Abstract
The [URE3] nonchromosomal genetic element is a prion of Ure2p, a regulator of nitrogen catabolism in Saccharomyces cerevisiae. Ure2p1–65 is the prion domain of Ure2p, sufficient to propagate [URE3] in vivo. We show that full length Ure2p–green fluorescent protein (GFP) or a Ure2p1–65-GFP fusion protein is aggregated in cells carrying [URE3] but is evenly distributed in cells lacking the [URE3] prion. This indicates that [URE3] involves a self-propagating aggregation of Ure2p. Overexpression of Ure2p1–65 induces the de novo appearance of [URE3] by 1,000-fold in a strain initially [ure-o], but cures [URE3] from a strain initially carrying the [URE3] prion. Overexpression of several other fragments of Ure2p or Ure2-GFP fusion proteins also efficiently cures the prion. We suggest that incorporation of fragments or fusion proteins into a putative [URE3] “crystal” of Ure2p poisons its propagation.
The term “prion” means “infectious protein,” a concept that originates in studies of the mammalian transmissible spongiform encephalopathies (1, 2). These diseases involve an amyloid form of the cell surface, PrP, first detected as a gene affecting scrapie incubation period (3) then as “scrapie-associated filaments” in the brains of affected animals (4) and as a protease-resistant protein specifically enriched in purified infectious material (5). PrP proved to be encoded by a chromosomal gene (6, 7) and is essential for propagation of the disease (8). The notion that propagation of the scrapie agent involves only the PrP protein and its altered form (reviewed in ref. 9) has been supported by genetic studies (e.g., refs. 10 and 11) and the development of an in vitro system with the specificity of the in vivo disease (12).
Yeast cells grown on a rich source of nitrogen, such as ammonia, repress transcription of genes needed for using a poor nitrogen source. Ure2p mediates this “nitrogen catabolite repression” by blocking the positive transcription regulator, Gln3p. Among the genes so regulated is Dal5p, a transporter for allantoate, a poor but usable nitrogen source. Ureidosuccinate (USA) is an intermediate in uracil biosynthesis whose chance resemblance to allantoate makes it a substrate for uptake by Dal5p. Thus, ure2 mutants can take up ureidosuccinate (USA+) on ammonia-containing media whereas wild-type cells cannot (USA−) (13–17).
[URE3] is a non-Mendelian genetic element that results in loss of nitrogen catabolite repression (the USA+ phenotype) (18, 19). Based on its genetic properties, it was suggested that [URE3] is a prion of Ure2p, a regulator of nitrogen catabolism (20). Although [URE3] could be efficiently cured by growth of cells on medium containing 5 mM guanidine, from these cured cells could again be isolated strains that had acquired [URE3] de novo (reversible curability) (20). Overproduction of Ure2p increased the frequency with which [URE3] arose by 100-fold (20). Finally, the phenotype of [URE3] strains is identical to that of ure2 mutants, and URE2 is necessary for propagation of [URE3] (19, 20), a relationship expected of a prion but not of a nucleic acid replicon. Similar results known for another yeast non-Mendelian genetic element, [PSI] (refs. 21–26; reviewed in refs. 27 and 28), led to the suggestion that [PSI] too was a prion form of Sup35p (20). In support of the prion hypothesis for [URE3], Ure2p is protease-resistant in [URE3] strains, compared with normal strains (29). Moreover, [URE3] really does arise de novo, and it is the Ure2 protein (not the mRNA or high copy gene) whose overproduction induces this event (30).
The N-terminal 65 residues of Ure2p comprise the “prion domain” of the molecule, being sufficient when overproduced to induce de novo formation of [URE3] and to propagate it and necessary in cis for a Ure2p molecule to be affected by the presence of [URE3] (29, 30). Deletion of the prion domain leaves a C-terminal fragment capable of carrying out nitrogen regulation (the “nitrogen-regulation domain”) (17, 29).
The protease resistance of Ure2p in [URE3] strains is reminiscent of the protease resistance of PrP in scrapie (5), which is probably caused by PrP forming a filamentous (4), amyloid structure (31) in diseased individuals. However, specific information on the aggregation state of Ure2p in strains carrying the [URE3] prion has not been available. In support of the prion model for [PSI], Sup35p has been shown to be aggregated specifically in extracts of [PSI]-carrying strains (32), and a fusion of the Sup35 prion domain to green fluorescent protein (GFP) is localized in aggregates in the cell specifically in [PSI] strains (33). We show here that Ure2-GFP fusion proteins form intracellular aggregates specifically in [URE3] strains. In the course of this work, we found that overexpression of such fusion proteins and certain fragments of Ure2p can cure the [URE3] prion, suggesting a means to approach treatment of other prion diseases.
METHODS
Strains.
Strain 3687 (MATa kar1 ura2 leu2 his- [URE3]) was isolated as a [URE3] derivative induced de novo by the overproduction of full length Ure2p in strain 3385 (20). Strain YHE64 (MATα trp1 ura2 leu2 [URE3]) was prepared by cytoduction of [URE3] from strain 3687 into strain 3686 [MATα trp1 ura2 leu2 (ure-o)].
Expression Vectors.
Two expression vectors were used, pH7 and pH312. The strong ADH1 promoter, as an SphI fragment from pVT103 (34), was blunt-ended with T4 polymerase and was inserted into PvuII-digested pRS425 (35), forming pH7. Thus, pH7 has the yeast 2μ origin of replication, LEU2 as yeast selective marker and the ADH1 promoter directed opposite to the LEU2 gene. To make an intermediate construct, the ADH1 promoter of pH7 was amplified by PCR by using Pfu polymerase (Stratagene) and oligos HE66 (5′-ACAGCTAGCATTACGCCAGCAACTTCT-3′) and HE67 (5′-ACAAGATCTTAATGCAGCCGGTAGAG-3′). This PCR product was cloned into PvuII-digested pRS315 (36) forming pH124. To construct pH312, the URE2 promoter was amplified by PCR from p530 (20) by using Taq polymerase and oligos HE109 (5′-CAAGCTAGCGAGGTTGAAAAGAATAGC-3′) and HE110 (5′-TCATCATTTGGGATCCAAC-3′). The ADH1 promoter in pH124 bordered by NheI (present in oligo HE66) and BamHI sites was replaced by the NheI-BamHI-bordered URE2 promoter, resulting in pH312. Thus, pH312 is a centromere (CEN) plasmid with a LEU2 selection marker and the weak URE2 promoter.
Ure2p Expression Plasmids.
URE2 and URE2C (the protein product of which starts at amino acid 66) were isolated as BamHI-EcoRI fragments from p576 (20) and pDM12 (30), respectively, and were ligated into the BamHI-EcoRI window of pBS KS+ (Stratagene), resulting in clones pH2 and pH1. Subcloning URE2 as a BamHI-XhoI fragment from pH2 into pH7 or pH312 resulted in the Ure2p expression plasmids pH14 and pVTG20, respectively. Subcloning URE2C as a BamHI-XhoI fragment from pH1 into pH7 or pH312 resulted in the Ure2Cp expression plasmids pH13 and pH323, respectively. Ure2Np (which stops at amino acid 65 by converting the asn66 to UAA) expression plasmids were created by digesting pH14 and pVTG20 with NotI and XhoI and ligating in the presence of oligos HE94 (5′-GGCCGCTAAGAGC-3′) and HE95 (5′-TCGAGCTCTTAGC-3′) resulting in pH288 and pH324, respectively.
URE2-GFP Expression Plasmids with GFP in the NotI Site of URE2.
GFP was cloned as a BamHI-XhoI fragment from pH163 (37) into the BamHI-XhoI windows of pH7 and pH312, resulting in the GFP expression plasmids pH199 and pVTG11, respectively. To create an Ure2N-GFP-Ure2C fusion protein, a GFP cassette was amplified from pYGFP (38) by PCR by using Taq polymerase and oligos G1(upstream) (5′-CAAGCGGCCGCATGTCTAAAGGTGAAGAA-3′) and G2 (downstream) (5′-GTTGCGGCCGCTTTTGTACAATTCATCCATACC-3′). The resulting PCR product was ligated as a NotI fragment into NotI-digested p530 to give p791. Transfer of the NotI-bordered GFP cassette from p791 to the NotI site of pH7 and pH312 resulted in pVTG1 and pVTG10, respectively. Next, the BamHI-NdeI fragment from pVTG1 was replaced with the similarly bordered fragment from pH163 resulting in pVTG5. Inserting the NdeI fragment from pVTG1 into the NdeI site of pVTG5 resulted in the high copy number vector pH198, which directs GFP-Ure2C protein expression by using the strong ADH1 promoter. Transfer of the BamHI-XhoI-bordered GFP-URE2C cassette from pH198 into the BamHI-XhoI window of pH312 resulted in pVTG13. A Ure2N-GFP expression plasmid (pVTG4) was created by replacing the NdeI-XhoI fragment from pVTG1 with the similarly bordered fragment from pH163. Subcloning the BamHI-XhoI-bordered URE2N-GFP cassette from pVTG4 into the BamHI-XhoI window of pH312 resulted in pVTG12.
Expression Plasmids with GFP at the 3′ End of URE2.
To fuse GFP to the 3′ end of URE2, a NotI site was created, replacing the stop codon of URE2 in pH14 by using oligo HE121 (5′-AAGGCATTGCGTGGTGAAGGCGGCCGCCTTTAAAAGCAAGAAAGAAAG-3′). The NotI fragment from the resulting plasmid pH325 then was ligated into NotI-digested pVTG4, creating pH326. The BamHI-XhoI-bordered URE2-GFP cassette from pH326 was ligated into the BamHI-XhoI window of pH312, resulting in pH327. Ure2C-GFP expression plasmids were created by ligating the HindIII fragment from pH326 into HindIII digested pH13 and pH323, resulting in plasmids pH328 and pH329, respectively. GPF was visualized as described (37). Growth and transformation of yeast cells was done according to standard protocols (39). Site-directed mutagenesis was performed by using a Bio-Rad kit.
RESULTS
Ure2p-GFP Fusion Proteins Aggregate in [URE3] Strains.
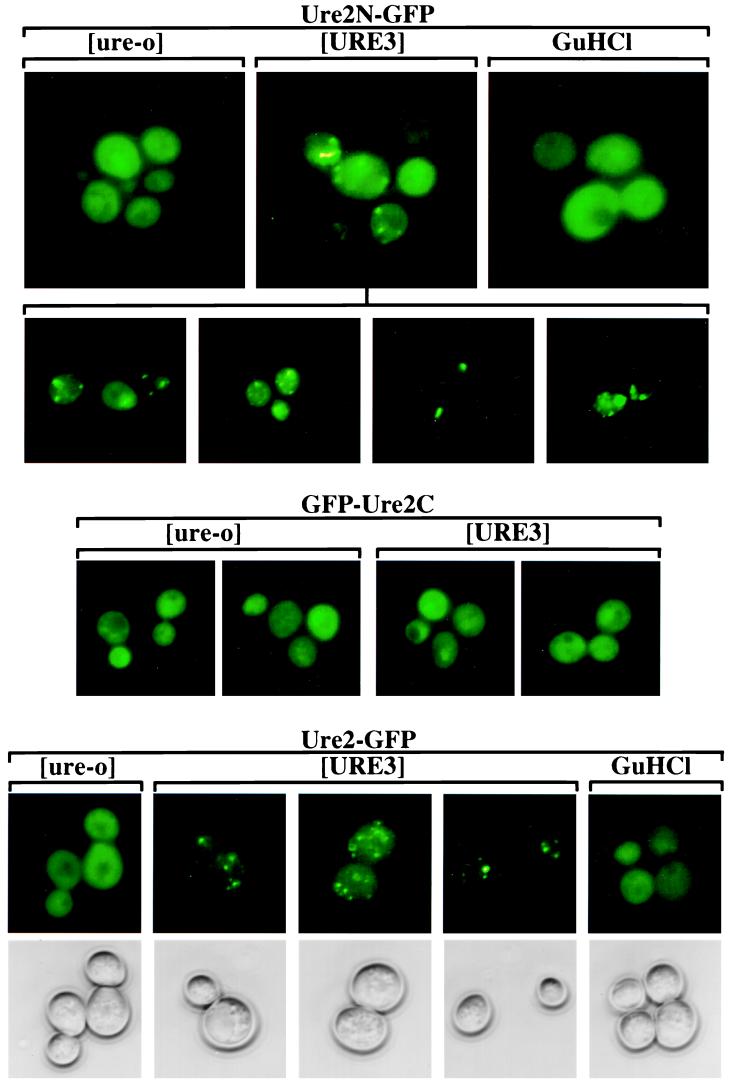
We expressed a fusion protein consisting of the Ure2p prion domain (Ure2p residues 1–65) and GFP (Ure2N-GFP) from a low copy vector with the weak URE2 promoter (PURE2) (pVTG12; Fig. 1 Top). In [URE3] cells, the fusion protein was seen as clumps whereas in [ure-o] cells, or cells cured of [URE3] by growth on low concentrations of guanidine, the fusion protein was distributed evenly throughout the cytoplasm. There were several clumps per cell, often only visible by focusing in different planes. Their distribution was apparently random and extended throughout the cell.
Figure 1.
Cellular distribution of Ure2-GFP fusion proteins. Into strains YHE64 (3686[URE3]) and 3686[ure-o] were introduced pVTG12 (CEN LEU2 PURE2 URE2N-GFP) (Top), pVTG13 (CEN LEU2 PURE2 GFP-URE2C) (Middle), and pH327 (CEN LEU2 PURE2 URE2-GFP) (Bottom). [ure-o], transformant of 3686[ure-o]; [URE3], transformant of YHE64 that retained [URE3]; GuHCl, transformant of YHE64 that was initially [URE3] but was cured by growth in the presence of 5 mM guanidine HCl. Phase contrast pictures of only the cells with pH327 (Bottom) are shown. The second row of pictures shows further examples of [URE3] cells with pVTG12.
Fusion proteins consisting of the C-terminal Ure2p nitrogen-regulation domain with GFP (GFP-Ure2C and Ure2C-GFP; pVTG13 and pH329) were expressed from the low copy PURE2 vector in isogenic [URE3] and [ure-o] cells. Both GFP-Ure2C and Ure2C-GFP complement a ure2Δ mutation (Table 1). The nitrogen regulation function of Ure2C is not inactivated by [URE3] because Ure2C does not have the prion domain (29, 30). Thus, we expected cells expressing GFP-Ure2C to be unable to take up ureidosuccinate (USA−) whether or not they had [URE3]. To test whether [URE3] was still present, we allowed loss of the plasmid from these cells. Nearly all colonies became USA+ (see below; Table 1), showing that these cells indeed had [URE3] while the plasmid was present. Both GFP-Ure2C (pVTG13, Fig. 1 Middle) and Ure2C-GFP (not shown) were distributed evenly in the cell whether or not the strain carried [URE3].
Table 1.
Curing of [URE3] by overexpression of Ure2-GFP fusion proteins
| Plasmid | Plasmid structure | Complementation of ure2Δ | Transformants USA+ of 18 | No. (of 18) USA+
after nonselective growth
|
|
|---|---|---|---|---|---|
| Plasmid retained | Plasmid lost | ||||
| pVTG1 | 2μ PADH1 URE2N-GFP-URE2C | ++ | 0 | 0 | 0 |
| pH198 | 2μ PADH1 GFP-URE2C | ++ | 0 | 0 | 0 |
| pVTG4 | 2μ PADH1 URE2N-GFP | − | 0 | 4 | 0 |
| pH199 | 2μ PADH1 GFP | − | 18 | 18 | 18 |
| pH326 | 2μ PADH1 URE2-GFP | ++ | 0 | 0 | 0 |
| pH328 | 2μ PADH1 URE2C-GFP | ++ | 0 | 0 | 0 |
| pVTG10 | CEN PURE2 URE2N-GFP-URE2C | + | 0 | 0 | 0 |
| pVTG13 | CEN PURE2 GFP-URE2C | + | 0 | 0 | 16 |
| pVTG12 | CEN PURE2 URE2N-GFP | − | 13 | 10 | 14 |
| pH327 | CEN PURE2 URE2-GFP | + | 1* | 0 | 0 |
| pH329 | CEN PURE2 URE2C-GFP | + | 0 | 0 | 10 |
| pVTG11 | CEN PURE2 GFP | − | 18 | 18 | 18 |
The haploid [URE3] strains YHE64 and 3687 were transformed with the expression plasmids. Eighteen transformants of each were assayed for USA phenotype. Twelve or eight of these transformants were grown in YPAD (39) liquid and were streaked to single colonies on YPAD (39). From each transformant, 18 colonies that had lost the plasmid and 18 that had retained the plasmid were tested for their USA phenotype. The average number of USA positive colonies is shown. Only data for YHE64 is shown, but similar results were obtained with strain 3687. URE2N, segment encoding Ure2p residues 1–65, the prion domain; URE2C, segment encoding Ure2p residues 66–354, the nitrogen regulation domain. Complementation of ure2Δ was done by using strain YHE311 (MATa trp1 his3 ura2 leu2 ure2∷URA3).
An additional hundred transformants were analyzed for their USA phenotype. One more transformant was found to be USA+. In both of these transformants, the Ure2-GFP fusion protein was localized in small spots inside the cells. In several USA− transformants tested, the Ure2-GFP fusion protein was distributed uniformly. This first-identified USA+ colony was grown on leucine dropout medium containing 5 mM GuHCl. All of the 15 colonies tested were USA− indicating that the USA+ cells were [URE3].
The full length Ure2p with GFP fused to its C-terminal end (Ure2-GFP; pH327) was clumped in [URE3] cells and was distributed evenly in [ure-o] cells or those cured of the prion by growth on guanidine (Fig. 1). These results again indicate that Ure2p is in an aggregated or clumped state specifically in [URE3] strains. The clumping is seen only when the strain carries [URE3] and the fusion protein includes the prion domain of Ure2p, previously shown to be necessary for inactivation of the nitrogen-regulation function of Ure2p by [URE3].
Nitrogen regulation is largely controlled at the transcriptional level in Saccharomyces cerevisiae in a process mediated in part by Ure2p (14, 16, 40). Ure2p is believed to interact directly with the transcription activator Gln3p, preventing its activating transcription without blocking binding to its DNA site (41). The Ure2-GFP, GFP-Ure2C, and Ure2C-GFP constructs are all active in nitrogen regulation, and each, expressed from a low copy CEN plasmid from the native URE2 promoter, is predominantly cytoplasmic in its distribution under growth conditions in which the protein is active (Fig. 1 and data not shown). Whether this reflects there being an excess of Ure2p, with the active portion in the nucleus, or a mechanism of action involving only transient entrance into the nucleus remains to be determined. In doing these studies, we noted that expression of Ure2N-GFP-Ure2C, with the GFP inserted between prion and nitrogen regulation domains, produced only USA− transformants of the [URE3] strain. On loss of the plasmid, all 144 colonies tested remained USA− and thus had lost [URE3].
Overexpression of Ure2-GFP Fusion Proteins Cures the [URE3] Prion.
On transforming high copy plasmids expressing Ure2-GFP fusion proteins from the strong ADH1 promoter into strains carrying [URE3], we found that all transformants were no longer USA+ (Table 1), even though some of these same proteins, when expressed from the weaker URE2 promoter on low copy plasmids, were compatible with [URE3]. Expressing GFP alone had no such effect (Table 1). For example, low-level expression of Ure2N-GFP gave mostly cells that remained [URE3] and showed aggregated fusion protein (Fig. 1 and Table 1). However, overexpression of the same Ure2N-GFP molecule produced clones, all of which were initially USA− ([ure-o]) and showed a uniform distribution of the fusion protein.
Because the constructs that include the C-terminal nitrogen-regulation domain complement ure2Δ, it was possible that some of these Ure2-GFP fusion proteins were not subject to the prion change and simply prevented detection of [URE3] without curing it. However, all 256 clones that had lost the plasmids from which the GFP-Ure2C fusion protein was expressed at high level and the 144 clones that had lost the plasmid similarly overexpressing Ure2C-GFP were USA− (Table 1), indicating that they had been cured of [URE3].
The level of expression of these fusion proteins was important in determining curing (Table 1). Expression of the same proteins from the weaker URE2 promoter on low copy (CEN) plasmids produced no curing or less efficient curing, particularly with GFP linked to fragments of Ure2p. This made possible the studies described above of the distribution of Ure2-GFP fusions in [URE3] cells. However, the lower level of expression did not mitigate curing by Ure2N-GFP-Ure2C in which GFP was inserted in-frame between the prion domain and the nitrogen regulation domain.
Overexpression of Ure2p Fragments Can Cure the [URE3] Prion.
Expression of the N-terminal prion domain or the C-terminal nitrogen-regulation domain of Ure2p, without attached GFP, from the strong ADH1 promoter on high copy plasmids resulted in efficient curing of [URE3] (Table 2). In contrast, expression of full length Ure2p from similar plasmids produced only occasional curing. We have detected no growth inhibition associated with overexpression of any of the Ure2-GFP fusions or Ure2p fragments in [URE3] or [ure-o] strains. Thus, this is curing of [URE3] rather than selection of cells that have lost the prion. Similar curing by Ure2N was observed in both haploid strains (Table 2) and in the diploid strain 3687 × 3686 (data not shown). Finding that overexpression of Ure2N cures [URE3] was certainly surprising because, as previously shown, this same fragment expressed in a [ure-o] cell also induces [URE3] (29, 30). This effect was reproduced for the constructs used in this study (Table 2).
Table 2.
Curing of [URE3] by overexpression of fragments of Ure2p
| Plasmid | Plasmid structure | Complementation of ure2Δ | Transformants USA+, of 18 | No. (of 18) USA+
after nonselective growth
|
[URE3] induction USA+/106 cells | |
|---|---|---|---|---|---|---|
| Plasmid retained | Plasmid lost | |||||
| pH14 | 2μ PAHD1 URE2 | ++ | 18 | 16 | 17.5 | 350 |
| pH13 | 2μ PADH1 URE2C | ++ | 0 | 0 | 0 | <0.1 |
| pH288 | 2 μ PADH1 URE2N | − | 0 | 0 | 0 | 11,000 |
| pH7 | 2μ PADH1 | − | 18 | 18 | 18 | 4 |
| pVTG20 | CEN PURE2 URE2 | ++ | 18 | 18 | 18 | |
| pH323 | CEN PURE2 URE2C | ++ | 0 | 0 | 18 | |
| pH324 | CEN PURE2 URE2N | − | 18 | 8 | 13 | |
| pH312 | CEN PURE2 | − | 18 | 18 | 18 | |
Experimental details are as in Table 1, except for the last column. The ability of Ure2p or its derivatives expressed from high copy number expression vectors with the ADH1 promoter to induce [URE3] formation was analyzed. Strain YHE 142 {3385 × 3686 (MATa/MATα ura2/ura2 leu2/leu2 trp1/+ his−/+ [ure-o]} was transformed with the plasmids. Four individual transformants were grown for 2 days in leucine dropout medium. The cell number was calculated based on the OD600, and 107, 106, 105, or 104 cells were plated on synthetic dextrose medium (ammonium nitrogen source) with leucine and USA.
DISCUSSION
The transmissible spongiform encephalopathies of mammals are associated with, and may be caused by, the formation of amyloid by PrP, a cell surface protein. [URE3] is a transmissible disease of the yeast S. cerevisiae, in which defective control of nitrogen catabolism results in slowed growth on the usual laboratory media. [URE3] is a self-propagating altered form of Ure2p, resulting in the partial protease-resistance of Ure2p in extracts of [URE3] strains. Using fusions of GFP to all or part of Ure2p, we show here that Ure2p is in intracellular aggregates, specifically in [URE3] strains. Ure2p in extracts of [URE3] strains is found in earlier fractions on Sephacryl S-400 column chromatography than in extracts of wild-type [ure-o] strains (D. C. Masison and R.B.W., unpublished work). This is consistent with our evidence that Ure2p is aggregated in [URE3] strains. The distribution is unlike that expected if Ure2p were associated, specifically in [URE3] strains, with an organelle such as the mitochondria, vacuole, nucleus, or endoplasmic reticulum, but we cannot completely rule out this type of explanation. Our results further the analogy of the [URE3] yeast disease with scrapie. Similar results have been reported for [PSI], a yeast prion based on an abnormal form of Sup35p (33).
The leading model for the mechanism of prion propagation is the “crystal seed” model, in which fragments of amyloid serve to prime the polymerization of the normal protein to make more amyloid. The energy of interaction of the normal form with the amyloid is used to drive the polymerization and a change in conformation of the protein. As with any crystal, the growth of the crystal of amyloid is poisoned by heterogeneity of the crystallizing molecules. Thus, transgenic mice expressing hamster PrP have a longer incubation period after inoculation with mouse PrPSc than do nontransgenic mice (11).
We have found that overexpression of Ure2p-GFP fusion proteins have a similar effect on propagation of the [URE3] prion, composed of propagating aggregates of Ure2p. We find that these fusion proteins cure [URE3], an indication of interference with the propagation mechanism. Fragments of Ure2p not including GFP also cure a [URE3] based on full length Ure2p, but overproduction of full length Ure2p itself does not cure. We hypothesize that it is incorporation of fusion protein molecules or fragments into the growing “crystal” that poisons its growth and thus cures [URE3]. Full length Ure2p is identical to the material already in the putative [URE3] crystal and so does not adversely affect its propagation, but the fusion proteins and fragments do interfere. Of course, although we have presented evidence here that Ure2p is aggregated in [URE3] strains, it remains to be shown that these aggregates are crystalline or amyloid in form.
It is striking that overexpression of the Ure2p prion domain fragment can both induce the de novo appearance of [URE3] in a strain that was previously [ure-o] and can cure [URE3] from a strain that began with the prion. We suggest that, in the first case, the prion domain fragment can easily form a crystal seed because it is not stabilized by the Ure2p C-terminal domain whereas, in the latter case, it interrupts the growth of the crystal formed of full length Ure2p. The “strain” of prion cured by the overproduced Ure2p1–65 was induced by overproduction of the full length Ure2p (20). It is possible that Ure2p1–65 induces a different prion strain and that they are, in some sense, incompatible. Although Ure2p1–65 induces de novo [URE3] formation 1,000-fold above the background rate, only ≈0.1% of cells acquire [URE3], too few to find among 18 clones we tested. Overexpression of the C-terminal part of Ure2p also cures, perhaps by association with the C-termini of intact molecules. We have detected interaction between C-terminal domains by using the yeast two-hybrid system (H.K.E. and R.B.W., unpublished work). Furthermore, the wild-type Ure2p is found as a dimer (K. Taylor and R.B.W., unpublished work).
Although we favor the crystal explanation for our curing results, there are other possibilities. The fact that overexpression of Hsp104 cures [PSI] (42) suggests the possibility that cells may react to the sudden overproduction of Ure2p fusions or fragments by producing some chaperone or other molecule that reverses the prion change. Alternatively, the overproduced molecule may consume a limiting factor that is necessary for [URE3] prion propagation. Could this curing of [URE3] by expressing fragments of the offending protein be applied to curing of other prion diseases or amyloid diseases in general? Peptides or other compounds that could fit in the crystal or filament growing points, but not provide a new growing point themselves, could poison amyloid propagation, just as fragments of Ure2p cure [URE3].
ABBREVIATIONS
- USA
ureidosuccinic acid
- CEN
centromere
- GFP
green fluorescent protein
References
- 1.Alper T, Cramp W A, Haig D A, Clarke M C. Nature (London) 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 2.Griffith J S. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 3.Dickinson A G, Meikle V M H, Fraser H. J Comp Path. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 4.Merz P A, Somerville R A, Wisniewski H M, Iqbal K. Acta Neuropathol. 1981;54:63–74. doi: 10.1007/BF00691333. [DOI] [PubMed] [Google Scholar]
- 5.Bolton D C, McKinley M P, Prusiner S B. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 6.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B, Aebersold R, Barry R A, Tempst P, Templow D B, Hood L E, et al. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 7.Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith J M, et al. Nature (London) 1985;315:331–333. doi: 10.1038/315331a0. [DOI] [PubMed] [Google Scholar]
- 8.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner S B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2901–2950. [Google Scholar]
- 10.Hsiao K, Baker H F, Crow T J, Poulter M, Owen F, Terwilliger J D, Westaway D, Ott J, Prusiner S B. Nature (London) 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 11.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 12.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 13.Schoun J, Lacroute F. C R Acad Sci. 1969;269:1412–1414. [Google Scholar]
- 14.Cooper T G. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern J N, Jones E W, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 39–99. [Google Scholar]
- 15.Turoscy V, Cooper T G. J Bacteriol. 1987;169:2598–2600. doi: 10.1128/jb.169.6.2598-2600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courchesne W E, Magasanik B. J Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coschigano P W, Magasanik B. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroute F. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aigle M, Lacroute F. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 20.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 21.Cox B S. Heredity. 1965;20:505–521. [Google Scholar]
- 22.Singh A C, Helms C, Sherman F. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund P M, Cox B S. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 24.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 25.Doel S M, McCready S J, Nierras C R, Cox B S. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ter-Avanesyan A, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox B S, Tuite M F, McLaughlin C S. Yeast. 1988;4:159–179. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 28.Cox B S. In: The Early Days of Yeast Genetics. Hall M N, Linder P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 219–239. [Google Scholar]
- 29.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 30.Masison D C, Maddelein M-L, Wickner R B. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prusiner S B, McKinley M P, Bowman K A, Bendheim P E, Bolton D C, Groth D F, Glenner G G. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 32.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 33.Patino M M, Liu J-J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 34.Vernet T, Dignard D, Thomas D Y. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 35.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edskes H K, Ohtake Y, Wickner R B. J Biol Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- 38.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 39.Sherman F. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. pp. 3–21. [Google Scholar]
- 40.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 283–317. [Google Scholar]
- 41.Blinder D, Coschigano P W, Magasanik B. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]