cycle, which relies on the chemical haploidization of artificial diploid strains (6, 50), can be used to demonstrate the essential function of A. fumigatus genes (17). In this setting, a heterozygous A. fumigatus diploid is generated by targeted gene replacement or by random insertional mutagenesis and is subjected to haploidization with or without the selective pressure corresponding to the introduced mutation. The absence of haploid progeny under selective conditions only is indicative of the inactivation of a gene essential for A. fumigatus growth (Fig. 1). By using this approach, it was demonstrated that the A. fumigatus FKS1 gene, encoding the 1,3-β-d-glucan synthase catalytic subunit, and the smcA gene, encoding a member of the SMC (structural maintenance of chromosome) protein family, are essential for A. fumigatus growth (17). However, it was also reported that the random insertional mutagenesis protocols currently used for A. fumigatus, which rely on integration of a heterologous DNA molecule into the fungal genome (4, 12), lead to frequent genomic rearrangements that hamper a high-throughput analysis (17, 48).
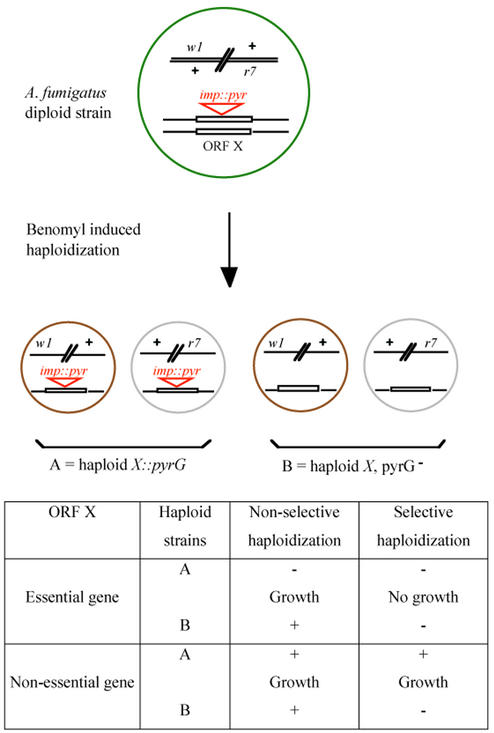
FIG. 1.
Strategy for identification of essential genes in A. fumigatus. A stable diploid strain heterozygous for spore color markers (w1 r7) is randomly mutagenized with the transposable element imp160::pyrG (imp::pyr). During haploidization on a benomyl containing medium, random loss of chromosomes gives rise to two subpopulations of colored haploid conidia (w1 or r7): one bearing the transposon-inactivated allele (population A) and one bearing the wild-type allele (population B). The ability to form haploid progenies on a nonselective haploidization medium and the inability to do so on a selective haploidization medium (without uridine and uracil) leads to identification of mutant strains with an insertion in an essential gene.
In order to perform genomewide identification of essential genes, we have developed an in vivo transposon mutagenesis system for A. fumigatus. Transposons are molecular tools widely used in vitro and/or in vivo for bacteria (30) and yeasts (31, 47), but only very recently have they been applied in the filamentous fungal kingdom (9, 24). In particular, impala160, a class II transposable element of the Tc1-mariner family (45), has been identified by transposon trapping in the phytopathogenic fungus Fusarium oxysporum (32) and has been shown to transpose efficiently in Fusarium species (28) as well as in Aspergillus nidulans (35) and Magnaporthe grisea (52). The results presented here show that impala160 is also functional in A. fumigatus and can be used to generate a collection of random heterozygous diploids. Screening of such a collection by parasexual genetics has resulted in the characterization, without prior sequence information, of A. fumigatus genes that are necessary for efficient fungal growth.
MATERIALS AND METHODS
A. fumigatus strain construction.
Media and growth conditions were as described previously (17). The A. fumigatus pyrG niaD haploid strain CEA113 is a chlorate-resistant derivative of strain CEA17 (11). The niaD mutation was confirmed by growth on minimal medium supplemented with different nitrogen sources (0.5 mM sodium glutamate, ammonium tartrate, sodium nitrate, sodium nitrite, or hypoxanthine) as previously described (7). Stable A. fumigatus diploids appropriate for transposon mutagenesis were obtained using the following procedure. Insertional mutagenesis of strain CEA17 has led to the isolation of spore color mutants CEA82 and CEA85 (17). The white strain CEA88 and the reddish strain CEA94 are chlorate-resistant derivatives of CEA82 and CEA85 with uncharacterized mutations in a gene involved in the biosynthesis of the molybdene cofactor (cnx) and the nitrate reductase gene (niaD), respectively. Strains CEA125 (w1 cnx1 pyrG1) and CEA129 (r7 niaD2 pyrG1) were obtained from strains CEA88 and CEA94 by growth on media containing 5-fluoroorotic acid (1 mg/ml), which selects for pyrG mutants. Simultaneous growth of CEA125 and CEA129 on minimal medium with nitrate as the sole nitrogen source yielded heterokaryons that produced grey-green spores similar to those of A. fumigatus haploid wild-type strains. This led to the isolation of the stable diploid strain CEA131 (w1/+ +/r7 cnx1/+ +/niaD2 pyrG1/pyrG1). A chlorate-resistant derivative of CEA131 that was unable to use nitrate as the sole nitrogen source and was defective at both niaD alleles was identified. This strain is referred to as CEA153 (w1/+ +/r7 cnx1/+ niaD4/niaD2 pyrG1/pyrG1). Spontaneous reversion of strain CEA153 was not observed on minimal medium containing nitrate as the sole nitrogen source.
Transformation and transposition.
pNIL160 has been described elsewhere (52). A 2.2-kb BamHI fragment from plasmid ppyrG containing the A. nidulans pyrG gene (41) was cloned at the NheI restriction site in impala160, yielding pNIpyr. NdeI-digested pNIpyr was introduced into haploid strain CEA113 and into diploid strain CEA153 by electroporation of intact conidia as described previously (4). Briefly, electroporation was carried out with 0.5 μg of linearized plasmid and 5 × 107 conidia in a 0.2-cm electroporation cuvette (Bio-Rad), and cells were subjected to a 1-kV pulse by using a Bio-Rad electroporation device (400 Ω, 25 μF). pyrG+ niaD− transformants CEA165 (haploid) and CEA225, CEA226, and CEA227 (diploid) were isolated for their high niaD+ reversion frequency and the integration of only one copy of pNIpyr into their genomes. Transposition of imp160::pyrG occurs after plating of serial dilutions of pyrG+ niaD− transformant conidia on minimal medium (without uridine and uracil) containing nitrate as the sole nitrogen source and supplemented with 0.02% Triton X-100. Haploid or diploid pyrG+ niaD+ revertants were isolated after 3 days at 37°C. Molecular analysis of A. fumigatus strains was performed by Southern blotting techniques after genomic DNA preparation as described previously (17).
Screening for essential genes by haploidization of somatic diploids.
Haploidization of A. fumigatus heterozygous strains was conducted on selective haploidization medium (rich medium containing 1.2 μg of benomyl [10 mg/ml in dimethyl sulfoxide; Aldrich]/ml) or on nonselective haploidization medium (selective haploidization medium plus uridine and uracil) for 5 days at 37°C (17). Haploid progenies are easily identified by the production of white and reddish sectors after haploidization of grey-green diploid strains.
Sequence determination.
Flanking sequence tags (FST) corresponding to genomic sequences bordering the 5′ end of imp160::pyrG were determined by adaptation of a two-step PCR strategy developed by Chun et al. (5). PCR conditions were as described previously (17), and semirandom primers were used in combination with 5′-end transposon-specific primers (Imp1 [ATGAAGGCGTAAGTTCCTTGC] and Imp2 [GTGTGGAGGAAGAAAGAGC]). Sequencing reactions of gel-purified PCR products were performed by ESGS (Evry, France) using primer Imp2. After removal of transposon sequences, FST were compared by BlastN analysis to the A. fumigatus genomic data from The Institute for Genomic Research (TIGR) (www.tigr.org/tdb/e2k1/afu1). The results presented in this study were obtained from the sequence release of 14 November 2001. At this time, the shotgun sequencing of A. fumigatus had progressed to 6× sequence coverage (28.7 Mb in 1,578 assemblies of more than 1,000 bp). Analyses of genomic sequences were carried out by Blast searches against public databases (National Center for Biotechnology Information [NCBI] nonredundant protein and expressed genomic tag sequence databases, available at www.ncbi.nlm.nih.gov/BLAST), and the annotation of S. cerevisiae genes given in Results was based on information from Stanford University (Saccharomyces Genome Database [SGD], available at http://genome-www.stanford.edu/Saccharomyces) and from the Munich Information Center for Protein Sequences (MIPS) (available at http://mips.gsf.de/proj/yeast/CYGD/db/index.html).
RESULTS
The transposable element impala is functional in A. fumigatus.
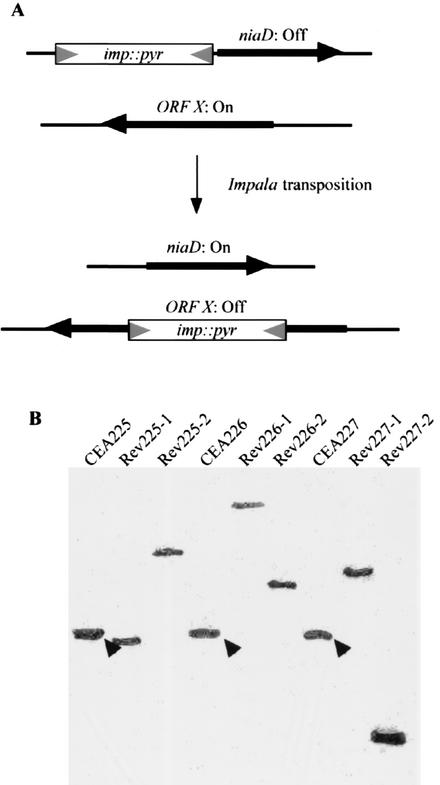
A prerequisite for the identification of A. fumigatus essential genes through a combination of random insertional mutagenesis and parasexual genetics is that the mutagenic molecules carry a selectable marker (e.g., the pyrG gene, encoding orotidine-5′-phosphate decarboxylase and required for uridine-uracil prototrophy), such that diploids can be distinguished for their ability to produce haploid progenies on selective and nonselective haploidization media (e.g., media that differ by the absence or presence of uridine and uracil, respectively) (Fig. 1). Therefore, we constructed plasmid pNIpyr by insertion of the A. nidulans pyrG gene between the 3′ end of the transposase-encoding gene and the 3′ inverted terminal repeat of impala160 in plasmid pNIL160 (52), which contains the A. nidulans niaD gene, encoding nitrate reductase, with a copy of impala160 inserted 10 bp upstream of the translation initiation codon of niaD (Fig. 2).
FIG. 2.
In vivo random transposon mutagenesis in A. fumigatus. (A) Schematic representation of imp160::pyrG transposition in an A. fumigatus strain transformed by pNIpyr. Expression of the nitrate reductase gene (niaD) is prevented by the presence of the transposable element imp160::pyG (imp::pyr) in the promoter region. Positive selection of transposition events is obtained by selection of nitrate-utilizing revertants, which appear as a result of the excision of imp::pyr and the restoration of a functional niaD promoter. Selection of imp::pyr reintegration events is ensured by the presence of pyrG in the transposable element when transposition events are induced in an A. fumigatus pyrG strain and in the absence of uridine and uracil. (B) Southern blot analysis of parental diploid pyrG+ niaD− transformants (CEA225, CEA226, and CEA227) and diploid pyrG+ niaD+ revertants (Rev225-1 and -2, Rev226-1 and -2, and Rev227-1 and -2). Hybridization with a probe for imp160::pyrG revealed integration of the transposable element into the promoter of the niaD gene in the three parental pyrG+ niaD− transfomants (arrowheads) and integration at apparently random sites in the genomes of the diploid pyrG+ niaD+ revertants.
Because of the insertion of imp160::pyrG into the niaD promoter, the niaD allele carried by pNIpyr is not functional (Fig. 2). Therefore, introduction of pNIpyr into the A. fumigatus haploid pyrG niaD strain CEA113 resulted in a haploid pyrG+ niaD− strain, referred to as CEA165. However, when CEA165 was grown on selective minimal medium with nitrate as the sole nitrogen source, pyrG+ niaD+ revertants were observed at frequencies of 10−4 to 10−5 (data not shown). Similar results were obtained when plasmid pNIpyr was introduced into A. fumigatus strain CEA153, a stable pyrG niaD diploid strain that is heterozygous for spore color markers. Three pyrG+ niaD− diploid transformants (namely, A. fumigatus strains CEA225, CEA226, and CEA227) gave pyrG+ niaD+ revertants on selective minimal medium with nitrate as the sole nitrogen source at frequencies of 10−5 to 10−6 (data not shown).
Southern blot analysis revealed that only one copy of imp160::pyrG was present in the genome of CEA165, CEA225, CEA226, and CEA227 and that all pyrG+ niaD+ revertants tested (n = 10) resulted from the transposition of imp160::pyrG from the A. nidulans niaD promoter to a single, apparently random site located elsewhere in the A. fumigatus genome (Fig. 2; also data not shown). Sequence analysis of the A. nidulans niaD promoter region in 10 haploid A. fumigatus pyrG+ niaD+ revertants revealed that an intact promoter had been restored except for a footprint of 5 bp (CAGTA [n = 8], TTGTA [n = 1], or CTGTA [n = 1]) which results from impala excision and does not significantly impair the transcription of the niaD gene. Characterization of the genome sequences flanking imp160::pyrG in pyrG+ niaD+ revertants by sequencing and comparison to public A. fumigatus genomic sequences (www.tigr.org/tdb/e2k1/afu1) showed that transposition of imp160::pyrG occurs at a genomic TA dinucleotide which is duplicated during the integration process, is apparently random without sequence preference (except for the TA), and is not associated with genomic rearrangements (Table 1; also data not shown). These three characteristics, as well as the 5-bp excision footprint, are marks of the transposition of members of the Tc1-mariner transposable element family (45) and were previously observed for impala160 transposition events in F. oxysporum (28), A. nidulans (35), and M. grisea (52).
TABLE 1.
Localization of imp160::pyrG in Aspergillus fumigatus heterozygous diploid strains with a haploid-lethal phenotype
| Strain | 5′ FST (bp) | Contiga
|
Localization of imp160::pyrG (bp) | |
|---|---|---|---|---|
| No. | Size (kb) | |||
| CEA228 | 97 | 960 | 5.1 | 223 |
| CEA229 | 435 | 221 | 76.1 | 60969 |
| CEA230 | 483 | 1754 | 6.3 | 603 |
| CEA231 | 114 | 131 | 27.2 | 15399 |
| CEA232 | 92 | 221 | 76.1 | 67069 |
| CEA233 | 447 | 164 | 45.3 | 40601 |
| CEA234 | 75 | 408 | 71.1 | 55903 |
| CEA254 | 199 | 43 | 93.1 | 72592 |
| CEA255 | 177 | 408 | 71.1 | 53190 |
| CEA256 | 411 | 110 | 207.8 | 105657 |
| CEA257 | 201 | 493 | 42.2 | 14886 |
| CEA258 | 558 | 190 | 22.1 | 7131 |
| CEA259 | 433 | 1327 | 4.7 | 2928 |
| CEA260 | 137 | 493 | 42.8 | 36145 |
| CEA261 | 830 | 93 | 53.7 | 11506 |
| CEA262 | 281 | 1366 | 3.6 | 1870 |
| CEA263 | 261 | 838 | 9.8 | 8261 |
| CEA264 | 119 | 846 | 16.7 | 3278 |
| CEA265 | 573 | 573 | 36.5 | 13244 |
| CEA266 | 423 | 585 | 39.5 | 24940 |
| CEA280 | 900 | 6 | 212.7 | 208370 |
| CEA281 | 306 | 792 | 13.7 | 1353 |
| CEA282 | 582 | 443 | 89.1 | 67662 |
| CEA283 | 339 | 716 | 14.3 | 9480 |
| CEA284 | 390 | 652 | 29.4 | 18411 |
| CEA285 | 222 | None | ||
Contigs obtained from http://www.tigr.org, release of 14 November 2001.
Screening for essential genes by haploidization of somatic diploids.
The three diploid transformants CEA225, CEA226, and CEA227 were used to generate a collection of 2,386 heterozygous A. fumigatus pyrG+ niaD+ revertants by direct selection on minimal medium lacking uridine and uracil. In this setting, the occurrence of revertants should result predominantly from excision and subsequent reintegration of imp160::pyrG. since transposition events associated with the loss of imp160::pyrG would result in uridine and uracil auxotrophs which cannot grow on minimal medium lacking uridine and uracil.
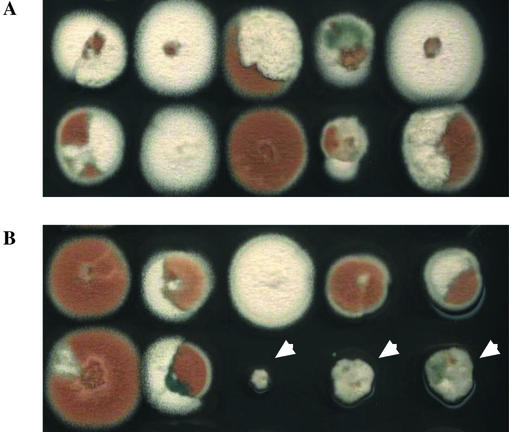
Haploidization of the 2,386 heterozygous diploid somatic strains was induced by the destabilizing reagent benomyl (6, 27). Strains CEA225, CEA226, and CEA227 and 97% of 2,386 pyrG+ niaD+ revertants showed no difference on selective and nonselective haploidization media in two independent tests, indicating that integration of pNIpyr into the genomes of the parental transformants and integration of imp160::pyrG in the majority of the pyrG+ niaD+ diploid revertants had not occurred in an essential locus (Fig. 3 and data not shown). In contrast, 73 mutants (3%) did not yield haploid conidia on selective haploidization medium, as indicated by the absence of colored sectors (Fig. 3 and data not shown).
FIG. 3.
Parasexual screening. Haploidization of 10 diploid pyrG+ niaD+ revertants on nonselective (A) and selective (B) media is shown. Random segregation of chromosomes is visualized by the production of differently colored haploid conidia. In the case of plasmid integration in an essential gene, residual growth is observed on selective haploidization medium (arrowheads). For these pyrG+ niaD+ revertants, haploid spores obtained on nonselective haploidization medium were tested for the absence of the transposable element in order to confirm the essential phenotype.
Diploid strains of A. fumigatus are hypersensitive to benomyl, and only haploid strains can grow at the benomyl concentration used (1.2 μg/ml) (27). However, transient formation of aneuploids during the haploidization of somatic diploids is often observed. These aneuploids display residual growth on selective haploidization medium and can to some extent overgrow haploid strains with morphological defects (17). In order to identify mutants which could have been retained in our screen because of the slow growth of haploid progenies rather than the lethality of the insertion, ca. 106 haploid progenies obtained by growth of the 73 mutants described above on nonselective haploidization medium were tested for the occurrence of imp160::pyrG by growth on selective medium. For 29 diploid revertants (29/2,386 = 1.2%), neither haploid nor aneuploid pyrG+ progeny could be obtained, suggesting that these diploids carry a copy of imp160::pyrG integrated into a chromosomal locus essential for A. fumigatus growth.
Characterization of A. fumigatus essential genes.
FST corresponding to genomic sequences at the 5′ end of imp160::pyrG were obtained for 26 of the 29 diploid strains mentioned above. Except for one strain (CEA285), corresponding genomic contigs were identified in the public preliminary sequence data for the A. fumigatus genome (Table 1). The genomic sequence was used to design specific primers that were used in standard PCRs to confirm the absence of genomic rearrangements after transposon integration and the occurrence of a wild-type chromosomal locus in each of the diploid revertants tested (data not shown). Similarity searches performed using the BLASTx algorithm identified three main categories of insertional mutants. The first category includes 15 strains with imp160::pyrG inserted into open reading frames (ORFs) with homologues in other fungal species (Table 2). The second category is composed of three strains with imp160::pyrG integrated into promoter regions. The third category includes seven strains with imp160::pyrG integrated into loci without homology to previously identified sequences in public databases.
TABLE 2.
S. cerevisiae homologues of A. fumigatus essential genes
| Strain | S. cerevisiae closest homologue at the imp160::pyrG integration locus | Protein length (aa) | Probability (e value) | Similarity | BDBHa | Essential in S. cerevisiae? | Functional category |
|---|---|---|---|---|---|---|---|
| CEA228 | Probable membrane protein (Yfl034wp) | 1,073 | 5e-42 | 55% on 286 aa | Nob | No | Unknown |
| CEA230 | Mitochondrial tryptophanyl-tRNA synthetase (Msw1p) | 379 | 2e-26 | 54% on 186 aa | Yes | No | Protein synthesis |
| CEA231 | Dead box protein 10 (Dbp10p) | 995 | e-166 | 55% on 949 aa | Yes | Yes | RNA processing |
| CEA232d | Ferrochelatase (Hem15p) | 393 | e-112 | 71% on 352 aa | Yes | Yes | Heme biosynthesis |
| CEA233 | Nuclear architecture-related protein (Nar1p) | 491 | 4e-59 | 48% on 465 aa | Yes | Yes | Nuclear architecture |
| CEA234d | Ribosomal protein [Rpl14(a/b)p] | 138 | 1e-24 | 63% on 130 aa | Yes | No | Protein synthesis |
| CEA254 | Guanylate kinase (Guk1p) | 187 | 2e-62 | 81% on 182 aa | Yes | Yes | Nucleotide metabolism |
| CEA255 | Signal recognition particle receptor alpha subunit (Srp101p) | 621 | e-102 | 62% on 431 aa | Yes | Yes | Protein transport |
| CEA256 | Oligosaccharyl transferase beta subunit (Wbp1p) | 430 | 2e-37 | 46% on 432 aa | Yes | Yes | Protein modification |
| CEA257 | Glutamate-tRNA synthetase (Ygl245wp) | 724 | 0.0 | 67% on 622 aa | Yes | Yes | Protein synthesis |
| CEA258 | Cell division control protein (Cdc27p) | 758 | 4e-71 | 63% on 304 aa | Yes | Yes | Cell cycle control |
| CEA259 | Remodels the structure of chromatin (Rsc9p) | 581 | 5e-29 | 45% on 414 aa | Yes | Yes | Chromatin structure |
| CEA260 | S-Adenosylmethionine decarboxylase (Spe2p) | 396 | 1e-54 | 51% on 462 aa | Yes | Yes | Metabolism |
| CEA261 | Ribosomal protein [Rpl17(a/b)p] | 136 | 5e-40 | 92% on 115 aa | Yes | No | Protein synthesis |
| CEA262 | Ribosomal protein [Rpl1(a/b)p] | 255 | 9e-86 | 86% on 238 aa | Yes | No | Protein synthesis |
| CEA263 | SNARE protein (Gos1p) | 223 | 8e-31 | 60% on 224 aa | Yes | No | Protein transport |
| CEA264 | Serine/threonine protein kinase (Rim11p) | 370 | e-104 | 77% on 323 aa | Yes/noc | No | Cell cycle control |
| CEA265d | Protoheme IX farnesyltransferase (Cox10p) | 462 | 1e-45 | 43% on 341 aa | Yes | No | Heme biosynthesis |
BDBH, Bi-Directional Best Hit; A. fumigatus closest homologue of the S. cerevisiae protein.
A. fumigatus orthologue is located on TIGR contig 428.
Several paralogues identified in the A. fumigatus genome.
Localization of imp160::pyrG in the promoter region of the gene (less than 250 bp of the putative translation initiation codon).
In 9 of the 15 strains of the first category, imp160::pyrG is integrated into ORFs with homology to genes previously demonstrated to be essential for S. cerevisiae growth (Table 2). These yeast genes encode proteins involved in a broad range of essential biological processes such as protein synthesis (YGL245W in CEA257), maturation (WBP1 in CEA256) and transport (SRP101 in CEA255), nuclear architecture (NAR1 in CEA233), RNA processing (DBP10 in CEA231), nucleotide metabolism (GUK1 in CEA254), chromatin structure (RSC9 in CEA259), and cell cycle control (CDC27 in CEA258). Interestingly, the gene interrupted by imp160::pyrG in strain CEA258 encodes an 809-amino-acid protein which is not only homologous to the S. cerevisiae Cdc27 protein but also shows 72% identity and 81% similarity to the A. nidulans BimA protein, a tetratricopeptide repeat motif containing protein which is essential for completion of mitosis, and hence growth, in A. nidulans (40, 43).
In six additional haploid-lethal strains, imp160::pyrG is localized into genes encoding homologues of nonessential S. cerevisiae proteins (Table 2). However, two of these insertions interrupt the A. fumigatus genes for ribosomal proteins Rpl1 (strain CEA262) and Rpl17 (strain CEA261), respectively, which are duplicated in the yeast genome and are not independently essential, although the double mutation is lethal. Another of these six insertions lies in the A. fumigatus homologue of MSW1 (strain CEA230), encoding the yeast tryptophanyl-tRNA synthetase that is localized to mitochondria. A null mutation in MSW1 leads to a slow-growth phenotype (“petite” phenotype) in yeast due to defects in mitochondrial protein synthesis and respiration (16, 20). That the A. fumigatus homologue of MSW1 is essential reflects the fact that, like most aspergilli, A. fumigatus is a strict aerobe that requires mitochondrial function for growth. In this regard, the search for chemically induced respiration-deficient mutants of A. nidulans has resulted only in the identification of conditional mutants, as would be expected if mitochondrial function is essential for growth in this species also (54). A fourth imp160::pyrG insertion is located in a homologue of the S. cerevisiae GOS1 gene (strain CEA263), which is dispensable for vegetative growth but required for ascospore germination at 37°C (39).
Two other genes with significant similarities to dispensable S. cerevisiae proteins, namely, Rim11 and Yfl034w, are essential for A. fumigatus growth (Table 2, strains CEA228 and CEA264). However, further characterization of the relationships between these A. fumigatus and S. cerevisiae proteins suggests that they are not orthologues, since the S. cerevisiae proteins have closer homologues encoded in the A. fumigatus genome than those identified through our insertional mutagenesis screen (Table 2, bidirectional best-hit analysis). Although the homology between these A. fumigatus and S. cerevisiae proteins is probably indicative of similar biochemical functions, their roles in the cell may differ significantly, as indicated by the difference in their contributions to fungal growth in these two species. In this regard, it should be noted that the A. fumigatus homologue of the S. cerevisiae protein Rim11 that we have identified is more closely related to the dispensable Schizosaccharomyces pombe kinase protein skp1, which might be involved in the control of septation and cytokinesis (46), and that genes involved in septation in A. nidulans are known to be essential for sustained growth (25).
The second class of mutants includes three pyrG+ niaD+ revertants, each with a transposon integration in the vicinity (<200 bp) of the deduced translation initiation codon of a gene which is likely to be essential for A. fumigatus growth, based on its homology to an S. cerevisiae gene: RPL14 (strain CEA234), encoding a duplicated ribosomal protein, and COX10 (strain CEA232) and HEM15 (strain CEA265), required for heme biosynthesis and respiration (Table 2). In these mutants, it is likely that imp160::pyrG integration prevents proper expression of these three genes. Yet the possibility that the haploid-lethal defect results from an additional effect on genes divergently transcribed from these intergenic regions cannot be excluded.
Insertions of imp160::pyrG in the latter seven diploid strains had occurred into an ORF of 2.9 kb (strain CEA229), an ORF of 1.6 kb (strain CEA266), or ORFs shorter than 500 bp without significant homology in public databases (five independent pyrG+ niaD+ revertants; strains CEA280 to CEA284 in Table 1). This result indicates that approximately 28% (7 of 25) of the essential loci identified through imp160::pyrG mutagenesis in A. fumigatus diploid strains are A. fumigatus specific. This is in the range of what has been observed in other species in which systematic searches for essential genes have been performed (15 to 20% in Haemophilus influenzae [1] and 38% in C. albicans [12]). However, while it is likely that the two ORFs inactivated by imp160::pyrG in strains CEA229 and CEA266 encode functions essential for A. fumigatus growth, the basis for the phenotype of the five remaining mutants remains to be investigated.
impala transposition characteristics.
The frequency of imp160::pyrG integrations in loci essential for A. fumigatus growth (1.2%) was lower than anticipated. Indeed, nearly 17% of the 6,200 S. cerevisiae genes are essential (20). Assuming a similar proportion of essential genes in S. cerevisiae and A. fumigatus and assuming that the A. fumigatus genome (32 Mb) encodes 10,000 genes, it can be estimated that 8 to 10% of the heterozygous diploids should have an integration of imp160::pyrG resulting in a haploid-lethal phenotype. In an attempt to explain the discrepancy between this estimate and the observed frequency of diploid strains with a haploid-lethal phenotype, 82 insertions of imp160::pyrG obtained in different backgrounds were investigated (Table 3). As mentioned above, integration of imp160::pyrG occurred systematically at a TA dinucleotide without any genomic sequence preference, as indicated from the comparison of the 40 nucleotides flanking the TA in these 82 insertions (data not shown). Insertion of the transposable element was also apparently not influenced by the chromosomal location, since the 82 insertions were distributed among 70 contigs ranging from 3.6 to 212.7 kb in size and representing 10% of the genome size (eight contigs were the sites of two integrations, and two contigs were the sites of three integrations) (Table 1 and data not shown).
TABLE 3.
Frequency of imp160::pyrG integration into coding and noncoding regions
| Genetic background | Phenotypea | No. of revertants analyzed | Localization of imp160::pyrG (%)
|
||
|---|---|---|---|---|---|
| Coding region | Promoter regionb | Other | |||
| Diploid | No growth | 25 | 68 | 12 | 20 |
| Altered growth | 14 | 21 | 43 | 36 | |
| Normal growth | 18 | 11 | 33 | 56 | |
| Haploid | Altered growth | 15 | 20 | 20 | 60 |
| Normal growth | 10 | 10 | 10 | 80 | |
For diploid revertants, growth after haploidization under selective conditions.
Defined as an insertion of imp160::pyrG localized less than 500 bp from a putative translation start codon.
Although these results were indicative of an absence of sequence preference for imp160::pyrG integration, the detailed characterization of the loci defined by the 82 insertions suggests that transposition of imp160::pyrG is not truly random. Indeed, analysis of 28 loci defined by insertions obtained in a diploid or haploid background and having no impact on the morphology or growth of A. fumigatus at the haploid stage revealed that a vast majority of these insertions lay in noncoding regions, with 7 located in intergenic regions and 18 located in regions where no ORF of a significant size (>500 bp) could be identified (Table 3). Among these 28 insertions, only 3 were located in ORFs; one of these ORFs encoded a homologue of the A. nidulans AmdA protein, which is not essential for A. nidulans growth (37), and two encoded proteins without homology in the databases (data not shown). Similarly, insertions that resulted in an altered growth phenotype at the haploid stage were mostly located in noncoding regions (20 of 29), although in this case a higher frequency of insertions in ORFs and promoter regions was observed (Table 3). Interestingly, an insertion was found located 237 bp upstream of the deduced initiation codon of a gene that encodes a homologue of an essential S. cerevisiae tRNA seryl transferase, suggesting that in this case the altered growth phenotype might result from an effect of the imp160::pyrG insertion on the transcription of this gene (data not shown). The data obtained from the characterization of imp160::pyrG insertions that result in a haploid-lethal phenotype (see the preceding section) contrast markedly with these observations, since in that group, a majority of insertions were located in ORFs (17 of 25). Taken together, these data suggest that imp160::pyrG has a tendency to insert preferentially into noncoding regions, generating phenotypically silent mutations, as already observed for other transposons (8). However, the proportion of integration of imp160::pyrG into an ORF is directly correlated to the stringency of the screen, as illustrated by our screening for essential genes by parasexual analysis, which enriches for insertions that lie predominantly in ORFs.
DISCUSSION
The goal of this study was to develop an efficient procedure for the identification of the entire set of essential genes in the most prevalent human filamentous fungal pathogen, A. fumigatus. Previously, it was demonstrated that the combination of insertional mutagenesis in somatic diploid strains and parasexual genetics can be used to determine whether a particular gene is essential for A. fumigatus or not, as illustrated by the essential nature of the A. fumigatus FKS1 gene, encoding the 1,3-β-d-glucan synthase catalytic subunit (17). However, due to the large number of genes encoded in the A. fumigatus genome (estimated at 10,000) and the relative poor efficiency of homologous recombination in this species, a random approach is necessary to define the set of essential genes. A random approach is also advantageous because no prior information on sequences or functions is necessary. Unfortunately, random insertional mutagenesis tools developed in A. fumigatus have so far been based on the integration of heterologous plasmid DNA into genomic DNA (3, 4), a process that results in significant genomic rearrangements that hamper a high-throughput analysis (17).
The functionality of the autonomous transposable element impala160 (32) in different filamentous fungi and its ability to sustain genetically engineered modifications (28, 35, 52) has led us to evaluate this heterologous in vivo transposition system in A. fumigatus. Our results demonstrate that an engineered impala160 element (imp160::pyrG) is active in both haploid and diploid strains of A. fumigatus and that the characteristics of the transposition process are identical to those observed after impala160 transposition in a Fusarium sp. (28), M. grisea (52), and A. nidulans (35). These results are in agreement with the proposed idea that transposition of members of the Tc1-mariner family of transposable element occurs through a conserved mechanism which is independent of host-specific factors (45).
Ideally, a random insertional mutagenesis tool should combine several characteristics: ease in the production of insertional mutants, absence of rearrangements associated with insertions, and random distribution of insertions along the genome. A major advantage of heterologous transposon mutagenesis compared to plasmid-based mutagenesis lies in the absence of DNA rearrangements, allowing rapid characterization of tagged loci. Moreover, the structure of the transposable element imp160::pyrG allows rapid generation of a large collection of insertional mutants from a single parental strain, since it can be positively selected for both excision (nitrate utilization) and integration (uridine-uracil prototrophy). However, our results show that, although integration of the imp160::pyrG element seems random at the nucleotide and chromosomal localization levels, this transposable element has a tendency to insert in noncoding regions, and therefore its mutagenesis potential is lower than expected. Li Destri Nicosia et al. (35) have used impala in A. nidulans to generate more than 104 niaD+ revertants. Upon visual screening of this collection, only two spore color mutants were identified. The nature of the spore color mutation in one of these mutants has been shown to be the consequence of the integration of the transposable element in the yB gene (35). The low frequency of visible mutations after impala transposition in A. nidulans was proposed to be due to the propensity of impala to land outside ORFs (35). We have also observed a low frequency of morphological mutations after transposition of imp160::pyrG in a haploid A. fumigatus strain (15 strains out of 106 pyrG+ niaD+ revertants). The characterization at the sequence level of the loci where imp160::pyrG is integrated in these strains, as well as in 42 additional pyrG+ niaD+ revertants which display altered or normal growth in the haploid state, confirms that imp160::pyrG inserts preferentially into noncoding regions. Despite this integration bias, impala was used efficiently in M. grisea to identify pathogenicity genes (52). In the sample analyzed, the frequency of nonpathogenic mutants obtained by transposition of impala160 was similar to that obtained by plasmid-based transformation. Moreover, impala integrations were observed in ORFs, and a linkage between the transposon insertion and the loss of pathogenicity was observed in most instances, which is not the case when nonpathogenic mutants of M. grisea obtained by DNA-mediated transformation are analyzed (52). Our results obtained using diploid strains of A. fumigatus confirm the usefulness of impala for generating tagged mutations predominantly in genomic coding sequences after a highly selective screen.
The analysis of 2,386 A. fumigatus diploid insertional mutants enabled us to identify 20 previously uncharacterized essential genes, 6 of which could not have been predicted as essential for A. fumigatus growth based on studies in other fungal species. The correlation between the growth characteristics of each mutant and the nature of the mutated genes supports the idea that genes genuinely essential for A. fumigatus growth can be identified among diploid heterozygous mutants. Furthermore, the identification of essential genes involved in very diverse cellular functions, from protein synthesis to cell cycle control, suggests that the screen that we have performed is not biased for a certain type of cellular function and that scaling it up should result in a wide compendium of A. fumigatus essential genes. That some of the homologues of nonessential yeast genes can be essential in A. fumigatus might be explained by their implication in additional essential pathways and/or by differences in the importance of similar biological pathways to the biology of A. fumigatus and other lower eukaryotes. For instance, we have identified several genes encoding components of the mitochondria which are essential in aspergilli, which are strict aerobes, but not in S. cerevisiae. The concordance of gene essentiality in different microorganisms is limited, and this fact is illustrated by a recent report by Elitra Pharmaceuticals (San Diego, Calif.), which has used a genomewide approach to identify essential genes conserved both in S. cerevisiae and C. albicans (26). Although the detailed results are not available, the investigators report that about half of the C. albicans genes that are orthologues of S. cerevisiae essential genes are not essential for C. albicans growth (26). This limited concordance between microorganisms highlights the need to work directly with relevant pathogenic species and is in favor for the systematic search of essential genes on a random basis. We have now under way a large-scale genomic approach for the identification of essential A. fumigatus genes by automation of the strategy using impala transposition and parasexual genetics. This represents an important step toward a better understanding of the biology of filamentous fungi and the identification of potential targets for novel antifungal agents. The definition of essential genes will be a functional complement to the A. fumigatus genomic sequence (14).
Acknowledgments
We thank Tony Pugsley, Marc-Henri Lebrun, Marie-Claire Grosjean-Cournoyer, Philipp Knechtle, and Maxime Schwartz for critical discussion of the work and reading of the manuscript. Preliminary sequence data were obtained from the TIGR website at http://www.tigr.org.
Sequencing of A. fumigatus was funded by the National Institute of Allergy and Infectious Disease, grant U01 AI 48830, to David Denning and William Nierman. This work was supported by a Ph.D. fellowship to A. Firon from the Association Nationale de la Recherche Technique (CIFRE-Ministère de la Recherche et de la Technologie) and by grants from the Institut Pasteur and Bayer CropScience to C. d'Enfert.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman, J. C., P. S. Hicks, M. B. Kurtz, H. Rosen, D. M. Schmatz, P. A. Liberator, and C. M. Douglas. 2002. The antifungal echinocandin caspofungin acetate kills growing cells of Aspergillus fumigatus in vitro. Antimicrob. Agents Chemother. 46:3001-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brookman, J. L., and D. W. Denning. 2000. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 3:468-474. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. S., A. Aufauvre-Brown, and D. W. Holden. 1998. Insertional mutagenesis of Aspergillus fumigatus. Mol. Gen. Genet. 259:327-335. [DOI] [PubMed] [Google Scholar]
- 5.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 6.Clutterbuck, A. J. 1992. Sexual and parasexual genetics of Aspergillus species. Bio/Technology 23:3-18. [PubMed] [Google Scholar]
- 7.Cove, D. J. 1976. Chlorate toxicity in Aspergillus nidulans. Studies of mutants altered in nitrate assimilation. Mol. Gen. Genet. 146:147-159. [DOI] [PubMed] [Google Scholar]
- 8.Craig, N. L. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437-474. [DOI] [PubMed] [Google Scholar]
- 9.Daboussi, M. J. 1996. Fungal transposable elements: generators of diversity and genetic tools. J. Genet. 75:325-339. [Google Scholar]
- 10.De Backer, M. D., B. Nelissen, M. Logghe, J. Viaene, I. Loonen, S. Vandoninck, R. de Hoogt, S. Dewaele, F. A. Simons, P. Verhasselt, G. Vanhoof, R. Contreras, and W. H. Luyten. 2001. An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans. Nat. Biotechnol. 19:235-241. [DOI] [PubMed] [Google Scholar]
- 11.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 12.d'Enfert, C., G. Weidner, P. C. Mol, and A. A. Brakhage. 1999. Transformation systems of Aspergillus fumigatus. New tools to investigate fungal virulence. Contrib. Microbiol. 2:149-166. [DOI] [PubMed] [Google Scholar]
- 13.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 14.Denning, D. W., M. J. Anderson, G. Turner, J. P. Latge, and J. W. Bennett. 2002. Sequencing the Aspergillus fumigatus genome. Lancet Infect. Dis. 2:251-253. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, M. 2002. Invasive fungal infections: evolving challenges for diagnosis and therapeutics. Mol. Immunol. 38:947-957. [DOI] [PubMed] [Google Scholar]
- 16.Entrup, R., W. Langgut, T. Lisowsky, and E. Schweizer. 1992. A yeast nuclear mutation conferring temperature-sensitivity to the mitochondrial tryptophanyl-tRNA synthetase. Curr. Genet. 21:281-283. [DOI] [PubMed] [Google Scholar]
- 17.Firon, A., A. Beauvais, J. P. Latgé, E. Couvé, M. C. Grosjean-Cournoyer, and C. d'Enfert. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus. Impact of genomic rearrangements associated with electroporation of DNA. Genetics 161:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firon, A., and C. d'Enfert. 2002. Identifying essential genes in fungal pathogens of humans. Trends Microbiol. 10:456-462. [DOI] [PubMed] [Google Scholar]
- 19.Georgopapadakou, N. H. 1998. Antifungals: mechanism of action and resistance, established and novel drugs. Curr. Opin. Microbiol. 1:547-557. [DOI] [PubMed] [Google Scholar]
- 20.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 21.Groll, A. H., A. J. De Lucca, and T. J. Walsh. 1998. Emerging targets for the development of novel antifungal therapeutics. Trends Microbiol. 6:117-124. [DOI] [PubMed] [Google Scholar]
- 22.Groll, A. H., S. C. Piscitelli, and T. J. Walsh. 1998. Clinical pharmacology of systemic antifungal agents: a comprehensive review of agents in clinical use, current investigational compounds, and putative targets for antifungal drug development. Adv. Pharmacol. 44:343-500. [DOI] [PubMed] [Google Scholar]
- 23.Groll, A. H., and T. J. Walsh. 2001. Caspofungin: pharmacology, safety and therapeutic potential in superficial and invasive fungal infections. Expert Opin. Investig. Drugs 10:1545-1558. [DOI] [PubMed] [Google Scholar]
- 24.Hamer, L., K. Adachi, M. V. Montenegro-Chamorro, M. M. Tanzer, S. K. Mahanty, C. Lo, R. W. Tarpey, A. R. Skalchunes, R. W. Heiniger, S. A. Frank, B. A. Darveaux, D. J. Lampe, T. M. Slater, L. Ramamurthy, T. M. DeZwaan, G. H. Nelson, J. R. Shuster, J. Woessner, and J. E. Hamer. 2001. Gene discovery and gene function assignment in filamentous fungi. Proc. Natl. Acad. Sci. USA 98:5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736-739. [DOI] [PubMed] [Google Scholar]
- 26.Haselbeck, R., D. Wall, B. Jiang, T. Ketela, J. Zyskind, H. Bussey, J. G. Foulkes, and T. Roemer. 2002. Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Curr. Pharm. Des. 8:1155-1172. [DOI] [PubMed] [Google Scholar]
- 27.Hastie, A. C. 1970. Benlate-induced instability of Aspergillus diploids. Nature 226:771.. [DOI] [PubMed] [Google Scholar]
- 28.Hua-Van, A., J. A. Pamphile, T. Langin, and M. J. Daboussi. 2001. Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264:724-731. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, B., H. Bussey, and T. Roemer. 2002. Novel strategies in antifungal lead discovery. Curr. Opin. Microbiol. 5:466-471. [DOI] [PubMed] [Google Scholar]
- 30.Judson, N., and J. J. Mekalanos. 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8:521-526. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., and M. Snyder. 2001. Emerging technologies in yeast genomics. Nat. Rev. Genet. 2:302-312. [DOI] [PubMed] [Google Scholar]
- 32.Langin, T., P. Capy, and M. J. Daboussi. 1995. The transposable element impala, a fungal member of the Tc1-mariner superfamily. Mol. Gen. Genet. 246:19-28. [DOI] [PubMed] [Google Scholar]
- 33.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latgé, J. P. 2001. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9:382-389. [DOI] [PubMed] [Google Scholar]
- 35.Li Destri Nicosia, M. G., C. Brocard-Masson, S. Demais, A. Hua Van, M. J. Daboussi, and C. Scazzocchio. 2001. Heterologous transposition in Aspergillus nidulans. Mol. Microbiol. 39:1330-1344. [PubMed] [Google Scholar]
- 36.Lin, S., J. Schranz, and S. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 37.Lints, R., M. A. Davis, and M. J. Hynes. 1995. The positively acting amdA gene of Aspergillus nidulans encodes a protein with two C2H2 zinc-finger motifs. Mol. Microbiol. 15:965-975. [DOI] [PubMed] [Google Scholar]
- 38.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33:641-647. [DOI] [PubMed] [Google Scholar]
- 39.McNew, J. A., J. G. Coe, M. Sogaard, B. V. Zemelman, C. Wimmer, W. Hong, and T. H. Sollner. 1998. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 435:89-95. [DOI] [PubMed] [Google Scholar]
- 40.Mirabito, P. M., and N. R. Morris. 1993. BIMA, a TPR-containing protein required for mitosis, localizes to the spindle pole body in Aspergillus nidulans. J. Cell Biol. 120:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oakley, B. R., J. E. Rinehart, B. L. Mitchell, C. E. Oakley, C. Carmona, G. L. Gray, and G. S. May. 1987. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene 61:385-399. [DOI] [PubMed] [Google Scholar]
- 42.Odds, F. C., N. A. Gow, and A. J. Brown. 2001. Fungal virulence studies come of age. Genome Biol. 2:1009.1-1009.4. [DOI] [PMC free article] [PubMed]
- 43.O'Donnell, K. L., A. H. Osmani, S. A. Osmani, and N. R. Morris. 1991. bimA encodes a member of the tetratricopeptide repeat family of proteins and is required for the completion of mitosis in Aspergillus nidulans. J. Cell Sci. 99:711-719. [DOI] [PubMed] [Google Scholar]
- 44.Perfect, J. R. 1996. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob. Agents Chemother. 40:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plasterk, R. H., Z. Izsvak, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326-332. [DOI] [PubMed] [Google Scholar]
- 46.Plyte, S. E., A. Feoktistova, J. D. Burke, J. R. Woodgett, and K. L. Gould. 1996. Schizosaccharomyces pombe skp1+ encodes a protein kinase related to mammalian glycogen synthase kinase 3 and complements a cdc14 cytokinesis mutant. Mol. Cell. Biol. 16:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 48.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 50.Timberlake, W. E. 1991. Cloning and analysis of fungal genes, p. 51-85. In J. W. Bennett and L. L. Lasure (ed.), More gene manipulations in fungi. Academic Press Inc., Oxford, England.
- 51.van Burik, J. A., and P. T. Magee. 2001. Aspects of fungal pathogenesis in humans. Annu. Rev. Microbiol. 55:743-772. [DOI] [PubMed] [Google Scholar]
- 52.Villalba, F., M. H. Lebrun, A. Hua-Van, M. J. Daboussi, and M. C. Grosjean-Cournoyer. 2001. Transposon impala, a novel tool for gene tagging in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 14:308-315. [DOI] [PubMed] [Google Scholar]
- 53.Walsh, T. J., and A. H. Groll. 1999. Emerging fungal pathogens: evolving challenges to immunocompromised patients for the twenty-first century. Transplant. Infect. Dis. 1:247-261. [DOI] [PubMed] [Google Scholar]
- 54.Waring, R. B., and C. Scazzocchio. 1980. Nuclear and mitochondrial suppression of a mitochondrially inherited cold-sensitive mutation in Aspergillus nidulans. J. Gen. Microbiol. 119:297-311. [DOI] [PubMed] [Google Scholar]