Abstract
Overexpression of the Arabidopsis thaliana vacuolar H+-pyrophosphatase (AVP1) confers salt tolerance to the salt-sensitive ena1 mutant of Saccharomyces cerevisiae. Suppression of salt sensitivity requires two ion transporters, the Gef1 Cl− channel and the Nhx1 Na+/H+ exchanger. These two proteins colocalize to the prevacuolar compartment of yeast and are thought to be required for optimal acidification of this compartment. Overexpression of AtNHX1, the plant homologue of the yeast Na+/H+ exchanger, suppresses some of the mutant phenotypes of the yeast nhx1 mutant. Moreover, the level of AtNHX1 mRNA in Arabidopsis is increased in the presence of NaCl. The regulation of AtNHX1 by NaCl and the ability of the plant gene to suppress the yeast nhx1 mutant suggest that the mechanism by which cations are detoxified in yeast and plants may be similar.
In Saccharomyces cerevisiae the primary pathway for Na+ extrusion is mediated by Ena1 (1, 2), the plasma membrane Na+-ATPase. Additional genes such as NHA1, NHX1, and YJLO94c that may contribute to Na+ tolerance have been uncovered by using genetic and physiological studies or through homology to known transporters in other organisms. Nha1 belongs to a family of fungi-specific Na+/H+ exchangers that is unrelated in sequence to the bacterial or mammalian Na+/H+ exchangers (3). The Nha1 homologue in Schizosaccharomyces pombe (Sod2) localizes to the plasma membrane, where it mediates a 1:1 Na+/H+ exchange driven by the electrochemical H+ gradient (4, 5). Nhx1 defines a new, rapidly growing subgroup of intracellular Na+/H+ exchangers related to the mammalian NHE family. In yeast, the Nhx1 protein localizes to a prevacuolar compartment, where it mediates Na+ sequestration (6, 7). The protein sequence encoded by YJL094c resembles that of the Enterococcus hirae Na+/H+ antiporter, Nap A, but is thought to be a K+/H+ antiporter (8).
Recent analyses of the genes involved in cation detoxification in yeast have led to a model in which the Nhx1 Na+/H+ exchanger acts in concert with the vacuolar ATPase and the Gef1 anion channel to sequester cations in a prevacuolar compartment (7, 9). This model posits that sequestration of sodium by Nhx1 depends on the vacuolar H+-ATPase and Gef1, the chloride channel. Gef1-mediated anion influx allows establishment by the vacuolar H+-ATPase of a proton gradient sufficient in magnitude to drive the uphill accumulation of Na+ via Na+/H+ exchange.
Here we provide evidence for the role of a prevacuolar compartment in salt tolerance. We show that this compartment contains both the chloride channel Gef1 and the Na+/H+ exchanger Nhx1 and that their functions, together with the proton gradient, are required for salt tolerance. By using yeast strains defective in nhx1, we have been able to clone the A. thaliana NHX1 homologue, AtNHX1. The ability of the Arabidopsis AtNHX1 gene to complement the yeast nhx1 mutant and its induction in plants by salt stress suggests that yeast and plants may achieve salt tolerance by a similar mechanism.
MATERIALS AND METHODS
Yeast Strains and Plasmids.
All strains used are isogenic to W303 (ura3–1 can1–100 leu2–3, 112trp1–1 his3–11, 15). Plasmids pRG52 (Δgef1∷HIS3) (9)and pRG197 (Δnhx1∷HIS3) were used to construct the deletions of GEF1 and NHX1 genes, yielding strains RGY85 and RGY296, respectively. The ena1∷HIS3 mutant was obtained from Fink Lab collection (L5709). Transformation was performed by using the lithium acetate method (10). Double mutants RGY324 (gef1∷HIS3 ena1∷HIS3), RGY326 (nhx1∷HIS3 ena1∷HIS3), and RGY343 (gef1∷HIS3 nhx1∷HIS3) were obtained by crossing the single-mutant strains. Double mutants were identified among the meiotic progeny by scoring for the phenotypes associated with each of the single mutants. Sporulation, tetrad dissection, and mating types were scored as described (11). Cells were grown in YPD (1% yeast/2% peptone/2% dextrose; Difco), YPGAL (1% yeast/2% peptone/2% galactose; Difco), SD (Difco; Synthetic medium with 2% Dextrose), or APG (APG is a synthetic minimal medium containing 10 mM arginine, 8 mM phosphoric acid, 2% glucose, 2 mM MgSO4, 1 mM KCl, 0.2 mM CaCl2, and trace minerals and vitamins) (12). MnCl2 (Sigma), tetramethylammonium chloride (Sigma), NaCl (Sigma), or hygromycin-B (Sigma) were added as indicated.
Wild type, L5709 (ena1∷HIS3), RGY324 (gef1∷HIS3 ena1∷HIS3), and RGY326 (nhx1∷HIS3 ena1∷HIS3) strains were transformed with pYES2 vector (Invitrogen) and plasmid pYES2-AVP1-E229D described in ref. 13. The strain RGY343 (gef1∷HIS3 nhx1∷HIS3), used for histochemical analysis, was transformed with pRG151 (GEF1-GFP) (9) and with pRin73 [NHX1-(HA)3] (7).
Wild-type and RGY296 (nhx1∷HIS3) strains were transformed with vector pAD4 (14). RGY296 (nhx1∷HIS3) was transformed with pRG308 (ADH1∷AtNHX1) (see Cloning of AtNHX1).
Determination of Intracellular Sodium and Potassium Content.
Cells were grown overnight in SD−ura medium (Difco; synthetic medium with 2% dextrose without uracil). YPGAL (1% yeast extract/2% peptone/2% galactose; Difco) media was inoculated with the overnight stocks and grow to an A600 of 0.6. At this OD, NaCl was added to a final concentration of 0.7 M. The cells incubated for 6 h, harvested by centrifugation, washed two times with 1.1 M sorbitol and 20 mM MgCl2, and extracted with water for 30 min at 95°C. The amount of Na+ and K+ in cells was determined at the University of Georgia Chemical Analysis Laboratory by an Inductively Coupled Plasma–MS (see http://www.rserv.uga.edu/rsnew/chemicalanalysis/). Intracellular cation concentrations were estimated as described (15) by using the intracellular water value calculated for cells grown in 1 M NaCl.
Immunofluorescence.
The strain RGY343 (gef1∷HIS3 nhx1∷HIS3) was grown in SD−ura, −leu medium (Difco; synthetic medium with 2% dextrose without uracil and leucine) to mid-logarithmic phase, 0.1 mg/ml hygromycin B was added, and the culture was incubated for 1 h at 30°C. Cells were fixed with 3.7% formaldehyde (Sigma) for 45 min at room temperature without agitation. Spheroplast formation, permeabilization, washing, and antibody incubation was performed as described (16). mAB HA.11 used as first antibody was from Babco (Richmond, CA). Cy3-conjugated goat antimouse IgG was from Jackson Immunoresearch. 4′,6-Diamidino-2-phenylindole (Sigma) was added to mounting medium to stain mitochondrial and nuclear DNA.
Subcellular Fractionation and Western Analysis.
The strain RGY343 (gef1∷HIS3 nhx1∷HIS3) was grown in APG medium (pH 7.0), and lysates fractionated on a 10-step sucrose density gradient as described (7). Aliquots of individual fractions (100 μg) were subjected to SDS/PAGE and transferred to nitrocellulose as described (7). Western blots were probed with monoclonal anti-GFP (green fluorescent protein) antibody (1:10,000 dilution; CLONTECH), anti-hemagglutinin antibody (1:10,000 dilution; Boehringer Mannheim), and peroxidase-coupled goat anti-mouse antibody (1:5,000;) and developed by using the ECL enhanced chemiluminescence system (Amersham Pharmacia).
Plant Strains, Growth Conditions, and RNA Preparation.
A. thaliana plants (ecotype Columbia) were grown aseptically on unsupplemented plant nutrient agar without sucrose (17) for 15 days at 19°C and under continuous illumination. NaCl or KCl was added to a final concentration of 250 mM, and the plants were incubated for 6 h. Total RNA from tissue of salt-treated and untreated plants was isolated (18). Hybond-N (Amersham) membranes were hybridized with a 32P-labeled DNA probe from plasmid pRG308. Hybridization was performed at 65°C overnight. Washes were performed at 65°C with 0.2% standard saline citrate (SSC)/0.1% SDS (19). 18S probe was used as loading control (20). macbas 2.4 program was used to quantify the relative amount of RNA.
Cloning of AtNHX1.
AtNHX1 was cloned from a phage cDNA library of A. thaliana (21) (obtained from the Arabidopsis Biological Resource Center) by probing with an expressed sequence tag (Arabidopsis Biological Resources Center, DNA Stock Center) containing a partial clone. A full-length clone (2.1 kB) was ligated into vector pSK2 (Stratagene) at the NotI site, generating plasmid pRG293. The AtNHX1 ORF was amplified via PCR by using pRG293 as template and GGCCCGGGATGGATTCTCTAGTGTCGAAACTGCCTTCG (italicized bases correspond to nucleotides 1–30 of the ORF) and T7 oligonucleotides. The PCR product was then digested with XbaI and SalI and ligated into pAD4 vector generating plasmid pRG308. The AtNHX1 ORF was sequenced to verify the fidelity of the PCR product. The full-length sequence is longer than the ORF reported by the Arabidopsis Genome Initiative (A TM021B04.4), and has been deposited in GenBank (accession no. AF106324).
RESULTS
The Arabidopsis Vacuolar H+-Pyrophosphatase (Avp1) Confers Salt Tolerance to Yeast ena1 Mutants.
To determine the components of the intracellular system required for sodium detoxification, we used an ena1 mutant that lacks the plasma membrane sodium efflux pump and therefore must rely on the internal detoxification system to overcome sodium toxicity. Growth of the ena1 strain is sensitive to low concentrations of sodium (200 mM), concentrations that do not inhibit the growth of wild-type strains. The sequestration model (7, 9) predicts that the ena1 strain would become salt tolerant if one could enhance the availability of protons in the postulated endosomal compartment. With increased influx of protons, cytoplasmic Na+ would be sequestered via the Nhx1 exchanger. The yeast vacuolar ATPase is a multisubunit protein, so it is difficult to increase its activity by overexpressing any one of its subunits. However, it is possible to increase the influx of protons by expressing the A. thaliana AVP1 gene in yeast. This gene encodes a single polypeptide that, when expressed in yeast, is capable of pumping protons into the lumen of the vacuole (22). To ensure maximum activity of this proton pump, we expressed the E229D gain-of-function mutant of the AVP1 gene (AVP1-D) that has enhanced H+ pumping capability (13).
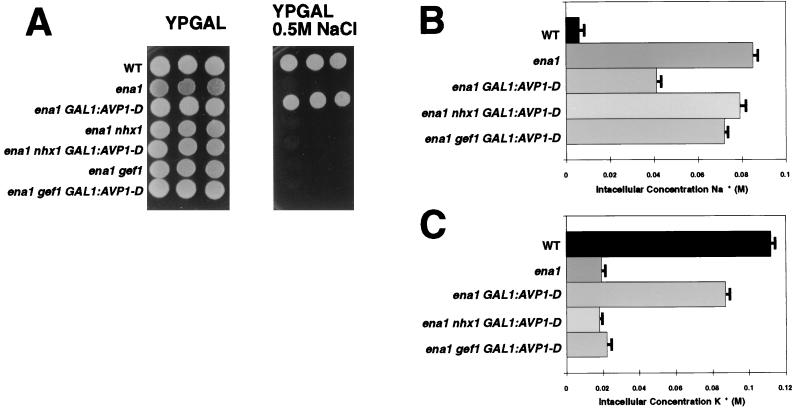
Overexpression of AVP1-D restored salt tolerance to salt-sensitive ena1 mutants (Fig. 1A). The restoration of salt tolerance to an ena1 strain by AVP1-D requires functional NHX1 and GEF1 genes: ena1nhx1 AVP1-D and ena1 gef1 AVP1-D strains are salt sensitive (Fig. 1A).
Figure 1.
Expression of Arabidopsis vacuolar pyrophosphatase AVP1 in ena1 mutants. (A) Vector pYES2 (Invitrogen) was introduced into wild-type, ena1, ena1 nhx1, and ena1 gef1 mutants. Plasmid pYes2-AVP1-D (13) was introduced into ena1, ena1 nhx1, and ena1 gef1 mutants. Five-fold serial dilutions (starting at 105 cells) of each strain were plated on YPGAL (1% yeast extract/2% peptone/2% galactose) with or without 0.5 M NaCl and incubated at 30°C for 2 days. (B and C) Intracellular concentrations of Na+ and K+. Exponentially growing cells (wild-type and ena1 transformed with pYES2 vector and ena1,ena1 nhx1, and ena1 gef1 mutants carrying pYes2-AVP1-D) were exposed to 0.7 M NaCl for 6 h. Total cell extracts were prepared (see Materials and Methods), and Na+ and K+ concentrations were determined. Values are the mean of two determinations, and bars represent the standard deviations. There is a consistent reduction in total cell Na+ in the ena1 AVP-D strain. The reason for this reduction is unknown.
The intracellular Na+ and K+ contents of wild-type strains and of strains carrying various mutations affecting sodium tolerance were determined after 6 h of exposure to media supplemented with 0.7 M NaCl (Fig. 1 B and C). The intracellular Na+ content in the ena1 mutant is 8-fold higher than in the wild-type strain. The ena1 AVP-D strain is salt-resistant, even though its intracellular Na+ content is 4-fold higher then that of the wild type. In ena1 AVP1-D strains lacking either gef1 or nhx1 (i.e., ena1 gef1 or ena1 nhx1), the Na+ content is not reduced to the extent that it is in GEF1 NHX1 strain. Taken together, the genetic and physiological data are consistent with the model that Nhx1, Gef1, and Avp1 cooperate to sequester sodium internally.
The intracellular K+ content correlates with salt tolerance and is inversely correlated with the Na+ content of our strains (Fig. 1C). The wild-type K+ concentration is ≈100 mM but is reduced to 20 mM in the ena1 mutant. Interestingly, in an ena1 strain that overexpresses the AVP1-D gene, the intracellular concentration of K+ is restored almost to wild-type levels (Fig. 1C). However, AVP1-D overexpression fails to restore wild-type levels of intracellular potassium unless both Nhx1 and Gef1 are functional (see the double mutants ena1 nhx1 or ena1 gef1 in Fig. 1C).
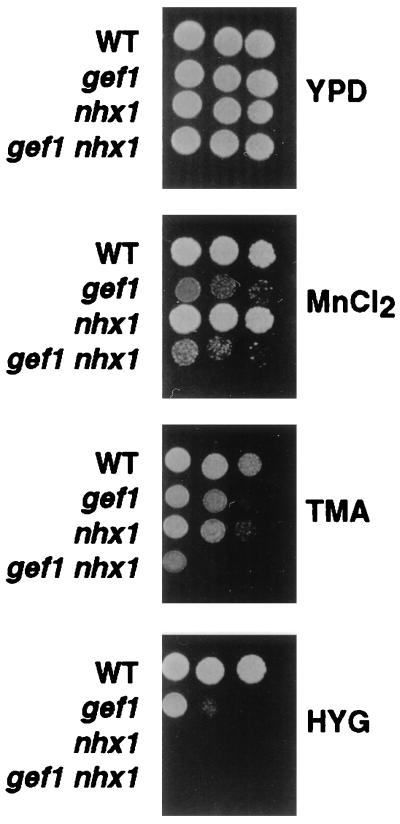
The NHX1 and GEF1 genes, which we have identified as important in sodium detoxification, are also required for the detoxification of other cations. For example, gef1 mutants are sensitive to 3 mM MnCl2, 0.45 M tetramethylammonium chloride and to 0.05 mg/ml hygromycin-B (Fig. 2). The nhx1 mutant is also sensitive to tetramethylammonium chloride and hygromycin. The extreme sensitivity of the nhx1 mutant to hygromycin (Fig. 2) provides an important tool for assaying nhx1 function.
Figure 2.
Growth of gef1 and nhx1 mutants in the presence of toxic cations. Five-fold serial dilutions (starting at 105 cells) of the indicated strains were grown at 30°C for 2 days on YPD (1% yeast extract/2% peptone/2% dextrose) with the addition of either 3 mM MnCl2, 0.45 M tetramethylammonium (TMA), or 0.05 mg/ml hygromycin B (HYG) as indicated.
Gef1p and Nhx1p Colocalize.
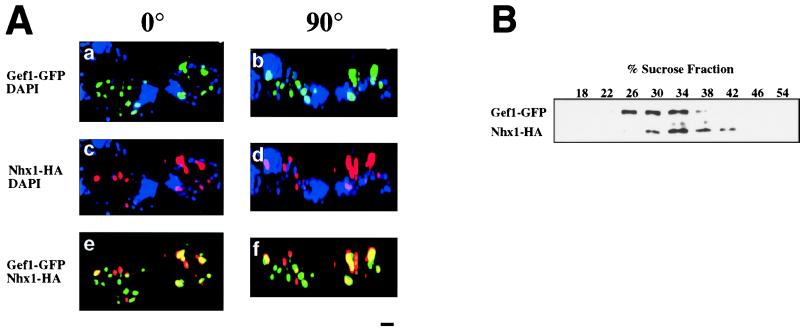
The sequestration model postulates not only a functional connection between the anion channel Gef1 and the sodium exchanger Nhx1 but also predicts that these two proteins colocalize within a common compartment. Because previous studies indicated that Nhx1 localizes to a prevacuolar compartment (7), we performed two types of experiments to determine whether Gef1 and Nhx1 proteins colocalize to this compartment. We found that hemagglutinin (HA)-tagged Nhx1 and a Gef1-GFP fusion protein colocalize as shown via epifluorescence deconvolution microscopy (Fig. 3A). Persistence of signal coincidence on 90° rotation of the image further supports colocalization of the two transporter proteins in these cells (Fig. 3A).
Figure 3.
Distribution of fluorescence and immunodetection of subcellular fractions in gef1 nhx1 cells transformed with two constructs: a GEF1-GFP fusion and a NHX1-(HA)3-tagged fusion. (A) The strain RGY419 (gef1 nhx1) was transformed with plasmids pRG151; GEF1-GFP and pRIN73; NHX1-(HA)3. Transformants were grown in SD (Difco; synthetic medium with 2% dextrose). When the cells reached OD600 = 0.5, hygromycin B (Sigma) was added to a final concentration of 0.1 mg/ml and the cells were incubated for 40 min at 30°C. Cells were fixed and stained with antibodies to HA epitope and 4′,6-diamidino-2-phenylindole (DAPI). Cells were viewed by charge-coupled device microscopy and optically sectioned by using a deconvolution algorithm (Scanalytics, Billerica, MA) (31). (Bar = 1 μm.) (a) Image obtained from Gef1-GFP fluorescence. (b) The same image rotated 90°. (c) Image obtained from the immunodetection of Nhx1-(HA)3. (d) The same image rotated 90°. (e) Image obtained from the superimposition of a and c. (f) Image obtained from the superimposition of b and d. 4′,6-Diamidino-2-phenylindole was omitted from images e and f. (B) The strain RGY419 (gef1 nhx1) transformed with plasmids pRG151; GEF1-GFP and pRIN73; NHX1-(HA)3 was grown in APG medium (12), converted to spheroplasts, lysed, and fractionated on a 10-step sucrose gradient (18–54%) as described (32, 33). Western blots show the distribution of Gef1-GFP and Nhx1-HA (see Materials and Methods).
The colocalization of Nhx1(HA)3 and Gef1-GFP is also supported by the comigration of the two proteins in sucrose density gradients of membrane preparations obtained from cells expressing the tagged proteins (Fig. 3B). The sedimentation behavior of the membrane fraction containing both proteins is consistent with that of a prevacuolar compartment (7). As can be seen, Gef1-GFP (but not Nhx1) is also present in Golgi fractions, consistent with previous studies (9, 23).
An A. thaliana Homologue of NHX1 Functions in Yeast.
The yeast strain we have built provides an important tool for identifying genes that mediate salt tolerance in other organisms. To test the utility of this system, we identified a sequence from Arabidopsis (see Materials and Methods) with very high homology to the S. cerevisiae NHX1 ORF and used an expressed sequence tag (see Materials and Methods) to obtain a full-length clone of this Arabidopsis gene. An alignment of the amino acid sequences of Nhx1 homologues from Arabidopsis (AtNhx1), human (HsNhe6), and yeast (ScNhx1) reveals segments of amino acid identity and similarity within predicted transmembrane domains (Fig. 4). However, it is important to note that despite these relationships, neither the N- nor the C-terminal regions of AtNhx1 and ScNhx1 show a high degree of homology (Fig. 4). A characteristic of mammalian Na+/H+ antiporters is their inhibition by amiloride. A putative amiloride binding site (163DVFFLFLLPPI173) has been defined via point mutants in the human NHE1 antiporter gene (24). AtNhx1, HsNhe-6 and ScNhx1 have an almost identical sequence (Fig. 4). However, our attempts to inhibit the activity of either Nhx1 or AtNhx1 in yeast cultures with amiloride were unsuccessful.
Figure 4.
Comparison of AtNHX1 with human and yeast homologues. Alignment of the deduced amino acid sequences of AtNHX1, HsNHE-6 and ScNHX1. Identical residues are in black boxes, and dashes indicate gaps in the sequence. ∗ above alignment denote putative amiloride binding site from human NHE1 (163DVFFLFLLPPI173).
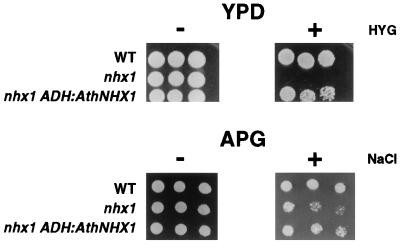
The extreme sensitivity of yeast nhx1 mutants to hygromycin (Fig. 2) permitted us to test whether the Arabidopsis AtNHX1 ORF we had cloned could provide Na+/H+ exchange function in yeast. The At NHX1 gene is capable of suppressing the hygromycin sensitivity of the nhx1 mutant. The AtNHX1 gene also suppressed the NaCl sensitivity of nhx1 mutants but only under conditions in which the K+ availability was reduced (Fig. 5). However, AtNHX1 was not capable of rescuing the Na+-sensitive growth phenotype of the double mutant ena1 nhx1 overexpressing the AVP1-D gene.
Figure 5.
Expression of A. thaliana NHX1 in nhx1 yeast mutants. (A) Vector pAD4 (14) was introduced into wild-type and nhx1 strains. Plasmid pRG308; ADH: AtNHX1 was introduced into nhx1 mutant as indicated. Five-fold serial dilutions (starting at 105 cells) of the indicated strains were grown at 30°C for 2 days on YPD (−) or on YPD supplemented with 0.05 mg/ml hygromycin (+). Serial dilutions of the same strains were grown on APG medium (see Materials and Methods) (−) or on APG supplemented with 0.4 M NaCl (+) (12).
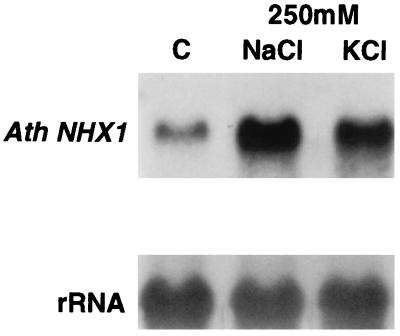
Further support for the role of the Arabidopsis AtNHX1 gene in salt homeostasis came from an analysis of its expression in salt-stressed plants (Fig. 6). Plants were grown for 15 days under standard conditions and then exposed for 6 h to either 250 mM NaCl or KCl. The NaCl stress increased AtNHX1 mRNA levels 4.2-fold, whereas KCl promoted only a 2.8-fold increase. This increase in mRNA level produced by sodium resembles that described for the yeast NHX1 gene (7).
Figure 6.
Analysis of AtNHX1 expression under salt stress. RNA tissue blot hybridized with AtNHX1. Ten micrograms of total RNA from 15-day-old plants exposed to 250 mM NaCl or KCl for 6 h and a control grown without salt was subjected to electrophoresis on a denaturing formaldehyde gel. The blot was hybridized with a probe internal to AtNHX1 ORF. An 18S ribosomal probe was used as loading control.
DISCUSSION
Our studies provide evidence for the importance of the prevacuolar pH gradient for intracellular Na+ sequestration in yeast. Overexpression of the plant H+-pyrophosphatase (Avp1) confers salt tolerance to yeast only in those strains containing a functional chloride channel (Gef1) and the Na+/H+ exchanger (Nhx1).
These data support a model in which the Nhx1 Na+/H+ exchanger acts in concert with the vacuolar ATPase and the Gef1 anion channel to sequester cations in a prevacuolar compartment. Several studies suggest that this prevacuolar compartment may be derived both from the plasma membrane and the late Golgi. These vesicles could be involved in the assembly of the vacuole or delivery of cargo to this organelle. We presume that these prevacuolar vesicles detoxify cations by sequestration, thereby lowering their concentrations in the cytoplasm and in other organelles.
The yeast system we have described permits the functional assessment of diverse heterologous proteins in salt tolerance: chloride channels, H+ pumps, and Na+/H+ exchangers. The system is robust and flexible. The function of the Arabidopsis chloride channels (9, 25), H+ pump, and Na+/H+ exchanger can be assayed in the corresponding yeast mutant. Despite the inability of At NHX1 to suppress all of the phenotypes of the yeast nhx1 mutant, the fact that it suppresses some phenotypes, coupled with the DNA homology between AtNHX1 and yeast NHX1, suggests that the plant gene carries out a similar function to that of the yeast homologue. The observation that the AtNHX1 gene suppresses the sensitivity of the nhx1 mutant to hygromycin but provides only a weak Na+ detoxification phenotype could be a consequence either of differential regulation of the transporters in the two organisms or of distinct cation transport selectivities.
The regulation of AtNHX1 by salt and the ability of the plant gene to suppress the yeast nhx1 mutant suggest that the mechanism by which cations are detoxified in yeast and plants may be similar. Indeed, previous work suggested that vacuolar sodium accumulation in salt-tolerant plants may be mediated by a tonoplast Na+/H+ antiporter that utilizes the proton-motive force generated by the vacuolar H+-ATPase (V-ATPase) and/or H+-translocating pyrophosphatase (V-PPase; refs. 26–28).
Our finding that both gef1 and nhx1 mutants are hypersensitive to hygromycin suggests that the level of resistance to hygromycin depends on the function of the vacuolar and prevacuolar organelles. Yeast mutants impaired in K+ uptake (trk1) are hypersensitive to hygromycin (29); reduced K+ uptake hyperpolarizes the plasma membrane potential and drives the uptake of alkali cations such as hygromycin. Mutations that reduce the H+ pumping activity of the plasma membrane H+-ATPase, Pma1, depolarize the plasma membrane potential and confer resistance to hygromycin (30). Thus, mutants such as gef1 or nhx1 that affect the pH or membrane potential of the vacuolar and prevacuolar compartments may be expected to affect hygromycin compartmentation.
Acknowledgments
We thank P. Rea for plasmid pYES2-AVP1-E229D; Ying Wei for excellent technical assistance; and A. Diener, T. Galitski, and H. Madhani for helpful discussions. R.R. was supported by National Institutes of Health Grants GM52414 and DK54214. R.A.G. and S.L.A. were supported by National Institutes of Health Grants DK43495, DK51059, and DK34854 (Harvard Digestive Diseases Center) and by a Grant-in-Aid from the American Heart Association. The work was supported by the following grants: National Science Foundation Grant MCB9317175 and National Institutes of Health Grant GM35010, both awarded to G.R.F.; S.L.A. is an Established Investigator of the American Heart Association. G.R.F. is an American Cancer Society Professor of Genetics. This work was conducted utilizing the W. M. Keck Foundation Biological Imaging Facility at the Whitehead Institute.
ABBREVIATIONS
- GFP
green fluorescent protein
- HA
hemagglutinin.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF106324).
References
- 1.Haro R, Garciadeblas B, Rodriguez-Navarro A. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 2.Rios G, Ferrando A, Serrano R. Yeast. 1997;13:515–528. doi: 10.1002/(sici)1097-0061(199705)13:6<515::aid-yea102>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Prior C, Potier S, Souciet J L, Sychrova H. FEBS Lett. 1996;387:89–93. doi: 10.1016/0014-5793(96)00470-x. [DOI] [PubMed] [Google Scholar]
- 4.Hahnenberger K M, Jia Z, Young P G. Proc Natl Acad Sci USA. 1996;93:5031–5036. doi: 10.1073/pnas.93.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dibrov P, Smith J J, Young P G, Fliegel L. FEBS Lett. 1997;405:119–124. doi: 10.1016/s0014-5793(97)00169-5. [DOI] [PubMed] [Google Scholar]
- 6.Nass R, Cunningham K W, Rao R. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 7.Nass R, Rao R. J Biol Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez J, Ramirez O, Saldana C, Coria R, Pena A. J Bacteriol. 1998;180:5860–5865. doi: 10.1128/jb.180.22.5860-5865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaxiola R A, Yuan D S, Klausner R D, Fink G R. Proc Natl Acad Sci USA. 1998;95:4046–4050. doi: 10.1073/pnas.95.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 12.Rodriguez-Navarro A, Ramos J. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhen R G, Kim E J, Rea P A. J Biol Chem. 1997;272:22340–22348. doi: 10.1074/jbc.272.35.22340. [DOI] [PubMed] [Google Scholar]
- 14.Ballester R, Michaeli T, Ferguson K, Xu H P, McCormick F, Wigler M. Cell. 1989;59:681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- 15.Gaxiola R, de Larrinoa I F, Villalba J M, Serrano R. EMBO J. 1992;11:3157–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pringle J, Adams A, Drubin D, Haarer B. In: Immunofluorescence Methods for Yeast. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. pp. 565–602. [DOI] [PubMed] [Google Scholar]
- 17.Haughn G W, Somerville C. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 18.Niyogi K K, Fink G R. Plant Cell. 1992;4:721–733. doi: 10.1105/tpc.4.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1988. [Google Scholar]
- 20.Unfried I, Stocker U, Gruendler P. Nucleic Acids Res. 1989;17:7513. doi: 10.1093/nar/17.18.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 22.Kim E J, Zhen R G, Rea P A. Proc Natl Acad Sci USA. 1994;91:6128–6132. doi: 10.1073/pnas.91.13.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwappach B, Stobrawa S, Hechenberger M, Steinmeyer K, Jentsch T J. J Biol Chem. 1998;273:15110–15118. doi: 10.1074/jbc.273.24.15110. [DOI] [PubMed] [Google Scholar]
- 24.Counillon L, Franchi A, Pouyssegur J. Proc Natl Acad Sci USA. 1993;90:4508–4512. doi: 10.1073/pnas.90.10.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hechenberger M, Schwappach B, Fischer W N, Frommer W B, Jentsch T J, Steinmeyer K. J Biol Chem. 1996;271:33632–33638. doi: 10.1074/jbc.271.52.33632. [DOI] [PubMed] [Google Scholar]
- 26.Barkla B J, Apse M P, Manolson M F, Blumwald E. Symp Soc Exp Biol. 1994;48:141–153. [PubMed] [Google Scholar]
- 27.Zhen R G, Kim E J, Rea P A. The Molecular and Biochemical Basis of Pyrophosphate-Energized Proton Translocation at the Vacuolar Membrane. San Diego: Academic; 1997. [Google Scholar]
- 28.Kirsch M, An Z, Viereck R, Low R, Rausch T. Plant Mol Biol. 1996;32:543–547. doi: 10.1007/BF00019107. [DOI] [PubMed] [Google Scholar]
- 29.Madrid R, Gomez M J, Ramos J, Rodriguez-Navarro A. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 30.McCusker J H, Perlin D S, Haber J E. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy B K, Gotta M, Sinclair D A, Mills K, McNabb D S, Murthy M, Pak S M, Laroche T, Gasser S M, Guarente L. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 32.Sorin A, Rosas G, Rao R. J Biol Chem. 1997;272:9895–9901. doi: 10.1074/jbc.272.15.9895. [DOI] [PubMed] [Google Scholar]
- 33.Antebi A, Fink G R. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]