Abstract
DNA chip technology enables simultaneous examination of how ≈6,200 Saccharomyces cerevisiae gene transcript levels, representing the entire genome, respond to environmental change. By using chips bearing oligonucleotide arrays, we show that, after exposure to the alkylating agent methyl methanesulfonate, ≈325 gene transcript levels are increased and ≈76 are decreased. Of the 21 genes that already were known to be induced by a DNA-damaging agent, 18 can be scored as inducible in this data set, and surprisingly, most of the newly identified inducible genes are induced even more strongly than these 18. We examined 42 responsive and 8 nonresponsive ORFs by conventional Northern blotting, and 48 of these 50 ORFs responded as they did by DNA chip analysis, with magnitudes displaying a correlation coefficient of 0.79. Responsive genes fall into several expected and many unexpected categories. Evidence for the induction of a program to eliminate and replace alkylated proteins is presented.
Exposure to DNA-damaging agents can increase DNA repair capacity and activate cell-cycle checkpoints. Such exposures may also induce enzymes that metabolize toxicants to facilitate their elimination from the organism or may activate programmed cell death (apoptosis) to eliminate highly damaged cells. Thus, it has long been known that cells induce the expression of a variety of genes after toxic exposure, and gene regulation in response to DNA-damaging agents has been well studied in many organisms (1–5).
Random lacZ gene fusions and differential hybridization previously have identified 21 Saccharomyces cerevisiae genes whose transcript levels are increased in response to DNA-damaging agents (1, 6–8). Both approaches produced catalogs of genes of known and unknown function, but the lack of redundancy with which they were identified indicates that the search for such inducible genes is far from complete (1, 8).
We previously studied the inducible transcription of an S. cerevisiae DNA repair gene (MAG1, encoding a 3-methyladenine DNA glycosylase) in response to simple alkylating agents such as methyl methanesulfonate (MMS; refs. 9–13). Upstream MAG1 regulatory elements were identified, and similar elements are found upstream of numerous DNA repair and metabolism genes, suggesting common transcriptional regulatory mechanisms (12–14). We therefore decided to identify all the genes that are regulated coordinately with MAG1. Here, we report that DNA chip analysis has expanded by more than 15-fold the catalog of genes that are inducible by a DNA-damaging agent. In addition, DNA chip analysis has identified a class of genes whose transcripts are repressed. Global responses to a DNA-damaging agent have now come into focus, and we present evidence that exposure to an alkylating agent elicits a program to eliminate and replace alkylated proteins from S. cerevisiae cells.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions.
S. cerevisiae strain DBY747 (MATa his3-Δ1 leu2-3,112 ura3-52 trp1-289a galS can1 CUPr) was used in this study and was grown in 1% yeast extract/2% peptone/2% glucose at 30°C. Cells were grown to a density of 5 × 106 cells per ml as measured by counting duplicated dilutions. Cultures were split into two; MMS (0.1%) was added directly to one culture, and both cultures were incubated at 30°C for 1 h. Cells were pelleted and washed once in distilled H2O and once in AE buffer (50 mM NaOAc, pH 5.2/10 mM EDTA) immediately before RNA extraction.
RNA Extraction.
Total RNA was isolated by using a hot-phenol method (15). Poly(A)+ RNA was purified from total RNA with Oligotex oligo(dT) selection step (Qiagen, Chatsworth, CA). Poly(A)+ RNA was amplified and biotin-labeled as follows. Poly(A)+ RNA (1 μg) was converted into double-stranded cDNA by using a modified oligo(dT) primer with a T7 RNA polymerase promoter sequence at the 5′ end and the Superscript Choice system for cDNA synthesis (GIBCO). Double-stranded cDNA was purified by phenol/chloroform extractions, precipitated with ethanol, and resuspended at a concentration of 0.5 μg/μl in diethyl pyrocarbonate-treated H2O. Phase-Lock Gel (5 Prime → 3 Prime) was used for all organic extractions to increase recovery. In vitro transcription was performed with T7 RNA polymerase (T7 Megascript kit, Ambion, Austin, TX) and with 0.5–1.0 μg of cDNA, 7.5 mM unlabeled ATP and GTP, 5.3 mM unlabeled UTP and CTP, and 1.9 mM biotin-labeled CTP and UTP (biotin-11-CTP, biotin-16-UTP, Enzo Diagnostics). Reactions were carried out for 6 h at 37°C, and cRNA was purified by RNA affinity resin (RNeasy spin columns, Qiagen). A sample was separated on a 1% agarose gel to check the size range, and then 10 μg of cRNA was fragmented randomly to an average size of 50 bases by heating at 94°C for 35 min in 40 mM Tris⋅acetate, pH 8.1/100 mM KOAc/30 mM MgOAc.
GeneChip Hybridizations.
A set of four oligonucleotide arrays (GeneChip Ye6100 arrays, Affymetrix, Santa Clara, CA) containing probes for 6,218 yeast ORFs were used for hybridizations. Hybridizations were done in 200 μl of AFFY buffer (Affymetrix) at 40°C for 16 h with constant mixing. After hybridization, arrays were rinsed three times with 200 μl of 6× sodium chloride/sodium phosphate/EDTA/Triton (SSPE-T; 1× 0.15 M NaCl/15 mM phosphate, pH 7.6/1 mM EDTA/0.005% Triton) and then washed with 200 μl of 6× SSPE-T (pH 7.6) for 1 h at 50°C. The arrays were rinsed twice with 0.5× SSPE-T (pH 7.6) and washed with 0.5× SSPE-T (pH 7.6) at 50°C for 15 min. Staining was done with 2 μg/ml streptavidin-phycoerythrin (Molecular Probes) and 1 mg/ml acetylated BSA (Sigma) in 6× SSPE-T (pH 7.6). The arrays were read at 7.5 μm with a confocal scanner (Molecular Dynamics) and analyzed with genechip software, version 3.0. The samples were normalized by using the total average difference between the perfectly matched probe and the mismatched probe (16).
Northern-Blot Analysis.
RNA was isolated from log-phase cells exposed to 0.1% MMS for 0, 30, 60, or 120 min by using a hot-phenol extraction method (15). Total RNA (25 μg) was fractionated in a 1% denaturing agarose gel, blotted, and probed with PCR-amplified labeled ORFs (Research Genetics, Huntsville, AL) by using standard methods (17).
RESULTS AND DISCUSSION
Global Expression Monitoring After Alkylation Damage.
The GeneChip methodology developed by Affymetrix was used to monitor global gene expression in S. cerevisiae. The 6,218 ORFs of this organism are represented on a set of four high density oligonucleotide arrays (16, 18, 19). Poly(A)+ mRNA was isolated from untreated cells and from cells exposed for 1 h to 0.1% MMS. These conditions were chosen, because they yield optimal MAG1 induction with minimal cell death (11). Poly(A)+ RNA was converted into double-stranded cDNA containing the T7 RNA polymerase promoter, and biotin-labeled cRNA was produced and hybridized to the GeneChip arrays. The hybridization-intensity information was gathered by scanning confocal microscopy and analyzed with genechip software, version 3.0 (16). Typical GeneChip-hybridization intensities for control and MMS-treated cells are shown in Fig. 1. As a guide, one MMS-induced, one MMS-repressed, and one nonresponsive ORF are indicated by arrows. It had been established that differences in hybridization intensity between the same ORFs on corresponding chips are proportional to changes in transcript levels and that intensity changes greater than 2.0-fold are both significant and accurate (16). It is important to note that 18 of the 21 genes previously reported to be induced by a DNA-damaging agent (not necessarily MMS) were found to be induced by 2.5- to 6.9-fold. This fact is remarkable, because our study is limited to a single agent (MMS) and a single time point. These genes (and their fold induction) are UBI4 (10.3), RNR3 (6.9), DDR48 (6.2), RNR1 (≈5.7), MAG1 (≈5.5), RAD7 (≈5.5), DDR2 (5.3), HIS4 (4.6), RNR2 (3.9), HIS3 (3.4), CDC8 (2.9), RAD2 (2.9), RAD54 (≈2.8), RAD23 (2.7), PHR1 (2.6), CDC9 (2.5), RAD51 (2.5), RNR4 (2.5), CDC17 (1), RAD6 (1), and RAD18 (1). (The complete data set can found at www.hsph.harvard.edu/geneexpression.)
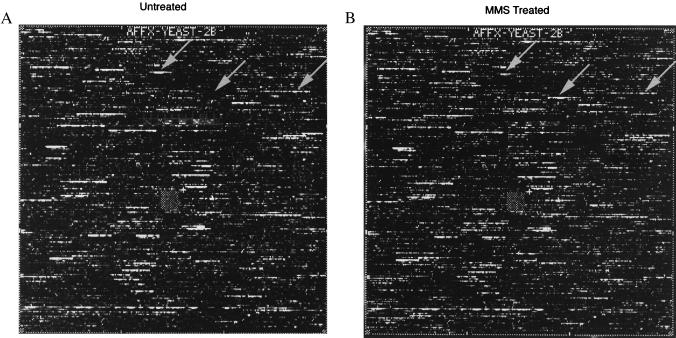
Figure 1.
Fluorescence image of S. cerevisiae cRNA hybridization to GeneChip oligonucleotide arrays corresponding to ORFs from YEL001c to YJL088w (chromosomes 5–10; ref. 30) probed with 10 μg of biotin-labeled cRNA prepared from S. cerevisiae DBY747 log-phase cells untreated (A) or treated (B) with 0.1% MMS for 1 h. At this dose and time, there was a >90% survival rate. Differential hybridization between A and B represents ORFs expressed at different levels before and after MMS exposure. Examples of repressed (left arrow), induced (middle arrow), and nonresponsive (right arrow) ORFs are indicated. These ORFs correspond to RPS26B, YFL061W, and ACT1 respectively, and the hybridization differences were 3.6-fold, 251-fold, and 1-fold, respectively.
Of 6,218 ORFs, 325 (5.2%) showed more than a 4-fold increase in transcript level (Table 1); 32 of these increased by greater than 10-fold, and the greatest increase was 251-fold (YFL061W). For reference, MAG1 was induced ≈5.5-fold in this particular experiment, and 115 ORFs were induced more highly than MAG1. MMS treatment also resulted in more than a 3-fold reduction in transcript levels for 75 of 6,218 ORFs (Table 2). Of these, 24 decreased by a factor of >5-fold; 10 decreased by a factor of >7.5-fold; 4 decreased by >10-fold.
Table 1.
ORFs whose transcripts are induced by >4-fold by MMS (n = 325*)
| ORF | Gene | Fold | Function† |
|---|---|---|---|
| Stress response/detoxification (25 ORFs‡) | |||
| YLL060C | GTT2 | 28.8 | Glutathione transferase |
| YER143W | DDI1 | 17.8 | DNA damage-inducible |
| YPL092W | SSU1 | 17.8 | Sulfite sensitivity |
| YBR008C§¶ | FLR1 | 15.1 | Fluconazole resistance |
| YLL039C§¶ | UBI4 | 10.3 | Ubiquitin |
| YBL064C | — | 8.5 | Antioxidant enzyme‖ |
| YML116W§¶ | ATR1 | 8.0 | Aminotriazole and 4-NQOR |
| YDR059C§ | UBC5 | 7.3 | E2 ubiquitin-conjugating enzyme |
| YAL005C | SSA1 | 6.4 | HSP70 family, cytoplasmic |
| YDL025C§ | — | 6.3 | Ser/Thr protein kinase |
| YML007W§¶ | YAP1 | 6.2 | Transcriptional activator |
| YMR173W¶ | DDR48 | 6.2 | DNA damage-inducible HSP |
| YNL241C§¶ | ZWF1 | 6.0 | Glucose-6-phosphate dehydrogenase |
| YOR162C§¶ | YRR1 | ∼5.7** | Transcription factor |
| YML070W§ | DAK1 | 5.4 | Dihydroxyacetone kinase |
| YOL052C-A | DDR2 | 5.3 | Heat-shock protein |
| YGR234W¶ | YHB1 | 5.2 | Flavohemoglobin |
| YMR174C | PAI3 | 5.1 | Protease A (ysca) inhibitor IA3 |
| YGL254W‡ | FZF1 | 4.9 | Sulfite resistance protein |
| YOL025W§ | LAG2 | ∼4.8 | Affects longevity |
| YIR038C | GTT1 | 4.6 | Glutathione transferase |
| DNA synthesis/repair (13 ORFs) | |||
| YAR007C¶ | RFA1 | 9.5 | DNA replication factor A |
| YAL015C¶ | NTG1 | ∼7.6 | DNA glycosylase |
| YIL066C | RNR3 | 6.9 | Ribonucleotide reductase |
| YER070W | RNR1 | ∼5.7 | Ribonucleotide reductase |
| YKL112W†† | ABF1 | ∼5.6 | ARS1 binding protein |
| YER142C | MAG1 | ∼5.5 | 3-Methyladenine DNA glycosylase |
| YJR052W¶ | RAD7 | ∼5.5 | Nucleotide excision repair protein |
| YMR228W§ | MTF1 | 5.2 | RNA polymerase specific factor |
| YIR008C¶ | PRI1 | 4.7 | DNA primase |
| Cell cycle (10 ORFs) | |||
| YLR299W‡‡ | CIS2 | 7.7 | γ-Glutamyltransferase |
| YDL132W§ | CDC53 | 6.1 | Controls G1/S transition |
| YLR178C§ | TFS1 | 5.7 | Cell-cycle regulator |
| YMR028W | TAP42 | 5.2 | Involved in Tor signaling |
| YKL179C | — | 4.5 | Kinesin‖ |
| Signaling/kinases/phosphatases (16 ORFs) | |||
| YPL150W | — | 7.2 | Ser/Thr protein kinases‖ |
| YDL025C§ | — | 6.3 | Ser/Thr protein kinase |
| YOR162C§¶ | YRR1 | ∼5.7 | Transcription factor |
| YLR178C§ | TFS1 | 5.7 | Cell-cycle regulator |
| YML112W§ | CTK3 | 5.5 | Carboxy-terminal domain kinase |
| YNL183C§ | NPR1 | 5.4 | Ser/Thr protein kinase |
| YPL152W | RRD2 | 5.1 | Phosphotyrosyl phosphatase‖ |
| YGR080W | TWF1 | 5.0 | Twinfilin A |
| YLR362W¶ | STE11 | 5.0 | Ser/Thr protein kinase |
| YJL164C | SRA3 | 4.7 | cAMP-dependent protein kinase 1 |
| YLL019C | KNS1 | ∼4.5 | Ser/Thr protein kinase |
| YOL100W | PKH2 | 4.5 | Ser/Thr protein kinase |
| Cell wall biogenesis (5 ORFs) | |||
| YKR076W | ECM4 | 17.7 | Cell wall biogenesis |
| YHL030W | ECM29 | ∼8.7 | Cell wall biogenesis |
| YLR299W‡ | CIS2 | 7.7 | γ-Glutamyltransferase |
| YKR009C | FOX2 | 6.5 | Hydratase-dehydrogenase-epimerase |
| YOL025W§ | LAG2 | ∼4.8 | Affects longevity |
| Membrane transport (13 ORFs) | |||
| YBR008C§¶ | FLR1 | 15.1 | Fluconazole resistance protein |
| YOR328W | PDR10 | 8.8 | ABC transporter proteins |
| YML116W§¶ | ATR1 | 8.0 | Aminotriazole and 4-NQOR |
| YOL119C | — | 7.6 | Monocarboxylate transporters‖ |
| YOR130C§ | ARG11 | 5.8 | Integral membrane protein |
| YMR060C | TOM37 | 5.6 | Outer membrane import receptor |
| YCR011C | ADP1 | 5.5 | ATP-dependent permease |
| YGL104C§ | — | 4.7 | Glucose transport proteins‖ |
| YGL186C | — | 4.5 | Purine cytosine permease‖ |
| Nitrogen and sulfur metabolism (12 ORFs) | |||
| YFL061W | — | 251 | Cyanamide hydratase‖ |
| YOL058W§ | ARG1 | 7.0 | Argininosuccinate synthetase |
| YFR030W§ | MET10 | 5.8 | Sulfite reductase, flavin-binding |
| YKL112W†† | ABF1 | ∼5.6 | ARS1 binding protein |
| YNL183C§ | NPR1 | 5.4 | Ser/Thr protein kinase |
| YDL215C§ | GDH2 | 5.2 | Glutamate dehydrogenase |
| YFL030W§ | — | 5.0 | Transaminases‖ |
| YGL254W‡‡ | FZF1 | 4.9 | Sulfite resistance protein |
| YJL060W | — | 4.9 | Glutamine transaminase‖ |
| YOR226C | — | 4.6 | Nitrogen fixation proteins‖ |
| YDR242W | AMD2 | ∼4.5 | Amidase |
| Carbohydrate metabolism/fermentation (28 ORFs) | |||
| YFL057C | — | 14.1 | Aryl-alcohol dehydrogenases‖ |
| YDL243C | — | ∼13.9 | Aryl-alcohol dehydrogenases‖ |
| YFL056C | — | 13.7 | Aryl-alcohol dehydrogenases‖ |
| YNL241C§¶ | ZWF1 | 6.0 | Glucose-6-phosphate dehydrogenase |
| YAL060W | FUN49 | 5.8 | Alcohol/sorbitol dehydrogenase‖ |
| YOR120W | GCY1 | 5.8 | Aldo/keto reductase |
| YEL020C | — | ∼5.7 | Oxalyl-CoA decarboxylase‖ |
| YGL062W | PYC1 | 5.6 | Pyruvate carboxylase 1 |
| YJL099W | CHS6 | ∼5.6 | Chitin biosynthesis protein |
| YML070W§ | DAK1 | 5.4 | Dihydroxyacetone kinase |
| YNL331C | — | 5.3 | Aryl-alcohol reductase‖ |
| YGR244C | LSC2 | 5.2 | Succinate-CoA ligase |
| YDL066W | IDP1 | 4.9 | Isocitrate dehydrogenase (NADP+) |
| YIR036C | — | 4.9 | E. coli FabD‖ |
| YDR001C | NTH1 | 4.8 | Neutral trehalase (α,α-trehalase) |
| YGL104C§ | — | 4.7 | Glucose transport proteins‖ |
| YLR164W | — | 4.7 | Sdh4p‖ |
| YPR184W | — | ∼4.7 | Human 4-α-glucanotransferase‖ |
| mRNA processing (15 ORFs) | |||
| YLR136C | TIS11 | 6.3 | Homolog of mammalian TIS11 |
| YML007W§¶ | YAP1 | 6.2 | Transcriptional activator |
| YKL070W | — | ∼6.1 | Transcriptional regulatory |
| YKL112W†† | ABF1 | ∼5.6 | Transcriptional activator |
| YML112W§ | CTK3 | 5.5 | Carboxy terminal domain kinase |
| YGL122C | NAB2 | 5.4 | Nuclear poly(A)-binding protein |
| YMR228W§ | MTF1 | 5.2 | RNA polymerase specific factor |
| YGL254W‡‡ | FZF1 | 4.9 | Sulfite resistance protein |
| YOR185C | GSP2 | 4.7 | GTP-binding protein |
| Others (57 ORFs) | |||
| YMR096W | SNZ1 | ∼65.8 | Stationary phase protein |
| YCL026C | FRM2 | ∼27.5 | Fatty acid regulation |
| YML131W | — | 15.2 | Leukotriene B4 12-hydroxydehydrogenase‖ |
| YIL164C | NIT1 | 12.8 | Nitrilase |
| YPL171C | OYE3 | ∼10.4 | NAPDH dehydrogenase |
| YNL335W | — | 10.3 | Cyanamide hydratase‖ |
| YLR214W | FRE1 | 8.9 | Ferric (and cupric) reductase |
| YBR256C | RIB5 | 6.4 | Riboflavin synthase a-chain |
| YNL036W | NCE10 | 6.4 | Protein export pathway |
| YBR046C | ZTA1 | 6.2 | ζ-crystallin homolog |
| YBR170C | NPL4 | 6.2 | Nuclear protein localization |
| YHR071W | PCL5 | 5.9 | PHO85 cyclin |
| YJL068C | — | 5.8 | Human esterase D‖ |
| YMR231W | PEP5 | ∼5.8 | Vacuolar biogenesis protein |
| YFR010W | UBP6 | 5.6 | Ubiquitin-specific protease |
| YGL194C | HOS2 | ∼5.4 | Putative histone deacetylase |
| YGR218W | CRM1 | 5.4 | Chromosome region maintenance |
| YKL073W | LHS1 | ∼5.3 | Chaperone of the ER lumen |
| YBL033C | RIB1 | 5.2 | GTP cyclohydrolase II |
| YHR016C | YSC84 | 5.2 | Hypothetical protein YFR024ca‖ |
| YLL063C | AYT1 | ∼5.0 | Transacetylase |
| YOR227W | — | 5.0 | Microtubule-interacting protein |
| YJL008C | CCT8 | 4.9 | Chaperonin-containing T complex |
| YMR004W | MVP1 | 4.9 | Sorting proteins to the vacuole |
| YJL041W | NSP1 | 4.8 | Nuclear pore protein |
| YLL001W | DNM1 | 4.8 | Dynamin-related protein |
| YNL237W | YTP1 | 4.8 | Mitochondrial electron transport proteins‖ |
| YCR029C | — | 4.7 | Hypothetical protein |
| YJL154C | VPS35 | 4.7 | Protein-sorting protein, vacuolar |
| YLR163C | MAS1 | ∼4.7 | Mitochondrial processing peptidase |
| YOR181W | LAS17 | ∼4.7 | Actin assembly factor |
| YKL173W | SNU11 | 4.6 | U5 snRNP-specific protein |
| YOR069W | VPS5 | 4.6 | Sorting nexin I homolog |
| Unknown/unclassified (112 ORFs) | |||
| These ORFs and their fold induction can be found on the World Wide Web at www.hsph.harvard.edu/geneexpression. | |||
| Degradation (15 ORFs) | |||
| YKL103C | LAP4 | 13.9 | Aminopeptidase yscI precursor |
| YLL039C§¶ | UBI4 | 10.3 | Ubiquitin |
| ORF | Gene | Fold | Function† |
| YDR059C§ | UBC5 | 7.3 | E2 ubiquitin-conjugating enzyme |
| YDR092W | UBC13 | 6.2 | E2 ubiquitin-conjugating enzyme |
| YDL132W§ | CDC53 | 6.1 | Controls G1/S transition |
| YMR304W | UPB15 | 5.7 | Ubiquitin-specific protease‖ |
| YHR027C | RPN1 | 4.7 | 26S proteasome regulatory subunit |
| YOR124C | UBP2 | 4.6 | Ubiquitin-specific protease |
| YJL001W | PRE3 | 4.5 | 20S proteasome subunit (b1) |
| Amino acid metabolism (41 ORFs) | |||
| YMR189W | GCV2 | 12.5 | Glycine decarboxylase complex |
| YJR109C | CPA2 | 12.3 | Carbamoyl phosphate synthetase |
| YJR130C | — | 11.5 | o-Succinylhomoserine (thiol)-lyase |
| YKL218C | — | 10.6 | Threonine dehydratases |
| YMR062C | ECM40 | 9.8 | Acetylornithine acetyltransferase |
| YER069W | ARG5,6 | 9.4 | Acetylglutamate kinase |
| YJL088W | ARG3 | 9.3 | Ornithine carbamoyltransferase |
| YLR160C | ASP3D | 8.4 | l-Asparaginase II |
| YHR018C | ARG4 | 7.8 | Arginosuccinate lyase |
| YIL116W | HIS5 | 7.8 | Histidinol-phosphate aminotransferase |
| YLR299W‡‡ | CIS2 | 7.7 | γ-Glutamyltransferase |
| YDR019C | GCV1 | 7.5 | Glycine decarboxylase |
| YLR158C | ASP3C | 7.1 | l-Asparaginase II |
| YOL058W§ | ARG1 | 7.0 | Argininosuccinate synthetase |
| YDR127W | ARO1 | 6.8 | Arom pentafunctional enzyme |
| YKL215C | — | ∼6.7 | P. aeruginosa hyuA and hyuB‖ |
| YFR055W | — | ∼6.6 | β-Cystathionases‖ |
| YLL058W | — | 6.4 | o-Succinylhomoserine (thiol)-lyase |
| YIR034C | LYS1 | 6.2 | Saccharopine dehydrogenase |
| YFR030W§ | MET10 | 5.8 | Sulfite reductase, flavin-binding |
| YHR037W | PUT2 | 5.8 | 1-Pyrroline-5-carboxylate dehydrogenase |
| YOR130C§ | ARG11 | 5.8 | Mitochondrial integral membrane protein |
| YKL112W†† | ABF1 | ∼5.6 | ARS1 binding protein |
| YNL104C | LEU4 | 5.6 | 2-Isopropylmalate synthase |
| YDL215C§ | GDH2 | 5.2 | Glutamate dehydrogenase |
| YER052C | HOM3 | 5.2 | l-Aspartate 4-p-transferase |
| YKL211C | TRP3 | 5.1 | Anthranilate synthase |
| YFL030W§ | — | 5.0 | Transaminases‖ |
| YDR354W | TRP4 | 4.8 | Anthranilate phosphoribosyltransferase |
| YDR035W | ARO3 | 4.7 | 2-Dehydro-3-deoxyphosphoheptonate |
| YER090W | TRP2 | 4.7 | Anthranilate synthase component I |
| YHR137W | ARO9 | 4.7 | Aromatic amino acid aminotransferase II |
| YJR025C | BNA1 | 4.7 | 3-Hydroxyanthranilic acid dioxygenase |
| YCL030C | HIS4 | 4.6 | Histidinol dehydrogenase |
| YLR155C | ASP3A | 4.6 | l-Asparaginase II |
Note that 35 ORFs fall into multiple categories.
Categories derived from MIPS database (Munich Information Center for Protein Sequences) (31).
Only ORFs induced >4.5 are listed. The remaining can be found on the World Wide Web at www.sph.harvard.edu/geneexpression.
ORFs falling into two categories.
ORFs known to be involved in resistance to DNA-damaging agents (31).
Predicted function based on sequence similarity.
Hybridization intensity in the untreated sample was below a certain threshold and was therefore increased to an arbitrary, low value for the purposes of this calculation. Values are therefore approximate.
ORFs falling into three categories.
ORFs falling into four categories.
Table 2.
ORFs whose transcripts are repressed by >3.0-fold by MMS (n = 76*)
| ORF | Gene | Fold | Function† |
|---|---|---|---|
| Ribonucleotide synthesis (7 ORFs) | |||
| YBL039C | URA7 | ∼11.6‡ | CTP synthase 1 |
| YNL141W | AAH1 | ∼9.4 | Adenosine deaminase |
| YDR399W | HPT1 | 8.5 | Hypoxanthine-guanine phosphoribosyltransferase |
| YML056C | — | 8.2 | IMP dehydrogenases¶ |
| YHR128W | FUR1 | 4.8 | Uracil phosphoribosyltransferase |
| YKL216W | URA1 | 3.4 | Dihydroorotate dehydrogenase |
| YMR217W | GUA1 | 3.4 | GMP synthase |
| rRNA synthesis (11 ORFs) | |||
| YNL112W | DBP2 | 18.1 | ATP-dependent RNA helicase |
| YGR159C | NSR1 | 7.9 | Nuclear localization sequence binding protein |
| YDL014W | NOP1 | 7.1 | Fibrillarin |
| YGL078C | DBP3 | 4.5 | ATP-dependent RNA helicase |
| YJR063W | RPA12 | 4.5 | RNA polymerase I, 13.7 kDa |
| YPL211W | NIP7 | 4.4 | 60S ribosome subunit biogenesis |
| YHR089C | GAR1 | 4.0 | Nucleolar rRNA processing |
| YOR310C | NOP5 | 3.8 | Nucleolar protein |
| YNL248C | RPA49 | 3.5 | RNA polymerase A, 46 kDa |
| YLR197W | SIK1 | 3.5 | Pre-rRNA processing |
| YNL113W | RPC19 | 3.1 | RNA polymeras I, III, 16 kDa |
| Ribosomal proteins (13 ORFs) | |||
| YEL026W§ | — | 6.2 | HMG-like protein Nhp2p¶ |
| YDL208W§ | NHP2 | 4.7 | HMG-like nuclear protein |
| YBR048W | RPS11B | 4.4 | Ribosomal protein S11B |
| YER131W | RPS26B | 3.9 | Ribosomal protein S26B |
| YKL009W | MRT4 | 3.8 | Acidic ribosomal protein PO¶ |
| YML026C | RPS18B | 3.8 | Ribosomal protein S18B |
| YNL301C | RPL18B | 3.8 | Ribosomal protein L18B |
| YLR009W | — | 3.5 | Ribosomal protein L24.e.B¶ |
| YDL130W | RPP1B | 3.4 | Ribosomal protein P1B |
| YLR048W | RPS0B | 3.4 | Ribosomal protein S0B |
| YPL198W | RPL7B | 3.3 | Ribosomal protein L7B |
| YEL054C | RPL12A | 3.0 | Ribosomal protein L12A |
| YDR025W | RPS11A | 3.0 | Ribosomal protein S11A |
| Phosphate regulation (2 ORFs) | |||
| YBR092C | PHO3 | 12.4 | Acid phosphatase, constitutive |
| YML123C | PHO84 | 5.3 | Inorganic phosphate transporter |
| Chromatin arrangement (5 ORFs) | |||
| YBL002W | HTB2 | 8.1 | Histone H2B.2 |
| YBL003C | HTA2 | 7.2 | Histone H2A.2 |
| YEL026W§ | — | 6.2 | HMG-like protein Nhp2p¶ |
| YDL208W§ | NHP2 | 4.7 | HMG-like nuclear protein |
| YDR225W | HTA1 | 4.2 | Histone H2A |
| Others (21 ORFs) | |||
| YJR047C | ANB1 | 9.5 | Translation initiation factor |
| YOR095C | RKI1 | 8.9 | d-Ribose-5-phosphate ketol-isomerase |
| YDL037C | — | 7.4 | Similarity to glucan 1,4-α-glucosidase |
| YNL111C | CYB5 | 5.8 | Cytochrome b5 |
| YMR290C | HAS1 | 5.7 | RNA-dependent helicase |
| YLR180W | SAM1 | 5.0 | s-Adenosylmethionine synthetase |
| YER043C | SAH1 | 4.4 | s-Adenosyl-l-homocysteine hydrolase |
| YNR053C | — | 4.3 | Tumor-associated autoantigen¶ |
| YDL051W | YLA1 | 4.2 | RNA binding protein |
| YLR045W | ERG3 | 4.2 | C-5 sterol desaturase |
| YDR144c | MKC7 | ∼3.9 | Aspartyl protease |
| YDL181W | INH1 | ∼3.8 | ATPase inhibitor |
| YJR048W | CYC1 | 3.7 | Cytochrome c isoform 1 |
| YNL289W | PCL1 | ∼3.5 | Cyclin, G1/S-specific |
| YKL081W | TEF4 | 3.4 | Translation elongation factor |
| YAL025C | MAK16 | 3.3 | Nuclear viral propagation protein |
| YCR034W | FEN1 | 3.3 | Probable β-1,3-glucan synthase |
| YJL121C | RPE1 | 3.3 | d-Ribulose-5-phosphate 3-epimerase |
| YDR044W | HEM13 | 3.2 | Coproporphyrinogen III oxidase |
| YGL055W | OLE1 | 3.0 | Stearoyl-CoA desaturase |
| YLR372W | SUR4 | 3.0 | Sterol isomerase |
| Unknown (19 ORFs) | |||
| These ORFs and their fold induction can be found on the World Wide Web at www.hsph.harvard.edu/geneexpression. | |||
Note that two ORFs fall into multiple categories.
Categories derived from MIPS database (Munich Information Center for Protein Sequences) (31).
Hybridization intensity in the untreated sample was below a certain threshold and was therefore increased to an arbitrary, low value for the purposes of this calculation. Values are therefore approximate.
ORFs falling into two categories.
Predicted function based on sequence similarity.
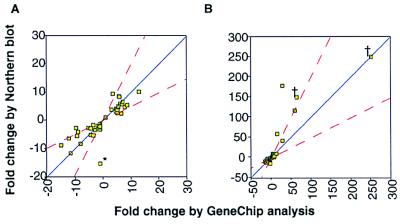
To establish that the information obtained from GeneChip analysis is accurate, we chose 50 ORFs to examine by conventional Northern-blot analysis; these included 26 inducible (3.1- to 251-fold), 16 repressible (3.1- to 18.1-fold), and 8 nonresponsive ORFs. Freshly isolated RNA from control and from MMS-treated cells was probed with each of the 50 ORFs. Hybridization signals, as measured by Northern-blot and GeneChip analysis, are shown for six of the ORFs (Fig. 2), and data for all 50 ORFs are compiled in Fig. 3 A and B. In terms of whether the ORFs were inducible, repressible, or nonresponsive, the Northern-blot and GeneChip data agreed for 48 of the 50 ORFs. Moreover, the data agreed remarkably well in terms of the extent of induction or repression (Fig. 3 A and B), displaying a correlation coefficient of 0.79 for the complete data set (Fig. 3B). For the majority of the ORFs, the fold change in transcript levels differed by no more than a factor of two between the Northern-blot and GeneChip data (Fig. 3A). However, for very highly induced ORFs (see Fig. 3B), the correlation weakens slightly, such that the Northern-blot analysis overestimates induction, the GeneChip analysis underestimates induction, or both. Note that for some highly induced ORFs, basal transcript levels are undetectable, making it difficult to calculate accurate fold-induction values. Further, some quantitative differences between GeneChip and Northern-blot data may not be unexpected given that the GeneChip analysis monitors hybridization of fragmented cRNAs to 20 oligonucleotide pairs per ORF (chosen for their uniqueness relative to the entire S. cerevisiae genome) and given that Northern blotting monitors hybridization of a complete ORF to immobilized full-length RNA transcripts separated on the basis of size. GeneChip analysis has the potential to discriminate between closely related genes, whereas Northern-blot analysis suffers from the potential that an ORF probe might hybridize to closely related RNAs. Indeed, the data point marked with an asterisk in Fig. 3A highlights this problem for the PHO3 and PHO5 genes. By GeneChip analysis, PHO3 was repressed 13-fold; PHO5 was scored as nonresponsive, because it was undetectable in both control and MMS-treated cells. One plausible explanation for PHO5 being undetectable by GeneChip analysis is that weak hybridization is caused by incorrect primer design. However, by using genomic DNA as a probe, the PHO5 gene can be detected on these chips (16). Because PHO3 and PHO5 have 87% nucleotide-sequence identity and are of similar size, PHO5 scored as repressible (15-fold) by Northern-blot analysis, presumably because the PHO5 probe hybridized to the PHO3 transcript.
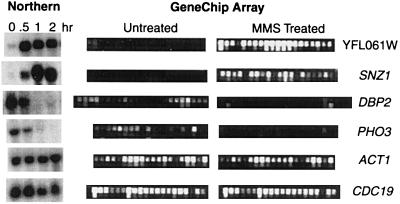
Figure 2.
Verification of GeneChip data by conventional Northern-blot analysis. Northern blots were prepared with total RNA isolated from untreated and 0.1% MMS-treated S. cerevisiae DBY747 log-phase cells grown at 30°C in rich media; RNA was from cells exposed to MMS for the indicated times. Blots were probed with 50 ORFs; 6 of them are shown, namely two induced [YFL061W (251-fold) and SNZ1 (≈65.8-fold)], two repressed [DBP2 (18.1-fold) and PHO3 (12.3-fold)] and two nonresponsive ORFs [CDC19 and ACT1], as determined by GeneChip analysis. The GeneChip data in this figure are taken from the experiment in Fig. 1. Each ORF is represented on the Ye6100 array by ≈20 oligonucleotide pairs. One member of each pair corresponds to a perfectly matched sequence from the ORF (top row); the other pair member contains a single-base mismatch (bottom row). The difference in intensity between the perfectly matched and the mismatched sequences is used to calculate an average intensity difference for each ORF.
Figure 3.
Correlation of GeneChip and Northern-blot data. The correlation between ORF induction and repression values (in response to MMS) obtained from the GeneChip analysis and Northern-blot data are compared for 50 ORFs. Northern-blot hybridization levels were determined with a Bio-Rad Molecular Imager, and the GeneChip data are from the experiment in Fig. 1. (A) Data plots for genes showing less than a 20-fold change in transcript level (by GeneChip). (B) Data plots for all 50 ORFs. The ORFs were selected to represent the entire range of change (as indicated by the GeneChip data) with changes from a 19-fold decrease to a 251-fold increase. The names and Northern-blot values are YFL061W (>150), SNZ1 (>150), GTT2 (180.0), YKL071W (118.0), SNO1 (60.0), YNR065C (42.0), YIL165C (17.3), SHM1 (9.8), ARG11 (8.7), GIN3 (8.6), ASP3 (6.7), NPL4 (6.1), RNR3 (6.0), ECM29 (5.5), YLR080W (5.3), HIS5 (5.1), YOR227W (5.1), MAG1 (5.0), NTH1 (4.5), YPR1 (4.0), HOM3 (4.0), LAG2 (3.5), CTK3 (3.0), YGR130C (2.8), LHE1 (1.4), YJL131C (1.2), SSA1 (1.0), PHO5 (−15.2), URA7 (−11.4), DBP2 (−8.7), NSB1 (−8.2), PHO3 (−6.2), AAH1 (−5.3), GUA1 (5.1), SAH1 (−5.0), RKI1 (−4.0), YDL213C (−3.1), RPS16A (−3.0), NIP7 (−2.8), YIL158W (2.6), ANB1 (−2.5), TCM1 (−2.3), ENO1 (−2.2), INH1 (−2.0), RPL16A (−2.0), SPE2 (−1.7), CDC19 (−1.6), RPL17 (−1.6), YLR009W (−1.4), and ACT1 (−1.1). The GeneChip values are in Table 1. Dashed red lines represent a factor-of-two difference from a perfect match between Northern-blot and GeneChip analysis. The asterisk in A represents the PHO5 ORF (discussed in Results and Discussion) and † marks transcripts that were undetectable by Northern-blot analysis in untreated cells and whose fold-induction is likely to be less accurate. Note that these points were not included in the calculation of the correlation coefficient.
Transcripts Induced by MMS.
Several types of defense mechanism would be expected to protect cells against an alkylating agent such as MMS, namely DNA repair and recombination, cell-cycle checkpoints, and pathways that somehow prevent alkylating agents from reacting with target molecules (e.g., by changing the cell wall, membrane permeability, or drug metabolism). The 325 ORFs that were induced greater than 4-fold by MMS are listed in Table 1; 4-fold was arbitrarily chosen as the cutoff and is more conservative than the cutoff (2-fold) recommended by Affymetrix. The first six groups of genes (stress response/detoxification, DNA repair/replication, cell cycle, signal transduction, cell wall biogenesis, and membrane transport) are not unexpected, because they could well be providing resistance to a chemical that damages DNA. However, with the exception of a few genes (e.g., MAG1), it remains to be determined whether and how each gene plays a protective role. Several other groups of genes are more difficult to rationalize, including those for nitrogen and sulfur metabolism, carbohydrate metabolism/fermentation, mRNA processing, “others,” and the largest group of MMS-inducible transcripts, 112 ORFs with no known biological function and no sequence similarity to known proteins (see Table 1).
In addition to finding inducible genes potentially involved in repairing, avoiding, or preventing DNA damage, we observed the induced expression of 15 genes involved in protein degradation. In fact, when we relaxed the induction criterion from 4-fold to 2.0-fold (intensity changes greater than 2.0 are considered significant), we observe that 91 of the 143 known protein degradation genes are induced after exposure to this relatively nontoxic MMS dose. Proteins are known substrates for MMS alkylation (20); the fact that genes involved in protein degradation are up-regulated in response to MMS suggests that alkylated proteins may be targeted for degradation and that the elimination of alkylated proteins may be important for cellular recovery. The selective removal of chemically damaged proteins is not unprecedented; recently, it was shown that oxidized proteins are targeted for ubiquitin-mediated degradation in eukaryotic cells (21, 22). It seems logical that cells must replace proteins that were degraded, and evidence for new protein synthesis is suggested by the increased expression of 41 genes involved in amino acid biosynthesis (Table 1). In fact, 91 of the known 194 ORFs involved in amino acid biosynthesis genes are induced greater than 2.0-fold. These data suggest that, in addition to inducing genes to promote recovery from DNA damage, cells also induce genes to promote recovery from protein damage. The relative importance of each pathway remains to be determined, and it should be noted that pathways that prevent DNA alkylation damage may also prevent protein alkylation damage.
Transcripts Repressed by MMS.
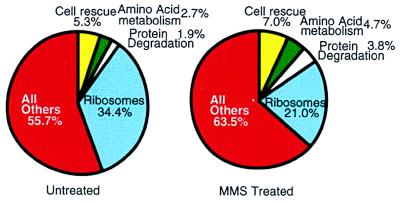
The 76 genes that are repressed after MMS exposure are listed in Table 2. The most notable groups include those involved in nucleotide and RNA synthesis and in the synthesis and assembly of ribosomal proteins. Repression of RNA synthesis and ribosomal genes might suggest that cells down-regulate de novo transcription and de novo protein synthesis in response to MMS, a suggestion that counters our proposal that MMS induces a program to degrade and replace alkylated proteins. However, an analysis of the global expression of S. cerevisiae genes might explain the apparent contradiction. In exponentially growing S. cerevisiae cells, ribosomal protein genes produce over one-third of the total poly(A)+ mRNA (Fig. 4); indeed, 75 of the 100 most highly transcribed genes in S. cerevisiae are ribosomal protein genes. These data are consistent with previous GeneChip and serial analysis of gene expression (16, 23). Overall, MMS treatment induces a modest decrease in the expression of all the ribosomal protein genes (average of 1.7-fold), such that after MMS exposure still one-fifth of all transcripts encode ribosomal proteins (Fig. 4). It seems likely that the modest reduction in ribosomal protein gene expression allows energy to be reshuffled for the increased expression of genes involved in protective responses, while maintaining a basal protein synthesis capacity (Fig. 4). Moreover, a transient but slight decrease in the manufacture of new ribosomes may serve to slow the global production of proteins until alkylation exposure is diminished. There may even be preferential translation of MMS-induced transcripts to promote recovery further, and such preferential translation is not unprecedented (24–26). It should be noted that not all ribosomal protein transcripts are repressed and that the mitochondrial ribosomal protein transcripts actually show a slight increase. (The complete data set can be found at www.hsph.harvard.edu/geneexpression.) In addition to the repression of transcripts involved in nucleotide, RNA, and protein synthesis, several other genes are repressed (Table 2), and as with the induced transcripts, ORFs of unknown function represent a large group.
Figure 4.
Changes in the ratios of mRNA subpopulation in response to MMS. The fraction of poly(A)+ mRNA transcripts from genes related to ribosomes (blue), protein degradation (white), amino acid metabolism (green), cell rescue (yellow), and all others (red) was calculated for untreated and MMS-treated cells by using the GeneChip hybridization data. Hybridization intensities are directly related to absolute poly(A)+ mRNA level (16). Therefore, hybridization intensities of a given mRNA reflect the abundance of a given mRNA relative to the total amount of mRNA. Genes are grouped into these categories according to the S. cerevisiae Genome Database and the Munich Information Centre for Protein Sequences (30, 31).
Recently, changes in global transcript levels during the cell-cycle progression of S. cerevisiae were reported (27). Of the 421 genes determined to have cell-cycle periodicity, only a small fraction (24 genes) are responsive to MMS, suggesting that changes in expression level after MMS treatment are not caused simply by changes in cell-cycle progression. Although over 60% of characterized genes showing cell-cycle periodicity have already been implicated in cell-cycle specific roles, very few of the characterized genes responsive to MMS are known to be involved in MMS resistance. One interpretation is that a majority of the responsive genes are not directly involved in the response to alkylation damage; another is that our study has shown that there are many more genes involved than previously known. The induction by another DNA-damaging agent of a large number of S. cerevisiae proteins involved in many different cellular processes was reported recently (28). Evidence obtained from two-dimensional gel electrophoresis indicated that at least 115 proteins are stimulated by H2O2 and that at least 52 are repressed. Of the inducible proteins, 71 have been identified, and previously only 12 were known to act directly in antioxidant defense. The other proteins include heat shock proteins; enzymes for carbohydrate metabolism, amino acid biosynthesis, and protein degradation; and a number of unclassified proteins or proteins with no known function. The induction of a large number of seemingly unrelated proteins after H2O2 exposure parallels the results presented here for transcript induction after MMS exposure.
We have made three major points. (i) The number of yeast genes known to be induced by a DNA-damaging agent has increased by at least 15-fold. (ii) A large number of genes are also repressed. (iii) There is evidence for the initiation of a program to eliminate and replace alkylated proteins from the cell. Whether all of the MMS-inducible and MMS-repressible ORFs listed in Tables 1 and 2 contribute significantly to protecting cells against alkylating agents must now be tested. For a few, their role is already well established, but for most, their role in MMS-resistance remains to be determined. The anticipated availability of thousands of new S. cerevisiae strains with null mutations in all nonessential ORFs (29) and with regulated expression of all the ORFs would be essential for such an analysis. Identification and characterization of the regulatory mechanisms for each regulon that contributes to the global response would be particularly important, and it would be important to establish the relative contributions of each type of protective mechanism in ameliorating alkylating-agent toxicity. It would be particularly interesting to determine how critical the turnover of damaged proteins is for the recovery of cells from alkylation damage.
Acknowledgments
We thank Drs. Armen Tashjian, Evelyn Waldstein, Graham Walker, and John Cairns for useful comments on the manuscript. We also thank Affymetrix for developing such outstanding technology and their Academic User Center Program for making this study possible. This work was supported by National Institutes of Health Grant CA55042 and by Harvard School of Public Health institutional funds. S.A.J. was supported by National Institutes of Health Training Grant CA09078, and L.S. is a Burroughs Wellcome Toxicology Scholar. The Affymetrix User Center is partially supported by National Institutes of Health Grant P01HG01323.
ABBREVIATIONS
- MMS
methyl methanesulfonate
- SSPE-T
sodium chloride/sodium phosphate/EDTA/Triton
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Friedberg E, Walker G, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Kiser G L, Weinert T A. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrlich P, Blattner C, Knebel A, Bender K, Rahmsdorf H J. Biol Chem. 1997;378:1217–1229. doi: 10.1515/bchm.1997.378.11.1217. [DOI] [PubMed] [Google Scholar]
- 4.Zhan Q, Carrier F, Fornace A J., Jr Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fornace A J, Jr, Alamo I, Jr, Hollander M C. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachant J B, Elledge S J. In: DNA Repair in Prokaryotes and Lower Eukaryotes, DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Totowa, NJ: Humana; 1998. [Google Scholar]
- 7.McClanahan T, McEntee K. Mol Cell Biol. 1984;4:2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby S W, Szostak J W. Mol Cell Biol. 1985;5:75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Derfler B, Maskati A, Samson L. Proc Natl Acad Sci USA. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Derfler B, Samson L. EMBO J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Samson L. Nucleic Acids Res. 1991;19:6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W, Singh K K, Chen B, Samson L. Mol Cell Biol. 1993;13:7213–7221. doi: 10.1128/mcb.13.12.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh K K, Samson L. Proc Natl Acad Sci USA. 1995;92:4907–4911. doi: 10.1073/pnas.92.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweet D H, Jang Y K, Sancar G B. Mol Cell Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt M E, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Cheng J, Sheldon E L, Wu L, Uribe A, Gerrue L O, Carrino J, Heller M J, O’Connell J P. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 20.Boffa L C, Bolognesi C. Carcinogenesis. 1985;6:1399–1401. doi: 10.1093/carcin/6.9.1399. [DOI] [PubMed] [Google Scholar]
- 21.Grune T, Reinheckel T, Davies K J A. J Biol Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- 22.Shang F, Gong X, Taylor A. J Biol Chem. 1997;272:23086–23093. doi: 10.1074/jbc.272.37.23086. [DOI] [PubMed] [Google Scholar]
- 23.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landers J E, Cassel S L, George D L. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- 26.Schmechel S, Chute M, Skinner P, Anderson R, Schiff L. Virology. 1997;232:62–73. doi: 10.1006/viro.1997.8531. [DOI] [PubMed] [Google Scholar]
- 27.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, et al. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 28.Godon C, Lagniel G, Lee J, Buhler J M, Kieffer S, Perrot M, Boucherie H, Toledano M B, Labarre J. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 30.Cherry J M, Adler C, Ball C, Chervitz S A, Dwight S S, Hester E T, Jia Y, Juvik G, Roe T, Schroeder M, et al. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mewes H W, Albermann K, Heumann K, Liebl S, Pfeiffer F. Nucleic Acids Res. 1997;25:28–30. doi: 10.1093/nar/25.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]