Abstract
We treated a male patient with Crimean-Congo hemorrhagic fever (CCHF). The diagnosis of CCHF was confirmed by reverse transcription-PCR and recombinant nucleoprotein (rNP)-based immunoglobulin G (IgG) and IgM capture enzyme-linked immunosorbent assays of serially collected serum samples. The patient was treated with intravenous ribavirin and recovered with no consequences. The study indicates that rNP-based CCHF virus antibody detection systems are useful for confirming CCHF virus infections. This case also suggests that intravenous ribavirin therapy may be promising for the treatment of CCHF patients.
Case report.
We treated a patient with Crimean-Congo hemorrhagic fever (CCHF). The diagnosis of CCHF was confirmed by reverse transcription-PCR (RT-PCR) and a recombinant CCHF virus (CCHFV) nucleoprotein (rNP)-based enzyme-linked immunosorbent assay (ELISA) for detection of immunoglobulin G (IgG) to CCHFV (6). Furthermore, an IgM capture ELISA using the CCHFV rNP was used as a serological tool for the diagnosis of this patient. This patient was successfully treated by intravenous administration of ribavirin.
The patient was a 28-year-old male shepherd who lived in an area of the western part of the Xinjiang Uygur Autonomous Region, People's Republic of China, where CCHF is endemic. Taking the day on which the fever first appeared as day 1, he visited a local clinic on day 1. He spent 3 nights at home but deteriorated abruptly. He was transferred to our hospital and hospitalized on day 4. He did not know whether he had been bitten by a tick, one of the main reservoirs of CCHFV. He presented on admission with low-grade fever, unconsciousness, and severe hemorrhage from the nostrils, gingiva, skin, and gastrointestinal tract. He had anemia, and the erythrocyte count and hemoglobin level were 3.41 × 1012 cells/liter (normal range, 4.5 × 1012 to 5.9 × 1012 cells/liter) and 10.0 g/dl (normal range, 13.5 to 17.5 g/dl), respectively. Thrombocytopenia was noticed, with a platelet count of 84 × 109/liter (normal range, 150 × 109 to 400 × 109/liter). The alanine transaminase, aspartate aminotransferase, and lactate dehydrogenase levels were 173 U/liter (normal range, 5 to 40 U/liter), 216 U/liter (normal range, 5 to 40 U/liter), and 268 U/liter (normal range, 114 to 240 U/liter), respectively, suggesting liver dysfunction. Mild hyperbilirubinemia, hypoproteinemia, and hypoalbuminemia were also present. Renal function was preserved. The patient was intravenously administered ribavirin (0.6 g/dose twice a day, 2-h drip infusion) from day 4 to 11. The symptoms improved gradually, and the patient recovered with no consequences.
Blood samples were drawn for diagnostic tests on days 1, 5, and 9. The blood sample drawn on day 1 was collected at the initial clinic visit and was brought to our hospital. Serum was separated and kept at −20°C until use. A nested RT-PCR was performed for amplification of the viral genome. Viral RNA was extracted from 200 μl of serum with a High Pure Viral RNA Kit (Roche Diagnostics GmbH, Mannheim, Germany) in accordance with the manufacturer's instructions. The primers were modified from those reported by other investigators (4) in accordance with the nucleotide sequence of Chinese CCHFV isolate 8402 (GenBank accession no. AJ010649). Five microliters of purified RNA was added to the Ready to Go RT-PCR mixture (0.5-ml tubes; Amersham Pharmacia Biotech Inc., Piscataway, N.J.) as a template, and then the primer set of 50 pmol each of CCHF/F2C (5′-TGGATACTTTCACAAACTC-3′) and CCHF/R3 (5′-GACAAATTCCCTGCACCA-3′) and an appropriate amount of water were added to the tube. The RT-PCR was performed in accordance with the manufacturer's instructions. The tube was kept at 42°C for 30 min for the RT reaction. The reverse transcriptase was then heat inactivated at 95°C for 5 min. The PCR conditions were as follows: 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s, and elongation at 72°C for 30 s, followed by an additional elongation at 72°C for 5 min. For the nested PCR, 1 μl of the first-round PCR product was added to a 0.5-ml Ready to Go PCR tube (Amersham Pharmacia Biotech Inc.) as a template and then 50 pmol each of primers CCHF/F3C (5′-GAGTGTGCCTGGGTTAGCTC-3′) and CCHF/R2C (5′-GACATTACAATTTCGCCAGG-3′) and an appropriate amount of water were added to the PCR tube. The second-round PCR was performed under the same conditions as described above. The PCR product was separated by electrophoresis in a 2% agarose gel and visualized by staining with ethidium bromide. The expected size of the PCR product was 262 bp.
IgG antibodies to CCHFV were detected by a CCHFV rNP-based IgG ELISA (6). IgM antibodies to CCHFV were detected by an IgM capture ELISA with the same antigen. The ELISA plate was coated with goat anti-human IgM antibody (μ chain specific; Zymed Laboratories Inc., South San Francisco, Calif.) at an approximate concentration of 100 ng/well at 4°C overnight. After the plate was washed with phosphate-buffered saline solution containing 0.05% Tween 20 (T-PBS), it was treated with the blocking reagent, 200 μl of T-PBS containing 5% skim milk (T-PBS-M) per well, at 37°C for 1 h. The plate was washed with T-PBS, and the top four and bottom four wells of the plate were inoculated with heat-inactivated test serum samples (100 μl/well), which were diluted twofold from 1:50 to 1:400 with T-PBS-M. The plate was incubated at 37°C for 1 h. After being washed with T-PBS, the top four wells of the plate were inoculated with the purified CCHFV rNP in T-PBS-M (100 μl/well) at a concentration of 1 μg/ml while the bottom four wells were also inoculated with T-PBS-M as a negative control. After being washed with T-PBS, all of the wells were inoculated with anti-CCHFV rNP rabbit serum at a dilution of 1:1,000 (6). After being washed with T-PBS, the plate was inoculated with goat anti-rabbit IgG antibody labeled with horseradish peroxidase (Zymed Laboratories Inc.) at a dilution of 1:1,000. The plate was washed with T-PBS, 2,2′-azino-di[3-ethylbenzthiazolin sulfate (6)] (ABTS) solution (Roche Diagnostics GmbH) was added to each well, and the plate was incubated at 37°C for 30 min. The optical density at 405 nm (OD405) was measured with a reference of 490 nm. The OD405 was adjusted by subtracting the OD405 of the non-antigen-inoculated well from that of the corresponding antigen-inoculated well. The cutoff value for the IgG ELISA at a dilution of 1:400 was defined as reported previously (6). The cutoff value that determined IgM positivity or negativity was calculated as the average plus 3 standard deviations of serum samples collected from 48 subjects with no history of CCHFV infection.
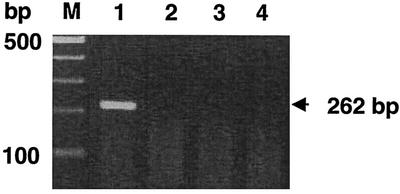
The viral genome was detected by nested RT-PCR in the day 1 serum sample but not in those from days 5 and 9 (Fig. 1). Neither IgM nor IgG antibodies to CCHFV were detected in the day 1 serum sample. On the other hand, IgM and IgG were both detected in the serum samples collected on days 5 and 9 (Table 1). The titers were higher on day 9 than on day 5.
FIG. 1.
Results of the nested RT-PCR assay of serum samples serially collected from the patient. Lanes: M, 100-bp markers; 1, 2, 3, and 4, day 1, 5, and 9 serum and negative control samples, respectively. A PCR product of the expected size (262 bp) was amplified only from the day 1 serum sample.
TABLE 1.
IgG and IgM antibodies to CCHFV detected by ELISAs in serum samples drawn sequentially
| Class of antibodies and dilution | OD405 of samples and antibodya
|
Cutoffb | ||
|---|---|---|---|---|
| Day 1 | Day 5 | Day 9 | ||
| IgM | ||||
| 1:50 | 0.000 | 2.692 | 2.709 | NDc |
| 1:100 | 0.020 | 2.672 | 2.711 | 0.205 |
| 1:200 | 0.044 | 2.528 | 2.767 | ND |
| 1:400 | 0.040 | 1.606 | 2.001 | ND |
| IgG | ||||
| 1:100 | 0.075 | 0.924 | 1.882 | ND |
| 1:400 | 0.031 | 0.486 | 0.972 | 0.213 |
| 1:1,600 | 0.000 | 0.152 | 0.384 | ND |
| 1:6,400 | 0.000 | 0.045 | 0.132 | ND |
The OD405 values shown here were adjusted as described in the text.
Cutoff values for IgM and IgG responses were defined at dilutions of 1:100 and 1:400, respectively, as described in the text. According to these cutoff values, both the IgM and IgG responses were demonstrated in the serum samples drawn on days 5 and 9 but not in the day 1 sample.
ND, not determined.
The virological and immunological status of CCHFV infection was closely followed by nested RT-PCR and the ELISAs. We previously reported the CCHFV rNP-based antibody detection systems (5, 6). In the present study, we used a CCHFV rNP-based IgM capture ELISA along with an IgG ELISA. We applied these assays to the serum samples serially collected from the patient. The results suggest that both the CCHFV rNP-based IgG ELISA and the IgM capture ELISA are useful for the diagnosis of CCHF. We believe that this is the first case of CCHF serologically diagnosed by CCHFV rNP-based antibody detection systems.
Ribavirin, an anti-RNA virus agent, has been reported to have an inhibitory effect on the replication of CCHFV in vitro and in vivo (2, 7, 9). There have been several reports on ribavirin therapy for CCHF (1, 3, 8). However, the efficacy of ribavirin for CCHF has not yet been proved. Oral ribavirin was used for postexposure prophylaxis of CCHFV infection with promising results, but its efficacy was not formally assessed (8). The patient in the present study was treated by intravenous administration of ribavirin. To our knowledge, this is the first case report of a CCHF patient treated with intravenous ribavirin therapy. The previous reports, along with the present study, suggest that ribavirin is a promising drug for the treatment of CCHF.
REFERENCES
- 1.Fisher-Hoch, S. P., J. A. Khan, S. Rehman, S. Mirza, M. Khurshid, and J. B. McCormick. 1995. Crimean Congo-haemorrhagic fever treated with oral ribavirin. Lancet 346:472-475. [DOI] [PubMed] [Google Scholar]
- 2.Huggins, J. W. 1989. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad spectrum antiviral drug. Rev. Infect. Dis. ii:S750-S761. [DOI] [PubMed] [Google Scholar]
- 3.Papa, A., B. Bozovi, V. Pavlidou, E. Papadimitriou, M. Pelemis, and A. Antoniadis. 2002. Genetic detection and isolation of Crimean-Congo hemorrhagic fever virus, Kosovo, Yugoslavia. Emerg. Infect. Dis. 8:852-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez, L. L., G. O. Maupin, T. G. Ksiazek, P. E. Rollin, A. S. Khan, T. F. Schwarz, R. S. Lofts, J. F. Smith, A. M. Noor, C. J. Peters, and S. T. Nichol. 1997. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 57:512-518. [DOI] [PubMed] [Google Scholar]
- 5.Saijo, M., Q. Tang, M. Niikura, A. Maeda, T. Ikegami, C. Prehaud, I. Kurane, and S. Morikawa. 2002. An immunofluorescence technique for detection of immunoglobulin G antibodies to Crimean-Congo hemorrhagic fever virus using HeLa cells expressing recombinant nucleoprotein. J. Clin. Microbiol. 40:372-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saijo, M., Q. Tang, M. Niikura, A. Maeda, T. Ikegami, C. Prehaud, I. Kurane, and S. Morikawa. 2002. Recombinant nucleoprotein-based enzyme-linked immunosorbent assay for detection of immunoglobulin G to Crimean-Congo hemorrhagic fever virus. J. Clin. Microbiol. 40:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tignor, G. H., and C. A. Hanham. 1993. Ribavirin efficacy in an in vivo model of Crimean-Congo hemorrhagic fever virus (CCHF) infection. Antivir. Res. 22:309-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van de Wal, B. W., J. R. Joubert, P. J. van Eeden, and J. B. King. 1985. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part IV. Preventive and prophylactic measures. S. Afr. Med. J. 68:729-732. [PubMed] [Google Scholar]
- 9.Watts, D. M., M. A. Ussery, D. Nash, and C. J. Peters. 1989. Inhibition of Crimean-Congo hemorrhagic fever viral infectivity yields in vitro by ribavirin. Am. J. Trop. Med. Hyg. 41:581-585. [DOI] [PubMed] [Google Scholar]