Abstract
To estimate patterns of molecular evolution of unconstrained DNA sequences, we used maximum parsimony to separate phylogenetic trees of a non-long terminal repeat retrotransposable element into either internal branches, representing mainly the constrained evolution of active lineages, or into terminal branches, representing mainly nonfunctional “dead-on-arrival” copies that are unconstrained by selection and evolve as pseudogenes. The pattern of nucleotide substitutions in unconstrained sequences is expected to be congruent with the pattern of point mutation. We examined the retrotransposon Helena in the Drosophila virilis species group (subgenus Drosophila) and the Drosophila melanogaster species subgroup (subgenus Sophophora). The patterns of point mutation are indistinguishable, suggesting considerable stability over evolutionary time (40–60 million years). The relative frequencies of different point mutations are unequal, but the “transition bias” results largely from an ≈2-fold excess of G⋅C to A⋅T substitutions. Spontaneous mutation is biased toward A⋅T base pairs, with an expected mutational equilibrium of ≈65% A + T (quite similar to that of long introns). These data also enable the first detailed comparison of patterns of point mutations in Drosophila and mammals. Although the patterns are different, all of the statistical significance comes from a much greater rate of G⋅C to A⋅T substitution in mammals, probably because of methylated cytosine “hotspots.” When the G⋅C to A⋅T substitutions are discounted, the remaining differences are considerably reduced and not statistically significant.
Modern evolutionary theory treats mutations as random with regard to the adaptive needs of the organism, but the theory makes no stipulation of randomness in other respects. Biased patterns of nucleotide substitution are one source of nonrandomness in the production of heritable variation. Although much attention has been devoted to estimating mutational patterns, such estimates have proven elusive. In multicellular organisms spontaneous mutation is generally extremely rare (on the order of 10−9 mutations per base pair per round of replication), and many mutations have negligible effects on phenotype. Direct investigation of spontaneous mutation is therefore very difficult and prone to experimental bias. A potentially more powerful approach is to infer mutational patterns from comparisons of homologous genes within and between different species. Unfortunately, such inferences are often compromised because patterns of DNA variation in functional genes are determined not only by mutation but also, often decisively, through the action of natural selection.
To avoid confounding mutation and natural selection, it is necessary to identify unconstrained, neutrally evolving DNA sequences in the genome. One approach makes use of pseudogenes (1); this method has been successfully applied to the study of mutational patterns in mammals, in which pseudogenes are both common and well studied (2–4). The use of pseudogenes creates a rather paradoxical situation in which there are more reliable estimates of spontaneous mutation patterns in mammals than there are in such genetically well known model organisms as Drosophila melanogaster and Caenorhabditis elegans, since these genomes are almost devoid of pseudogenes (5, 6).
Although the genomes of many organisms lack large numbers of pseudogenes, they usually do contain other kinds of unconstrained sequences. In particular, eukaryotic genomes often contain non-long term repeat (LTR) retrotransposable elements (also called LINE elements), most copies of which are “dead-on-arrival” (DOA) elements, truncated at the 5′ end and nonfunctional. Being nonfunctional, DOA elements are predicted to evolve essentially as pseudogenes. Unlike pseudogenes, however, non-LTR elements are both ubiquitous (with a few exceptions, such as Saccharomyces cerevisiae) and are readily identified through a number of highly conserved signature motifs in their sequences (7–9).
To study the unconstrained evolution of DOA copies of non-LTR elements, it is necessary to distinguish nucleotide substitutions that occur in transpositionally active lineages that give birth to DOA elements from those substitutions that occur in each individual DOA copy after its creation. We have demonstrated that this distinction can be accomplished through sampling a large number of independently transposed elements and separating the substitutions into those that are shared between two or more elements and those that are found in single elements only (10, 11). The reasoning is based on the fact that active non-LTR elements can generate new copies of themselves through transposition, and therefore substitutions in active lineages have a chance of being incorporated in multiple independent insertions. On the other hand, most DOA copies cannot transpose, and therefore (except for parallel mutations) a substitution that occurs in a DOA element will appear only in a single copy.
In our previous studies we applied the non-LTR approach to the study of length mutations—deletions and insertions— and discovered that the profile of length mutations varies dramatically among species. In particular, deletions in Drosophila are about three times more frequent and eight times longer than those in mammals, leading to an approximate 24-fold increase in the rate of spontaneous DNA loss (10, 12, 13). In the present study, we direct our attention to the study of relative frequencies of different kinds of point mutations (simple nucleotide substitutions). We demonstrate that, in contrast to length mutations, the pattern of point substitutions is quite similar in Drosophila and mammals, provided that the methylated C⋅G mutational hotspots in mammalian DNA are discounted.
MATERIALS AND METHODS
Phylogenetic Analysis.
We analyzed two sets of data consisting of DNA sequences of multiple copies of the non-LTR retrotransposable element Helena (14). The first data are a region of 363 bp from each of 18 Helena copies isolated from eight species in the Drosophila virilis species group (10). The second data are a region of 1,357 bp from each of 22 Helena copies from seven species in the D. melanogaster species subgroup (13). The sequenced regions are both in the ORF for reverse transcriptase, and are partially overlapping. The sequences were initially aligned with the aid of sequencher 2.0 (Gene Codes, Ann Arbor, MI) and then adjusted, by using as a guide the relatively highly conserved amino acid sequence of reverse transcriptase. Phylogenetic analysis of the sequences was carried out by maximum parsimony as implemented in the paup software package (15), with all characters in the nucleotide alignment assigned equal weight. Deletions were treated as missing data. Analysis and manipulation of the Helena gene trees was aided by the macclade software package (16).
Statistical Methods.
Each nucleotide A, T, G, or C can be substituted in three distinct ways, yielding a matrix of 12 different substitutions. In our data (see Results), complementary mutations are found in equal frequency; for example, the number of instances of A changing to G is not significantly different from the number of instances of T changing to C. Hence the complementary substitutions were added together to yield counts for nucleotide-pair substitutions, for example, A⋅T changing to G⋅C. The pooling of the complementary changes yields a set of six possible nucleotide-pair substitutions. There are five degrees of freedom in the comparisons of relative frequencies of point substitution, which were used to support three maximum likelihood tests. The first test (using two degrees of freedom) was whether the relative frequencies of A⋅T → G⋅C, A⋅T → C⋅G, and A⋅T → T⋅A are different between D. melanogaster and D. virilis or between Drosophila and mammals. The second test (also using two degrees of freedom) was whether the relative frequencies of G⋅C → A⋅T, G⋅C → T⋅A, and G⋅C → C⋅G are different between D. melanogaster and D. virilis or between Drosophila and mammals. And the third test (using the remaining degree of freedom) was whether the relative substitution rate of A⋅T base pairs differs from that of G⋅C base pairs between D. melanogaster and D. virilis or between Drosophila and mammals. The log-likelihood ratios of the three tests were summed and used in an omnibus χ2 test with five degrees of freedom.
RESULTS
Pseudogene Evolution of Helena in Drosophila.
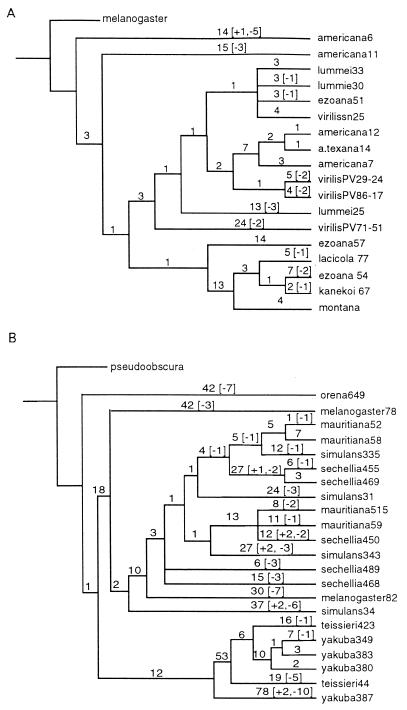
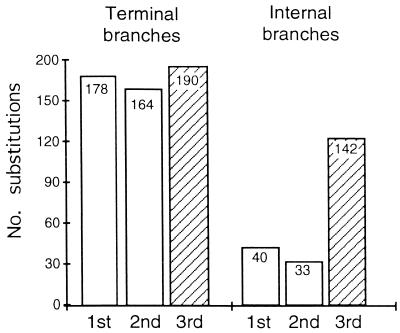
We examined two partially overlapping regions of the Helena ORF for reverse transcriptase in 18 copies from the D. virilis species group and 22 copies from the D. melanogaster species subgroup (10, 13). Maximum parsimony estimates the evolutionary history of the sampled Helena insertions (Fig. 1). The method separates all substitutions into shared substitutions mapping to internal branches of the gene tree (mostly representing the evolution of active Helena lineages) and substitutions that appear in a single sequence that map to a terminal branch (mostly representing the pseudogene-like evolution of nonfunctional DOA copies). Both sets of data admit of multiple maximum parsimony trees that are equally good, but alternative trees differ almost exclusively in placement of the long branches, not in swapping internal branches with terminal branches. As shown in Fig. 2 and refs 10 and 13, the prediction of constrained evolution in the internal branches (active lineages) and unconstrained evolution in the terminal branches (DOA copies) is confirmed by the observation that evidence of purifying selection is limited to the internal branches of the trees. Substitutions in the internal branches are found mostly in synonymous third positions of codons, whereas substitutions in the terminal branches are distributed equally among the synonymous and nonsynonymous codon positions. Moreover, types of mutation that close or shift the reverse transcriptase ORF (e.g., deletions, insertions, and stop codons) are restricted almost exclusively to terminal branches. The only exception was observed in the D. melanogaster dataset (Fig. 1B), where deletions, insertions, and equal frequencies of mutations in all three codon positions are present on the internal branches connecting five of the Helena sequences (mauritiana52, mauritiana58, sechellia455, sechellia469, simulans335) sampled from three closely related species (Drosophila simulans, Drosophila sechellia, and Drosophila mauritiana). The most likely explanation of this finding is that these sequences correspond to a DOA element that transposed in the ancestor of these species and was then propagated vertically through speciation (13). The internal branches connecting these five sequences would therefore correspond to the pseudogene-like evolution of a single DOA element and accordingly show no signs of purifying selection; this prediction is consistent with the observed pattern.
Figure 1.
Phylogenetic analysis of the Helena sequences (A), the D. virilis species group, and (B), the D. melanogaster species subgroup. The number of unambiguous nucleotide substitutions is shown above each branch. Numbers in square brackets refer to the numbers of indels (insertions or deletions) mapping to each branch, with plus signs standing for insertions and minus signs for deletions.
Figure 2.
The codon distribution of internal-branch and terminal-branch substitutions in Helena in the pooled data from the D. melanogaster species subgroup and the D. virilis species group. The distribution is uniform for terminal substitutions (P = 0.38) but strongly skewed toward the mostly synonymous third codon positions for the internal substitutions (P = 2.8 × 10−23). Although the overall pattern of terminal substitutions is not different from random, there is a slight excess of third position substitutions, indicating a possible minor effect of purifying selection along the terminal branches. Note, however, that the excess accounts for less than 4% of all substitutions in the dataset.
Pseudogene Substitutions in Helena.
Having separated the evolution of active lineages of Helena (most of the internal branches) from the evolution of individual DOA, pseudogene-like copies (terminal branches and internal branches connecting the mauritiana52, mauritiana58, sechellia455, sechellia469, simulans335 sequences), we compiled a set of “pseudogene” substitutions by combining only the substitutions along branches corresponding to the evolution of DOA elements. These substitutions should reflect the relative rates of different kinds of nucleotide substitution, unbiased by natural selection of the active Helena lineages.
An additional potential complication in the analysis comes from the possibility of multiple independent mutations at the same site, known as the problem of multiple hits. This problem is significant only when the proportion of sites substituted in a sequence is high, more than about 10%. This is because, when the probability of substitution per site is P1 = 0.1, the probability of two independent substitutions at same site equals (P1)2 = 0.01, hence when the proportion of substituted sites is less than 10%, the correction for multiple hits is less than 1%. In the set of Helena pseudogene substitutions, no correction for multiple hits is necessary because the proportion of substituted sites along individual pseudogene branches is very low, averaging 1.65% with a range from 0.03% to 6.6%. In spite of the low proportion of substitutions in each pseudogene branch, the total number of substitutions is quite large (585 total substitutions), which assures good statistical power.
Complementary Substitutions Are Equally Frequent.
Statistical analysis indicates that, in the Helena datasets, complementary nucleotide substitutions are equally frequent; for example, the number of A → G substitutions is within sampling error of the number of T → C substitutions. For the D. virilis species group, a G test yields P = 0.78, and for the D. melanogaster species subgroup a G test yields P = 0.87. The equality of complementary substitutions in the Helena data justifies combining the complementary classes into six types of nucleotide-pair substitutions, hereafter called “point mutations.”
The Profile of Point Mutations in Drosophila.
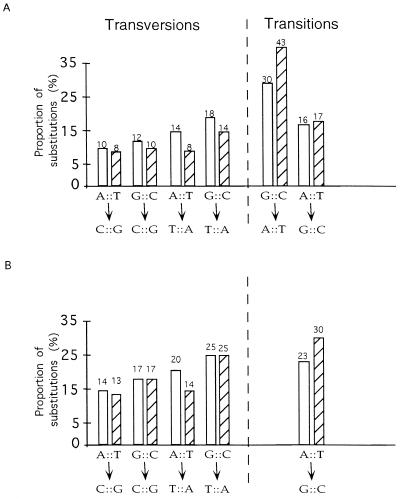
The relative frequencies of the six types of point mutation do not differ between the Helena data collected from the D. virilis species group and those collected from the D. melanogaster species subgroup (P = 0.21 in the maximum likelihood analysis). We have therefore combined the two sets of pseudogene substitution. The patterns of point mutation, normalized for the different frequencies of nucleotides in Helena sequences, are shown in Fig. 3. There are several noteworthy features of this pattern. First, the frequencies of different types of mutation are clearly not equal (G test, P = 6 × 10−14). In particular, the transition from a G⋅C pair to a A⋅T pair is by far the most prevalent mutation, averaging a 2.2-fold higher frequency than any other mutation. Note that the reverse mutation from A⋅T to G⋅C is much less frequent than that from G⋅C to A⋅T.
Figure 3.
The relative rates of nucleotide pair substitutions. The Drosophila pattern is shown with open bars and the mammalian pattern, with hatched bars. The rates were normalized to take into account the unequal numbers of different nucleotides in the Drosophila (Helena) and the mammalian pseudogene sequences. Estimation of the mammalian pattern was done by using the dataset of 572 substitutions in 15 mammalian pseudogenes compiled in refs. 3 and 4, whereas the Drosophila dataset consists of 585 substitutions in 40 independent insertions of Helena in 15 different Drosophila species. (A) The complete pattern of relative rates. (B) The relative rates of all point mutations excluding G⋅C → A⋅T transition.
A second noteworthy feature of the mutation pattern is that it is biased toward A and T nucleotides, as compared with G and C nucleotides. Assuming that the pattern of point mutation is stable through time, we can estimate that the expected base composition of unconstrained Drosophila DNA sequences, at equilibrium, would be 64.9% A + T, with a 95% confidence interval from 60% A + T to 69% A + T.
The Estimate of the Mutational Profile Is Robust.
The set of point substitutions used in our analysis was derived from the maximum parsimony Helena gene trees and the subsequent mapping of characters onto these trees. It is important to evaluate how sensitive our estimates may be to any particular phylogenetic reconstruction and the pattern of inferred character change. We have addressed the question of robustness in two different ways. First we used two alternative methods of phylogenetic reconstruction—maximum likelihood and neighbor joining—to estimate Helena trees. Although the trees differ in their details, the pattern of point substitutions remains practically unaffected. In particular, the mutational profiles derived from both the maximum likelihood and the neighbor-joining trees are not statistically different from the one derived from the maximum parsimony trees (P = 0.98 in both cases). We also estimated the profile of point substitutions in a manner completely independent of phylogeny by analyzing only unique nucleotide substitutions. By a “unique substitution” we mean a difference that occurs in a single sequence at a nucleotide site at which the remaining sequences are identical to each other. The relative frequencies of the unique substitutions are also not statistically different from the pattern of point substitutions in the “pseudogene” branches of the maximum parsimony tree (P = 0.69).
DISCUSSION
Comments on Complementarity.
The Helena data indicate that complementary nucleotide substitutions take place at approximately equal relative rates, for example A → G occurs about as often as T → C. Equality in complementary nucleotide substitutions would not necessarily be expected. For example, there could be significant differences in the substitution pattern in the leading strand vs. the lagging strand in DNA replication, or in the transcribed vs. the nontranscribed strand in transcription (17). On the other hand, such equality is expected in the case of the DOA Helena elements, because most of the DOA copies are expected to be nontranscribed (except possibly for readthrough transcription from nearby promoters), and most DOA copies are expected to be oriented at random relative to the direction of movement of the nearest replication fork. Therefore, while our data are relevant to overall patterns of base pair substitutions, the approach is not sensitive to differences in substitution pattern that may be correlated with strand identity in transcription or replication. On the contrary, our method supplies the “baseline data” against which strand-specific mutational patterns may be compared.
Relative Evolutionary Stability of Mutation Patterns.
The D. virilis species group and the D. melanogaster species subgroup represent both parts of the two great subgenera in the Drosophila radiation. The D. melanogaster species subgroup is in the subgenus Sophophora, whereas the D. virilis species group is in the subgenus Drosophila. These subgenera are estimated to have diverged 40–60 million years ago (18), yet their patterns of point mutation are indistinguishable as judged from the relative frequencies of the six types of point mutation in the two Helena datasets. This finding is reassuring, because it indicates a certain stability in the point mutation pattern through time.
Biases in Drosophila Mutation.
The frequencies of different nucleotide substitutions are far from equal. The greatest bias is the high frequency of transition from G⋅C → A⋅T, which is on average 2.2 times more frequent than any other substitution. The other possible transition, A⋅T → G⋅C, is much less frequent and is, in fact, less frequent than some of the transversions. The “transition bias” in Drosophila therefore is a bias only toward the G⋅C → A⋅T transition.
Assuming homogeneity in the mutation pattern through time, the expected equilibrium A + T content of unconstrained Drosophila DNA sequences is 64.9% A + T. This value is close to the 60% A + T to 65.5% A + T content actually observed in introns in D. melanogaster (19–21). The good agreement supports the view that mutational pressure is primarily responsible for the elevated A + T content of Drosophila introns, and that most sites in introns are indeed unconstrained by selection (or only very weakly selected). On the other hand, our data also confirm the view that mutation alone cannot explain codon usage bias in Drosophila genes (22), since most of the overrepresented codons in Drosophila end in G or C (20, 23), whereas mutation bias should favor codons ending in A or T.
Patterns of Point Mutation in Drosophila vs. Mammals.
The present analysis of spontaneous point mutation in Drosophila affords the first opportunity to compare mutational patterns in Drosophila with those observed in mammals (3, 4). The mutational patterns are shown in Fig. 3. They are quite similar despite the great evolutionary distance separating these animals, although the patterns are by no means identical (P = 1 × 10−4 in a maximum likelihood test). The primary cause of the difference is the much higher relative rate of G⋅C → A⋅T transition in mammals. When G⋅C → A⋅T transitions are excluded from the comparison, the differences remaining between the Drosophila and mammalian patterns are much reduced and, in fact, are not statistically significant (P = 0.11 in a maximum likelihood test). The basis of the G⋅C → A⋅T difference may relate to cytosine methylation. Cytosines are often methylated in mammalian DNA, which sharply increases the probability of C to T transitions through deamination of 5-methyl cytosine (24). Because Drosophila DNA lacks methylation (25), this difference alone could account for much of the discordance in the patterns of point mutation.
Discounting the difference in G⋅C to A⋅T transitions, the otherwise similar patterns of base substitution between Drosophila and mammals is unexpected. Why should they be so similar? One possibility is that the similarity in patterns of point mutation is a sheer coincidence, which implies that other metazoan organisms will be found to exhibit a great diversity of different patterns. On the other hand, if there is an underlying similarity in patterns of point mutation in diverse organisms, this may mean that the pattern is conserved by purifying selection, since a transition bias is expected to reduce the incidence of potentially harmful mutations (4). This hypothesis would imply that the pattern of point mutation is adapted to the nature of the genetic code. An alternative hypothesis to explain a conserved pattern of point mutation is that the pattern is mainly determined by properties intrinsic to the chemical composition, structure, and metabolism of DNA, and less so by natural selection. This alternative would imply that the genetic code is adapted to the pattern of point mutation rather than the other way around.
Generality of the Experimental Approach.
At this point there is far too little data to ascertain whether different metazoan organisms have similar or distinct patterns of point mutation. Part of the reason for the paucity of data is the lack of a suitable experimental approach for estimating mutational patterns. The approach to studies of mutation illustrated in the present paper should be applicable to any organism that contains non-LTR retrotransposable elements. The rationale of the approach is to use phylogenetic analysis to separate lineages of the non-LTR retrotransposable elements into either internal branches, representing mostly active and evolutionarily constrained lineages, or else into terminal branches, representing mostly nonfunctional DOA copies that evolve like pseudogenes. Because non-LTR retrotransposable elements are widespread in the genomes of most metazoans, this method opens an experimental pathway to the comprehensive study of patterns of spontaneous mutation in a wide variety of organisms. One limitation of the approach is that possible strand-specific mutational effects correlated with transcription or replication are obscured, because of the averaging of mutational patterns across many copies at multiple positions in the genome. In principle, with additional study of each individual DOA copy, it might be possible to separate the DOA sequences according to, for example, strand polarity in replication, but for comparative purposes the overall average pattern of point mutation is the issue of greatest interest anyway. The key questions that now become available for experimental study are: (i) To what extent do patterns of point mutation differ from one metazoan organism to the next? and (ii) What are the chemical and biological determinants of these differences?
Acknowledgments
We thank Alexander Petrov, Mark Siegal, Chao-Ting Wu, Walter Gilbert, Hiroshi Akashi, and Simon Easteal for valuable discussions and the members of the Hartl laboratory for advice and support. This work was supported in part by National Institutes of Health Grant HG01250 to D.L.H. and the William F. Milton Fund Award to D.A.P.
ABBREVIATIONS
- DOA
“dead-on-arrival”
- LTR
long terminal repeat
- A⋅T
C⋅G, Watson–Crick base pairs in DNA
References
- 1.Li W-H, Gojobori T, Nei M. Nature (London) 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- 2.Graur D, Shuali Y, Li W-H. J Mol Evol. 1989;28:279–285. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- 3.Gojobori T, Li W-H, Graur D. J Mol Evol. 1982;18:360–369. doi: 10.1007/BF01733904. [DOI] [PubMed] [Google Scholar]
- 4.Li W-H, Wu C-I, Luo C-C. J Mol Evol. 1984;21:58–71. doi: 10.1007/BF02100628. [DOI] [PubMed] [Google Scholar]
- 5.Jeffs P, Ashburner M. Proc R Soc London, Ser B. 1991;244:151–159. doi: 10.1098/rspb.1991.0064. [DOI] [PubMed] [Google Scholar]
- 6.Weiner A M, Deininger P L, Efstratiadis A. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClure M. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 425–444. [Google Scholar]
- 9.Wright D A, Ke N, Smalle J, Hauge B M, Goodman H M, Voytas D F. Genetics. 1996;142:569–578. doi: 10.1093/genetics/142.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrov D A, Lozovskaya E R, Hartl D L. Nature (London) 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- 11.Petrov D A, Hartl D L. Gene. 1997;205:279–289. doi: 10.1016/s0378-1119(97)00516-7. [DOI] [PubMed] [Google Scholar]
- 12.Petrov D A, Chao Y-C, Stephenson E C, Hartl D L. Mol Biol Evol. 1998;15:1562–1567. doi: 10.1093/oxfordjournals.molbev.a025883. [DOI] [PubMed] [Google Scholar]
- 13.Petrov D A, Hartl D L. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- 14.Petrov D A, Schutzman J L, Hartl D L, Lozovskaya E R. Proc Natl Acad Sci USA. 1995;92:8050–8054. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford D L. paul: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1991. Version 3.0s. [Google Scholar]
- 16.Maddison W P, Maddison D R. macclade. Sunderland, MA: Sinauer Associates; 1992. Version 3. [Google Scholar]
- 17.Francino M P, Ochman H. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 18.Powell J R, DeSalle R. Evol Biol. 1995;28:87–138. [Google Scholar]
- 19.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 20.Moriyama E N, Hartl D L. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kliman R M, Hey J. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 22.Akashi H. Gene. 1997;205:269–278. doi: 10.1016/s0378-1119(97)00400-9. [DOI] [PubMed] [Google Scholar]
- 23.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 25.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]