Abstract
The human immunodeficiency virus (HIV) replicates its genome and mutates at exceptionally high rates. As a result, the virus is able to evade immunological and chemical antiviral agents. We tested the hypothesis that a further increase in the mutation rate by promutagenic nucleoside analogs would abolish viral replication. We evaluated deoxynucleoside analogs for lack of toxicity to human cells, incorporation by HIV reverse transcriptase, resistance to repair when incorporated into the DNA strand of an RNA⋅DNA hybrid, and mispairing at high frequency. Among the candidates tested, 5-hydroxydeoxycytidine (5-OH-dC) fulfilled these criteria. In seven of nine experiments, the presence of this analog resulted in the loss of viral replicative potential after 9–24 sequential passages of HIV in human CEM cells. In contrast, loss of viral replication was not observed in 28 control cultures passaged in the absence of the nucleoside analog, nor with other analogs tested. Sequence analysis of a portion of the HIV reverse transcriptase gene demonstrated a disproportionate increase in G → A substitutions, mutations predicted to result from misincorporation of 5-OH-dC into the cDNA during reverse transcription. Thus, “lethal mutagenesis” driven by the class of deoxynucleoside analogs represented by 5-OH-dC could provide a new approach to treating HIV infections and, potentially, other viral infections.
Current therapies for the treatment of HIV infection include combinations of inhibitors of the viral reverse transcriptase (RT) and protease. Drugs that target the viral RT either are nucleosides that terminate viral DNA synthesis, such as zidovudine (ZDV), dideoxyinosine (ddI), and dideoxycytidine (ddC), or are nonnucleoside analogs that bind to a hydrophobic cavity adjacent to the polymerase active site, such as nevirapine (1). Unfortunately, the rapid evolution of HIV in vivo results in the emergence of viruses resistant to each of these agents. Combination therapy involving RT and protease inhibitors has been very successful in reducing viral loads and, in principle, should reduce the outgrowth of resistant viruses. Even this approach, however, is limited by drug availability, patient compliance, and the likelihood that virus populations harbored by treated individuals will eventually develop drug resistance.
The development of HIV resistance to host immunity or chemotherapy results both from the high replication rate of the virus and from the infidelity of the HIV RT. HIV-1-infected individuals produce approximately 1010 virions per day (2), and the HIV RT produces one error per 2,000–5,000 nucleotides polymerized (3–7). As a result, HIV genomes within an infected individual do not exist as a homogeneous nucleotide sequence, but rather as a “quasispecies” (8, 9), an ensemble of related genomes in which selection operates at the level of the structure of the population. Within the quasispecies are drug-resistant viruses that are present early in infection before exposure to a drug, as well as mutant viruses that can easily acquire additional mutations that render them drug resistant (10).
The exceptionally high rate of mutagenesis of RNA viruses (11, 12), coupled with the finding that most HIV virions in the blood appear to be nonviable (13), suggest that the HIV genome is unable to tolerate many additional mutations without a loss of viability. Thus, even a small increase in mutation rate might result in the virus population reaching an error threshold beyond which the population cannot be sustained because of a loss of viral replication capacity and infectivity. Here, we examined the capacity of mutagenic deoxynucleoside analogs to modify the fidelity of replication of HIV during sequential passages in culture and to induce lethal mutagenesis.
MATERIALS AND METHODS
Serial Transfer Experiments.
Stock preparations of HIV-1LAI containing approximately 106 infectious units/ml were titered by syncytium induction (14). HIV was added at a multiplicity of 0.01 to 1-ml aliquots of medium containing 2 × 105 CEM cells that were previously incubated for 1 hr with or without different deoxynucleoside analogs. After 4 hr at 37°C, the infected cells were washed twice with PBS (without Mg2+ or Ca2+) and resuspended in 1 ml of medium with or without the analog in a 48-well plate. Half the volume of the medium with fresh analog was replenished at 2 days. After 4–6 days, the indicated amount of supernatant was transferred to fresh cells that were preincubated with or without analog for 1 hr. This procedure was iterated for the indicated number of cycles; aliquots of both cells and supernatants were frozen at each passage. Virus production was monitored by measuring HIV p24 core antigen in culture supernatants with the Abbott Antigen ELISA Kit. All experiments were carried out with a double-blind protocol.
Insertions of dNTPs by HIV RT.
A 5′-32P-end-labeled 15-mer DNA primer was hybridized to the 3′ end of a 46-mer DNA template containing either dG or dA at position 16 from its 3′ end. The reaction mixture contained 25 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 40 mM KCl, 2 mM DTT, 0.1 mg/ml BSA, 50 nM template-primer, and the stated amount of either dCTP or its 5-hydroxy analog 5-OH-dCTP. The concentration of HIV RT was adjusted such that the reaction was linear with time and <20% of the primer was extended. After incubation at 37°C, the reactions were terminated by the addition of an equal volume of denaturing sample buffer. The samples were boiled and the 16-mer product was resolved from the 15-mer substrate by electrophoresis through a 14% polyacrylamide/urea gel. The amount of product generated was quantitated by PhosphorImager (Molecular Dynamics) analysis and the kinetic constants were calculated from Hanes–Woolf plots (15).
Nucleoside Analogs.
The nucleoside analogs initially tested included the following compounds. 5-Hydroxydeoxycytidine (5-OH-dC) is formed in DNA on exposure to reactive oxygen species (16). 5-OH-dCTP is incorporated into DNA by DNA polymerases and HIV RT (17, 18) and yields predominantly G⋅C → A⋅T substitutions, although other mispairings have been observed. When present in double-stranded DNA, 5-OH-dC is excised by Escherichia coli endonuclease III (Nth protein) or formamidopyrimidine-DNA glycosidase (19, 20) or by homologous enzymes in eukaryotic cells (21). In contrast, when present in an RNA⋅DNA hybrid, 5-OH-dC is resistant to digestion (22). O4-Methyl-dTTP is incorporated efficiently into DNA by a number of DNA polymerases, including RTs (23, 24). Incorporation into HIV cDNA followed by methyltransferase-mediated DNA repair would yield G → A substitutions (25). Even though there is evidence for repair of O4-methyl-dT by several pathways (26, 27), the rate of removal from rat liver DNA (t1/2 of 20–60 hr) is very slow. O6-Methyl-dG is produced in DNA by alkylating agents and base-pairs with thymidine at high frequency (28–30), and the triphosphate is incorporated into DNA by HIV RT as well as by DNA polymerases. While O6-methyl-dG residues are repaired in double-stranded DNA by O6-methylguanine-DNA methyltransferase (31), repair is inefficient within a DNA⋅RNA duplex (22). 8-Amino-dG is an inhibitor of purine nucleoside phosphorylase (32) and has been evaluated for toxicity as a potential chemotherapeutic agent (33). It is incorporated efficiently by HIV RT (34) and can be excised from DNA by formamidopyrimidine-DNA glycosidase (22). Last, 8-oxo-dG was included on the basis of its mutagenic potency, incorporation by HIV RT, and the multiple mechanisms for its excision from DNA in mammalian cells (35).
Toxicity was assessed by culturing CEM cells in graded amounts of each analog for 15 days. Every 2 days, medium was replaced with and without the analog. Viability was determined by counting both the total number of cells and the percent that exclude trypan blue after each transfer.
Cloning and Sequencing of Proviral DNA.
Cells were lysed and viral DNA was amplified by nested PCR using Pfu DNA polymerase (36, 37). The outer primers were complementary to nucleotides spanning positions 2127–2156 and 2990–3020 in the HIV RT gene (LAV-1, K0213⋅gb-vi); amplification consisted of 30 cycles. A 1-μl aliquot of each reaction mixture was diluted into a second 50-μl PCR mixture containing inner primers corresponding to nucleotides 2281–2318 and 2881–2916 and amplified for an additional 20 cycles. The amplified DNA was ligated into the PCR II-TOPO TA cloning vector (Invitrogen). The nucleotide sequence of the HIV inserts was determined by sequencing both strands, using forward and reverse primers complementary to the adjacent M13 DNA.
RESULTS
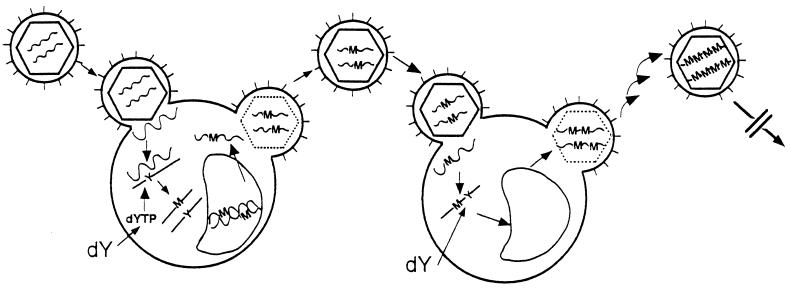
Lethal mutagenesis results from a progressive accumulation of mutations in the HIV genome because of the incorporation of mutagenic deoxynucleoside analogs during each round of viral replication (Fig. 1). The analog is taken up by cells as a nucleoside and is phosphorylated by cellular kinases to the corresponding deoxynucleoside triphosphate. The mutagenic nucleoside triphosphate then can be incorporated during HIV RNA template-directed synthesis of the minus DNA strand. We postulate that mutagenesis would occur more frequently in the HIV genome because of the incorporation of a mutagenic analog into an RNA⋅DNA hybrid for the following reasons. First, reverse transcription occurs in the cytoplasm, whereas repair of cellular DNA is a nuclear process. Second, DNA repair enzymes have evolved to utilize double-stranded DNA that is present in a B-type structure, whereas RNA⋅DNA hybrids are in an A-type structure (38). In particular, E. coli DNA methyltransferase, formamidopyrimidine-DNA glycosidase, and endonuclease III fail to repair efficiently altered substrates when present in the DNA strand of an RNA⋅DNA hybrid (22). As a result, altered nucleotide residues in the DNA strand are not excised, and they pair with noncomplementary nucleotides.
Figure 1.
Scheme for the progressive accumulation of mutations in HIV RNA as a result of repetitive infection of host cells. dY and dYTP refer to nucleoside and nucleoside triphosphate analogs. Y and M are analogs and mutations, respectively, present in the HIV genome. The wavy lines represent viral RNA and the solid lines represent viral DNA.
Upon synthesis of a double-stranded viral DNA intermediate, the mutations are fixed; excision of the altered nucleotide would not obliterate the mutation. In contrast, incorporation of the analog into the host cell genome is subject to removal during DNA repair. After integration of the double-stranded viral DNA containing the mutation into the host genome, transcription results in corresponding base substitutions in the viral RNA. Iteration of this process by sequential HIV infections of host cells will result in the progressive accumulation of mutations throughout the HIV genome; some of these mutations would diminish viral replication and fitness. Eventually the mutations would exceed the error threshold for maintenance of the quasispecies, resulting in a precipitous decline in viral replication (39).
On the basis of the above considerations, we envision that a small increase in mutagenesis may be adequate to abolish viral replication, the resultant mutants should be predictable from the base-pair properties of the nucleoside analog, and escape variants should contain few mutations and continue to be susceptible to lethal mutagenesis upon further replication in the presence of the analog.
Screening of Analogs.
We standardized a protocol for passaging HIV (HIV-1LAI) during the peak of virion production that surrounds the acute cytopathic phase of infection. Viral replication was assessed by measuring p24 in culture medium after growth of human CEM cells for 4–6 days in the presence or absence of different mutagenic deoxynucleoside analogs. Five deoxynucleoside analogs initially were selected for studying lethal mutagenesis; the rationale for their selection is presented in the legend to Table 1. The most efficacious analogs would be those that diminish HIV replication by increasing the number of mutations to the point of nonviability and yet are not toxic to human cells.
Table 1.
Testing of nucleoside analogs
| Analog | Conc., μM | Sequential transfers | Mutations/nucleotides sequenced | % Mutations |
|---|---|---|---|---|
| 5-Hydroxydeoxycytidine (5-OH-dC) | 1,000 | 7 | 31/7,379 | 0.92 |
| O4-Methylthymidine (O4-methyl-dT) | 1,000 | 10 | 109/8,202 | 1.33 |
| O6-Methyldeoxyguanosine (O6-methyl-dG) | 1 | 7 | 12/3,997 | 0.31 |
| 8-Aminodeoxyguanosine (8-amino-dG) | 0.5 | 7 | 9/3,378 | 0.27 |
| 8-Oxodeoxyguanosine (8-oxo-dG) | 10 | 7 | 3/2,252 | 0.12 |
| Controls | (0) | 7 | 29/10,097 | 0.29 |
The concentrations studied were those that failed to reduce growth of CEM as determined by cells that excluded trypan blue. After the indicated number of serial transfers, the proviral DNA was cloned and sequenced. The number of mutations and the number of nucleotides sequenced are given.
Studies were first carried out to establish nontoxic concentrations of each of the candidate analogs. The concentration used was 1 mM; if toxic, the concentration used was 1/10 of that which reduced growth by >30%. After seven sequential passages, a segment of HIV RT was amplified from the cellular DNA, cloned in E. coli, and sequenced (Table 1). After this limited number of transfers, viral titer was not significantly reduced. An increase in mutations per nucleotide sequenced was observed only with O4-methyl-dT and 5-OH-dC. In the case of O4-methyl-dT, multiple mutations were observed in only a few of the clones sequenced, and viral replication was not diminished even after 24 passages (data not shown). With 5-OH-dC, however, mutations were present in many clones. In addition, exposure of human cells to 4 mM 5-OH-dC reduced cell proliferation by less than 30%. On the basis of these findings, we selected 5-OH-dC for further studies.
Incorporation of 5-OH-dCTP by HIV RT.
The ability of 5-OH-dCTP to substitute for dCTP was assessed under steady-state conditions by measuring single nucleotide additions (15). A labeled DNA primer was hybridized to a series of DNA templates, each of which contained a different nucleotide immediately downstream from the 3′-OH primer terminus. The primers were extended by HIV RT in the presence of either dCTP or 5-OH-dCTP, resolved on denaturing gels, and quantitated (Table 2). 5-OH-dCTP efficiently substitutes for dCTP. When 5-OH-dCTP was introduced opposite a template G, the Km for the incorporation of a single molecule by HIV RT is only 5-fold more than for incorporating the complementary dCTP and the Vmax is approximately identical. The mispairing properties based on Km values indicate that 5-OH-dC is most likely to base pair with the noncomplementary A residues. This preference for A residues is in accord with the types of mutations formed in copying templates containing residues of 5-OH-dC (18, 19, 40). Thus, 5-OH-dC should substitute for dCTP opposite template dGs and miscode for A during subsequent replication of the altered DNA.
Table 2.
Kinetic constants for single nucleotide addition of dCTP and 5-OH-dCTP by HIV RT
| Template nucleotide | dCTP
|
5-OH-dCTP
|
||
|---|---|---|---|---|
| Km, μM | Vmax, min−1 | Km, μM | Vmax, min−1 | |
| dG | 0.84 ± 0.16 | 540 ± 180 | 5 ± 2 | 1700 ± 280 |
| dA | 1800 ± 850 | 94 ± 13 | 8 ± 3 | 1.2 ± 1.5 |
| dT | 1800 | 130 | 810 | 11 |
| dC | 1700 | 0.43 | 130 | 0.05 |
Insertion of dCTP and 5-OH-dCTP was determined by measuring the rate of single nucleotide extension. The rates of incorporation of 5-OH-dCTP were normalized to reflect the concentration of enzyme in the reaction with dCTP. Km values for dCTP >1 mM are extrapolations from Hanes–Woolf plots. Experiments with dG and dA as the next template nucleotide were carried out on four separate occasions and the results are stated ± one standard deviation. Experiments with dT and dC are the averages of duplicate determinations.
Serial Transfer Experiments.
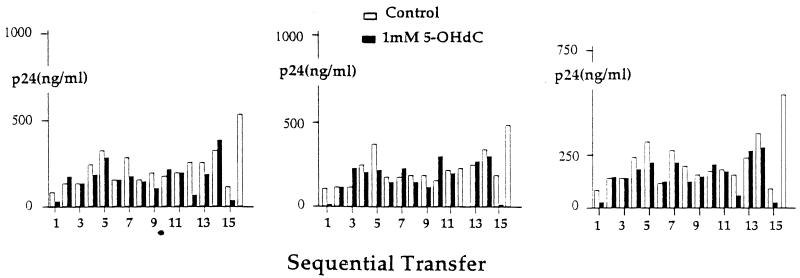
Modulations of viral production in culture supernatants over the course of 18 transfers of HIV in the absence or presence of 1 mM 5-OH-dC are shown in Fig. 2. In this experiment, viral antigen (p24) titer was determined after each transfer and the volume of the subsequent inoculum was adjusted so that the same amounts of virus in control and 5-OH-dC-treated cells were added at each transfer step. The decrease in viral titer observed after the first passage in the presence of 5-OH-dC was not sustained. However, a sustained loss of viral titer was observed after 16 sequential transfers, but not in any of the controls. This loss in viral titer was confirmed by infecting fresh cells with frozen supernatants obtained at the 14th passage; a similar loss of p24 in 5-OH-dC-containing cultures but not in controls was observed in subsequent passages (data not shown).
Figure 2.
Production of HIV-1LAI in the course of sequential passage of cell culture supernatants in the absence or presence of 5-OH-dC. Cultures containing 2 × 105 CEM cells were maintained for 88 hr; thereafter, supernatant containing the same amount of p24 was transferred to fresh cells. Three replicates are shown.
Alterations in the nucleotide sequence of HIV-1LAI in individual genomes were determined by cloning and sequencing a 674-nucleotide segment of the HIV RT gene from viral DNA obtained at the 16th passage, immediately prior to the loss of p24 production (Fig. 2). The number of clones sequenced, the mutations per base pair, and the types of mutations observed are presented in Table 3. We used uncloned virus as our inoculum; as a result the mutations in the controls could have been present in the initial inoculum or could have arisen during serial transfer. The standard sequence used for comparison was that from a clonal isolate of HIV-1LAI, and mutations present in more than one of the control clones were excluded. HIV-1-infected control clones contained 18 single base substitutions and one deletion among 4,306 sequenced nucleotides. In a total of 28 5-OH-dC-derived clones, we detected 72 mutations among 15,060 nucleotides sequenced. In this experiment, HIV infection in the presence of 5-OH-dC did not significantly increase the overall percent of mutants/base pairs at the 16th passage. However, there was a 5.6-fold increase in G → A substitutions in the 5-OH-dC-treated cells.
Table 3.
Mutations in HIV RT
| Analog | No. of clones | Mutations/nucleotides | % | No. of
mutations
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G→A | A→G | C→T | T→C | T→A | A→C | A→T | G→T | T→G | C→A | G→C | ||||
| Controls and 5-OH-dC-induced mutations | ||||||||||||||
| Control | 8 | 18/4,306 | 0.42 | 2 | 7 | 2 | 5 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| 5-OH-dC | 28 | 72/15,060 | 0.48 | 39 | 18 | 4 | 4 | 1 | 1 | 1 | 1 | 2 | 0 | 1 |
| Controls and lethal and breakthrough mutations | ||||||||||||||
| Control | 85 | 99/47,855 | 0.21 | 34 | 30 | 15 | 19 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 5-OH-dC (lethal) | 95 | 196/53,485 | 0.37 | 94 | 26 | 33 | 27 | 1 | 7 | 3 | 2 | 0 | 2 | 1 |
| 5-OH-dC (breakthrough) | 23 | 24/12,949 | 0.18 | 4 | 13 | 0 | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 |
DNA sequence analysis of a segment of the HIV-1LAI RT gene spanning nucleotides 2193–2867. The segment of the HIV RT gene was amplified from proviral DNA obtained from frozen cells after the number of serial transfers indicated below. In the first two lines, the proviral DNA was amplified from control and 5-OH-dC-treated cells obtained at the 16th passage (see Fig. 2). In the bottom three lines, proviral DNA was obtained at passages indicated in Fig. 3. The method for amplification and cloning is similar to that described above, except that proviral DNA was purified by Isoquick Kit (Orca Research, Bothell, WA) and amplification was performed only with inner primers. The amplified DNA was ligated into the PCR II TOPO TA cloning vector (Invitrogen). The arrows in Fig. 3 indicate the cultures sampled and combined prior to obtaining viral DNA for amplification, cloning, and sequencing.
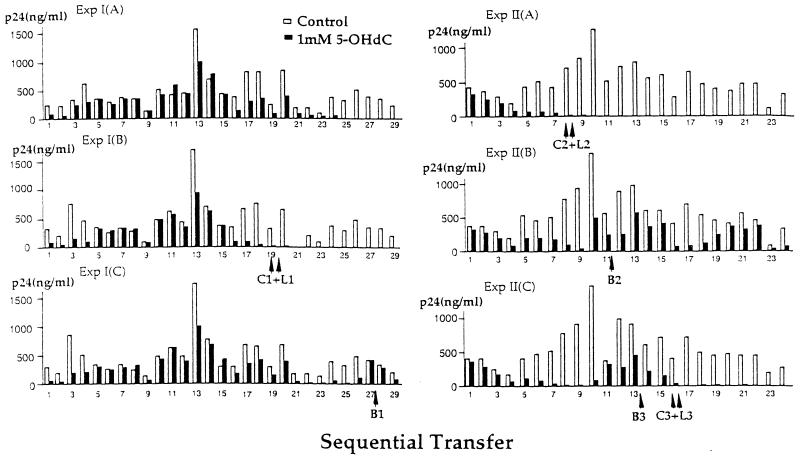
Fig. 3 shows the effect of 5-OH-dC on HIV replication after sequential transfer in two additional serial transfer experiments, each one carried out in triplicate. A constant amount of supernatant from the control and 5-OH-dC cultures was transferred at each step. In control cultures, there was considerable fluctuation in p24 titers from one infectious cycle to the next, while within an experiment, control cultures were nearly identical. In contrast, each lineage of virus was different in the 5-OH-dC-treated cultures. In four of the six 5-OH-dC-treated cultures, p24 titers in the supernatant were rendered undetectable after passages 9–24 (quantitative PCR indicated less than 10 copies of proviral DNA per 105 cells), whereas in the remaining two cultures p24 titers were still detectable after 29 and 24 passages, respectively. The loss in viral detectability was maintained for the additional 7–13 passages conducted.
Figure 3.
Sequential passage of HIV. Experiments I and II each contain data on three sets of control and 5-OH-dC-treated cultures. The arrows indicate the passage number for which aliquots were obtained for the preparation of DNA for control (C), 5-OH-dC-treated (L), and breakthrough (B) viruses. Results are expressed as the amount of p24 in culture supernatants. In Exp. I, cultures were maintained for 29 passages, and 5 μl of the supernatant was transferred at each step irrespective of the p24 titer. In experiment II, cultures were maintained for 24 passages, and 1 μl of the supernatant was transferred.
The mutation frequency and spectrum in controls, 5-OH-dC-induced lethal cultures, and putative breakthrough cultures (i.e., those demonstrating the reemergence of the virus after a diminution in titer), are presented in the bottom three lines of Table 3. The frequencies of mutations in the control and 5-OH-dC-induced lethal cultures were 0.21% and 0.37%, respectively; there was a 2.5-fold increase in G → A substitutions in the 5-OH-dC-induced lethal cultures. The second most frequent substitutions in the 5-OH-dC-treated culture were C → T, and these could have resulted from 5-OH-dCTP incorporation into plus strand DNA. The average number of mutations per clone in the control cultures was 1.16; the distribution is not significantly different from a Poisson distribution, suggesting that the mutations resulted from random events. The average number of mutations per clone in the 5-OH-dC-induced lethal cultures was 2.06, significantly different from controls at the 0.01 level using the likelihood ratio test. The mutation frequency and spectrum of new mutations obtained from control cells that escaped lethal mutagenesis (breakthrough viruses) are also shown. Here the average number of mutations per clone and the proportion of G → A substitutions are similar to the controls.
DISCUSSION
Our results indicate that growth of HIV in tissue culture in the presence of 5-OH-dC results in the loss of the HIV population and in the accumulation of G → A substitutions. In seven of nine serial experiments, a precipitous decline in viral infectivity occurred over serial passage. This result contrasts with simultaneous incubations carried out in the absence of a nucleoside analog; in a total of 28 control cultures in which the supernatant was serially transferred from 7 to as many as 34 times, the replication of HIV was never abolished, nor was the viral titer diminished by more than 90%. Furthermore, abolishment of HIV titer was not observed with nine other deoxynucleoside analogs so far tested. We detected 97 new 5-OH-dC-induced mutations in 53,000 nucleotides (bottom of Table 3). Assuming that these mutations were evenly distributed throughout an HIV genome containing 10,000 nucleotides, then each proviral DNA obtained immediately prior to lethality contains approximately 18 5-OH-dC-induced mutations. This may be an underestimate, because the viral DNA contains both lethal and viable mutations and HIV RT is the most conserved element in the HIV genome. Some of the 5-OH-dC-treated cultures exhibited a loss of viral titer followed by recovery; sequence analysis of viral DNA suggests that virus recovery reflected outgrowth of nonmutated wild-type genome. Thus escape from lethal mutagenesis may result from persistence of the wild-type species. With continued infectious cycles, these escape viruses should also accumulate lethal mutations.
Our finding, that a limited number of mutations in the HIV genome after exposure to 5-OH-dC has a disproportionately large effect on viral lethality, substantiates the concept that the mutation frequency of HIV is close to the error threshold for the viability of the quasispecies (41). A similar result was obtained in studies with vesicular stomatitis virus (VSV), in which a 2-fold increase in mutation frequency at a single site was obtained by growth in 5-fluorouracil or 5-azacytidine or when purified virions were treated with ethyl methanesulfonate or nitrous acid (42). In each case the increase in VSV mutagenesis was paralleled by a much larger increase in lethality, 9-fold. The fact that only a 2-fold increase in HIV mutations results in viral lethality provides a mechanism for the success of combined drug therapy for HIV. During viral replication, lethal mutations are more likely to be generated than specific mutations required for resistance to each of the drugs.
The most direct explanation for the observed lethality, alterations in mutation frequency, and the types of mutations observed with 5-OH-dC is the incorporation of the analog into viral DNA by HIV RT. Once it is incorporated into the DNA strand of an RNA⋅DNA hybrid, repair is inefficient (22). As a result, mutations would be produced during the copying of the minus DNA strand containing the analog. 5-OH-dC has been shown to base pair with dATP during DNA synthesis (18, 40). Incorporation of 5-OH-dC opposite dG residues during minus DNA strand synthesis would account for the disproportionate increase in G → A substitutions that we observed during the serial transfer experiments.
Explanations involving oxidation of 5-OH-dC in cells cannot be ruled out by the data presented. Indeed, evidence has been presented suggesting that mutagenesis by 5-OH-dC may be due to deamination to either 5-hydroxyuracil or uracil glycol, both of which have been reported to be highly mutagenic and also result in G → A transitions (43). The enhanced mutagenesis by these latter analogs may be due to decreased repair of the uracil derivatives in DNA. This decrease in repairability may be akin to what we observe when 5-OH-dC is incorporated into the DNA strand of an RNA⋅DNA hybrid (22). Last, it is possible that 5-OH-dC alters the nucleotide pools within cells, and thus causes mutagenesis indirectly. There is a bias for G → A substitutions to occur at the 5′ position at template GpA similar to the multiple mutations that characterize “hypermutations” (44, 45). Among the G → A substitutions in the 5-OH-dC-treated cultures, the frequencies of the next nucleotide were A, 55%; G, 22%; C, 15%; and T, 7%. However, we have not observed a significant increase in multiple mutations in the 5-OH-dC-treated cultures.
The rapid emergence of HIV resistance is a major concern when considering any chemotherapeutic or immunological approach to AIDS therapy. Resistance to mutagenic analogs seems less likely than other single approaches for the treatment of HIV. First, substituents that alter base pairing are located between the DNA template and the incoming deoxynucleoside triphosphate and may not associate directly with the RT. Thus, mutant HIV RTs that exhibit resistance to mutagenic nucleotides are likely to exhibit simultaneously reduced affinity for the complementary nucleoside triphosphates and thus to reduce viral fitness. Second, considering the size of the HIV genome (10,000 base pairs) compared with the small number of nucleotide changes that yield resistant mutants, it seems reasonable that a far larger proportion of mutagenic changes would result in lethality than in resistance. Third, it is unlikely that an immediate selective advantage for resistance to mutagenic nucleotides would arise. Incorporation of mutagenic deoxynucleotides is in contrast to incorporation of chain terminators or to the use of inhibitors of viral replication, where resistance mutations afford an immediate selective growth advantage.
The use of mutagenic deoxynucleoside analogs to inactivate HIV provides a different approach to AIDS therapy. The concept is to employ a nucleoside analog to drive the intrinsically high mutation rate of HIV over the threshold of viral viability with minimal toxicity to human cells. The concentration of 5-OH-dC used in these studies was close to the maximum amount that did not elicit cellular toxicity; thus, it is possible that the same effects might be observed at reduced concentrations. The results we present provide evidence to validate the concept of lethal mutagenesis of HIV.
Acknowledgments
We thank A. Hizi, M. H. S. Horwitz, A. Kamath-Loeb, R. Monnat, P. Patel, and R. Prehn for critical comments, A. Rodrigo for statistical analyses, and M. Eigen for many stimulating and encouraging discussions. This work was supported by National Institutes of Health Grants OIG R35-CA-39903 (L.A.L), AI-42570 (L.A.L), OIG R35-CA52127 (J.M.E.), AI-33773 (J.I.M.), and CA59042 (J.I.M.)
ABBREVIATIONS
- RT
reverse transcriptase
- 5-OH-dCTP
5-hydroxydeoxycytidine triphosphate
- 5-OH-dC
5-hydroxydeoxycytidine
References
- 1.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 2.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 3.Preston B D, Poiesz B J, Loeb L A. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 4.Roberts J D, Bebenek K, Kunkel T A. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S S, Grosse F. J Biol Chem. 1991;266:20483–20490. [PubMed] [Google Scholar]
- 6.Ricchetti M, Buc H. EMBO J. 1990;9:1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansky L M, Temin H M. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eigen M, Schuster P. Naturwissenschaften. 1979;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 9.Eigen M. Sci Am. 1993;269(July):42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 10.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preston B D, Dougherty J P. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 12.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 13.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 14.Chesebro B, Wehrly K. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boosalis M S, Petruska J, Goodman M F. J Biol Chem. 1987;262:14689–14699. [PubMed] [Google Scholar]
- 16.Wagner J R, Hu C-C, Ames B N. Proc Natl Acad Sci USA. 1992;89:3380–3384. doi: 10.1073/pnas.89.8.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feig D I, Loeb L A. Biochemistry. 1993;32:4466–4473. doi: 10.1021/bi00067a040. [DOI] [PubMed] [Google Scholar]
- 18.Purmal A A, Kow Y W, Wallace S S. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 20.Klein J C, Bleeker M J, Roelen H C P F, Rafferty J A, Margison G P, Brugghe H F, van den Elst H, van der Marel G A, van Boom J H, Kriek E, Berns A J M. J Biol Chem. 1994;269:25521–25528. [PubMed] [Google Scholar]
- 21.Hilbert T P, Boorstein R J, Kung H C, Bolton P H, Xing D, Cunningham R P, Teebor G W. Biochemistry. 1996;35:2505–2511. doi: 10.1021/bi952516e. [DOI] [PubMed] [Google Scholar]
- 22.Kamath-Loeb A S, Hizi A, Tabone J, Solomon M S, Loeb L A. Eur J Biochem. 1997;250:492–501. doi: 10.1111/j.1432-1033.1997.0492a.x. [DOI] [PubMed] [Google Scholar]
- 23.Preston B D, Singer B, Loeb L A. Proc Natl Acad Sci USA. 1986;83:8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer B, Kusmierek J T, Folkman W, Chavez F, Dosanjh M K. Carcinogenesis. 1991;12:745–747. doi: 10.1093/carcin/12.4.745. [DOI] [PubMed] [Google Scholar]
- 25.Altshuler K B, Hodes C S, Essigmann J M. Chem Res Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 26.Sassanfar M, Dosanjh M K, Essigmann J M, Samson L. J Biol Chem. 1991;266:2767–2771. [PubMed] [Google Scholar]
- 27.Becker R A, Montesano R. Carcinogenesis. 1985;6:313–317. doi: 10.1093/carcin/6.2.313. [DOI] [PubMed] [Google Scholar]
- 28.Basu A K, Essigmann J M. Chem Res Toxicol. 1988;1:1–18. doi: 10.1021/tx00001a001. [DOI] [PubMed] [Google Scholar]
- 29.Snow E T, Foote R S, Mitra S. J Biol Chem. 1984;259:8095–8100. [PubMed] [Google Scholar]
- 30.Hizi A, Kamath-Loeb A, Rose K D, Loeb L A. Mutat Res. 1997;374:41–50. doi: 10.1016/s0027-5107(96)00217-5. [DOI] [PubMed] [Google Scholar]
- 31.Lewis J G, Swenberg J A. Nature (London) 1980;288:185–187. doi: 10.1038/288185a0. [DOI] [PubMed] [Google Scholar]
- 32.Sircar J C, Suto M J, Scott M E, Doug M K, Gilbertsen R B. J Med Chem. 1986;28:1804–1806. doi: 10.1021/jm00159a045. [DOI] [PubMed] [Google Scholar]
- 33.Shewach D S, Chern J W, Pillote K E, Townsend L B, Daddona P E. Cancer Res. 1986;46:519–523. [PubMed] [Google Scholar]
- 34.Kamath-Loeb A S, Hizi A, Kasai H, Loeb L A. J Biol Chem. 1997;272:5892–5898. doi: 10.1074/jbc.272.9.5892. [DOI] [PubMed] [Google Scholar]
- 35.Newcomb T G, Loeb L A. In: Oxidative DNA Damage and Mutagenesis. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Totowa, NJ: Humana; 1998. pp. 1–18. [Google Scholar]
- 36.Fitzgibbon J E, Mazar S, Dubin D T. AIDS Res Hum Retroviruses. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- 37.Delwart E L, Herring B, Rodrigo A G, Mullins J I. PCR Methods Appl. 1995;4:202–216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 38.Leveillard T, Andera L, Bissonnette N, Schaeffer L, Bracco L, Egly J M, Wasylyk B. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 39.Eigen M, Winkler-Oswatitsch R. Methods Enzymol. 1990;183:505–530. doi: 10.1016/0076-6879(90)83034-7. [DOI] [PubMed] [Google Scholar]
- 40.Feig D I, Sowers L C, Loeb L A. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eigen M. Gene. 1993;135:37–47. doi: 10.1016/0378-1119(93)90047-7. [DOI] [PubMed] [Google Scholar]
- 42.Holland J J, Domingo E, de la Torre J C, Steinhauer D A. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreutzer D A, Essigmann J M. Proc Natl Acad Sci USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temin H M. Cancer Res. 1988;48:1697–1701. [PubMed] [Google Scholar]
- 45.Martinez M A, Vartanian J P, Wain-Hobson S. Proc Natl Acad Sci USA. 1994;91:11787–11791. doi: 10.1073/pnas.91.25.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]