Abstract
Intracisternal A-type particles (IAP) are defective endogenous retroviruses that accumulate in the endoplasmic reticulum (ER) of rodent cells. The enveloped particles are produced by assembly and budding of IAP Gag polyproteins at the ER membrane. In this study, we analyzed the specific ER transport of the Gag polyprotein of the IAP element MIA14. To this end, we performed in vitro translation of Gag in the presence of microsomal membranes or synthetic proteoliposomes followed by membrane sedimentation or flotation. ER binding of IAP Gag occurred mostly cotranslationally, and Gag polyproteins interacted specifically with proteoliposomes containing only signal recognition particle (SRP) receptor and the Sec61p complex, which form the minimal ER translocation apparatus. The direct participation of SRP in ER targeting of IAP Gag was demonstrated in cross-linking and immunoprecipitation experiments. The IAP polyprotein was not translocated into the ER; it was found to be tightly associated with the cytoplasmic side of the ER membrane but did not behave as an integral membrane protein. Substituting the functional signal peptide of preprolactin for the hydrophobic sequence at the N terminus of IAP Gag also did not result in translocation of the chimeric protein into the ER lumen, and grafting the IAP hydrophobic sequence onto preprolactin failed to yield luminal transport as well. These results suggest that the N-terminal hydrophobic region of the IAP Gag polyprotein functions as a transport signal which mediates SRP-dependent ER targeting, but polyprotein translocation or integration into the membrane is prevented by the signal sequence itself and by additional regions of Gag.
The last step in the formation of enveloped viruses is budding of the viral core containing the genome through a cellular membrane, which can be different depending on the respective virus. Most retroviruses bud predominantly at the plasma membrane, while many other enveloped viruses form at the membranes of the endoplasmic reticulum (ER), the Golgi apparatus, or vesicles of the endosomal pathway (reviewed in reference 30). However, the transport route of the internal structural proteins of most viruses is currently not known. Retroviral assembly and budding are directed by the viral Gag polyprotein, which is transported to the cell membrane leading to formation of an immature noninfectious virion, still mostly consisting of uncleaved polyproteins during its release from the host cell. Subsequent proteolysis of the Gag polyproteins by the viral proteinase (PR) gives rise to the individual structural proteins matrix (MA), capsid, and nucleocapsid and several additional proteins and smaller peptides, depending on the virus. This process is termed maturation and leads to morphological condensation of the inner core, thereby rendering the virion infectious (reviewed in reference 30).
Gag alone is sufficient for the formation of immature particles, and neither the viral glycoproteins nor any other viral proteins are needed in most cases. Transport of the Gag polyprotein to the cell membrane is currently incompletely understood. Numerous studies have shown that the N-terminal MA domain of Gag, which is myristoylated in most retroviruses (26), is crucial for membrane transport and mutations of the myristoylation signal abolish virus release and tight membrane association (6, 22, 54). Besides myristoylation, clusters of basic amino acids appear to be important in membrane transport as well (48, 76). Structural analysis of MA proteins from several retroviruses revealed that basic amino acids are clustered at the presumed membrane-apposed side of the trimeric MA complex and may form ionic interactions with the acidic head groups of phospholipids on the inner leaflet of the membrane (28, 52). Accordingly, mutation of basic residues has been shown to reduce the membrane binding potential of the respective protein and to affect virus release (48, 76). While these results clearly imply that the bipartite signal of myristoylation and positively charged regions is important for stable membrane binding, they do not determine the transport pathway used. Gag may be actively transported to the membrane or may localize there by diffusion and then be retained. Various studies implicated the cytoskeleton or the vesicular transport route in Gag transport to the plasma membrane (24, 36), but no clear picture regarding their respective role has emerged so far. Moreover, no specific transport receptor binding to the membrane targeting region of Gag has been identified to date. The alternative model of diffusion and retention does not easily explain the specificity of membrane targeting, because negatively charged lipids are found on the cytoplasmic faces of all membranes. Conceivably, differences in lipid composition may play a role in targeting, and recent studies implicated detergent-resistant membrane subdomains termed lipid rafts in Gag polyprotein transport and budding, at least in the case of human immunodeficiency virus type 1 (HIV-1) (42, 45).
Retrovirus budding can be redirected to a different membrane by alteration of the targeting signal. Deleting a large part of the globular MA domain of HIV-1 Gag led to budding of immature virus particles into the lumen of the ER with almost complete loss of extracellular virion formation (16). This phenotype was reminiscent of intracisternal A-type particles (IAP), a class of endogenous retroviruses which bud exclusively into the cisternae of the ER (reviewed in reference 32). These noninfectious, membrane-enveloped particles retain their spherical immature (A-type) cores consisting of uncleaved polyproteins and remain sequestered in the ER. Several IAP genomes have been fully or partially sequenced (14, 17, 38, 46, 55), and most contain multiple stop codons in the putative env region, likely reflecting the lack of an extracellular phase. IAP genomes documented significant homologies to B-type and D-type retroviruses extending through almost the entire gag-pol region, with the notable exception of the 5′-terminal region of gag, where the intracellular targeting domains are thought to reside (70). The presence of alternative targeting signals at the N terminus of Gag was confirmed in a previous series of cell transfection experiments, which showed that the ER transport of IAP Gag polyproteins is governed by an N-terminal hydrophobic sequence situated in the MA-analogous region of IAP Gag (70). Substituting a heterologous plasma membrane targeting signal for this domain redirected budding to the plasma membrane and led to release of extracellular particles and activation of proteolysis. However, as in the case of other retroviruses, it is currently not clear whether IAP Gag transport to the ER membrane is an active process or occurs by diffusion with membrane-apposed polyproteins being retained at the membrane by hydrophobic and possibly also by ionic interactions.
In principle, there are two active transport pathways which specifically target proteins to ER membranes (reviewed in reference 68). The most common transport occurs cotranslationally and is mediated by a signal sequence on the respective protein which is recognized by the cytosolic signal recognition particle (SRP) (67). SRP directs the nascent chain together with the ribosome to its receptor, located in the ER membrane. There, the nascent chain is inserted into the Sec61p complex, forming the channel through which the polypeptide traverses the membrane (13, 20). A second pathway occurs posttranslationally and is mediated by different signals but does not involve the SRP. Examples for proteins making use of the latter pathway are the two membrane proteins cytochrome b5 (2) and M13 preprocoat protein (69) and the presecretory protein honeybee prepromellitin (41). Posttranslational transport of proteins into the ER appears to be rare in mammalian cells but is common and well characterized in Saccharomyces cerevisiae (12, 47), and the latter is true also for the related process in bacteria (57, 71).
In this study, we show that the IAP Gag polyprotein is transported to the ER membrane via SRP, requiring SRP receptor and Sec61p complex for membrane binding. The Gag polyprotein is not translocated into the lumen but stays tightly bound to the ER membrane. These properties are mediated both by the hydrophobic signal sequence as well as by the remainder of the protein.
MATERIALS AND METHODS
Expression plasmids.
In vitro translation vectors were derived from the expression plasmid pTM1 (40), which contains the internal ribosome entry site of encephalomyocarditis virus and a T7 RNA polymerase promoter. Plasmid pTM1-MIA2 containing almost the entire gag-pol region (nucleotide 594 to 4102) of the murine IAP MIA14 (38) has been described previously (17). Plasmids pTM1-MIA4 and pTM1-MIA5 were derived from pL-MIA4 and pL-MIA5, respectively (70). In pTM1-MIA5 the first 28 codons of the MIA14 gag gene have been deleted. In the case of pTM1-MIA4 these 28 codons have been replaced by the membrane targeting signal of the Src protein.
pTM1-ppl was constructed by cloning the entire coding region of bovine preprolactin (ppl) from pSPB4 (59) into pTM1. Plasmid pTM1-SPpplMIA, in which the first 28 codons of the MIA14 gag gene have been replaced by the first 33 codons of ppl (coding for the signal peptide of the ppl protein and 3 amino acids of the mature protein) was generated by PCR. The ppl signal peptide was amplified from pTM1-ppl, using the forward primer T7 universal primer and the reverse primer 5′-TGCGAATTCTAGAGCGCTGACGGGGGTGGAGACCACACC-3′. The amplified fragment was cleaved with ApaI (in the internal ribosome entry site of encephalomyocarditis virus) and at the newly introduced XbaI site (underlined) and ligated together with the XbaI-BamHI fragment (nucleotides 677 to 4102 of the MIA14 sequence) into pTM1 opened with ApaI and BamHI.
To construct plasmids pTM1-plMIA28 and pTM1-plMIA23 in which the signal sequence of ppl was exchanged for the first 28 or 23 codons of the MIA14 gag gene, the prolactin gene lacking its own signal sequence was amplified by PCR from pTM1-ppl, using the forward primer 5′-AGCGAATTCACTAGTGTCCACCCCCGTCTGTCCC-3′ and the reverse primer: 5′-TAATACGACTCACTATAGGG-3′, which binds to the pTM1 sequence (new SpeI site underlined). This fragment was inserted into a modified MIA14 subclone (pTM-L4) that contains the 5′ fragment of pTM1-MIA2 amplified with the T7 universal primer and the reverse primer 5′-TCGCTAGCCGTCCCTCGATTCGAACTAGTTGATAACATGTGAAAAGG-3′ to introduce an SpeI restriction site downstream of codon 23 and an NheI restriction site downstream of codon 28 of the MIA14 gag gene (both underlined). To generate pTM1-plMIA28 the ppl-derived PCR fragment was cleaved at the newly introduced SpeI site and with SalI and ligated into pTM-L4, which was opened with NheI (compatible to SpeI) and SalI. To generate pTM1-plMIA23, pTM-L4 was cut with NheI and SalI prior to inserting the prolactin fragment. The sequences derived by PCR were confirmed by DNA sequence analysis of the respective plasmids. All molecular biological manipulations and sequence analysis were carried out using standard methods (56).
In vitro transcription and translation.
Coupled transcription and translation reactions were performed according to the manufacturer's instructions with T7 RNA polymerase in the presence of [35S]methionine (Tran35S-label [>1,000 Ci/mmol; >10 mCi/ml]; ICN Pharmaceuticals) using a coupled system based on rabbit reticulocyte lysate (TNT; Promega). Transcription-translation reactions were usually performed for 90 min at 30°C, unless otherwise indicated. Where indicated, canine pancreas microsomes, liposomes, or reconstituted proteoliposomes were present. Canine pancreas microsomes were prepared as described previously (58), and reconstituted proteoliposomes were prepared according to the protocol of Görlich and Rapoport (21). For posttranslational binding, translation in the absence of membranes was terminated after 60 min at 30°C by addition of puromycin (final concentration, 1.25 mM; Sigma) followed by further incubation for 10 min at 30°C. Subsequently, microsomes were added and the sample was incubated for an additional hour at 30°C.
Analysis of membrane-associated IAP Gag polyproteins. (i) Sedimentation.
Following transcription-translation, the samples were diluted with NTE buffer (100 mM NaCl; 10 mM Tris-HCl, pH 8.0; 1 mM EDTA) to an appropriate volume. The membrane fractions were then sedimented at 12,000 × g for 15 min at 4°C in a Beckman tabletop centrifuge. After removing and collecting the supernatants, the pellets were resuspended in NTE buffer, and adequate volumes of pellets and supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
(ii) Sucrose flotation gradient centrifugation. For membrane flotation, the coupled transcription-translation reaction was performed as described above. Subsequently, 25 μl of the sample was mixed with 425 μl of 68% sucrose (wt/wt), placed at the bottom of a Beckman TLS 55 centrifuge tube and overlaid with 400 μl 65% (wt/vol) and 150 μl 10% (wt/vol) sucrose in NTE buffer. The step gradient was then centrifuged to equilibrium at 43,000 rpm for 18 h at 4°C in a Beckman Optima TLX Ultracentrifuge. Eight fractions were collected from the top and analyzed by SDS-PAGE and autoradiography.
Membrane extraction.
Completed transcription-translation reactions containing microsomes were treated with carbonate buffer (pH 11.5) or adjusted to either 1 M NaCl, 50 mM EDTA, or 0.1% Triton X-100 in NTE buffer and incubated for 1 h at 0°C. Control samples were treated with NTE buffer. The membrane fractions were sedimented as described above. Pellets and supernatants were analyzed by SDS-PAGE.
Proteinase protection assay.
Following transcription-translation, reaction mixtures were chilled on ice and divided into three fractions. One fraction was left untreated, and the others were treated with proteinase K (Boehringer Mannheim) at a final concentration of 50 μg/ml for 1 h at 0°C in the absence or presence of 0.1% Triton X-100. Proteinase K was inactivated by addition of phenylmethylsulfonyl fluoride (final concentration, 10 mM; Serva), and incubation at 0°C for 10 min. Aliquots of each sample were analyzed by SDS-PAGE.
Cross-linking and immunoprecipitation. (i) Generation of ribosome-nascent-chain complexes.
The plasmid pTM1-MIA2 was linearized with the restriction enzymes StuI, EcoRI, or NgoMI to produce mRNAs lacking a stop codon and coding for the first 58 (58-mer), 122 (122-mer), or 262 (262-mer) amino acids of the IAP Gag polyprotein, respectively. The linearized plasmids were subjected to coupled transcription-translation reactions for 30 min at 30°C, producing C-terminally truncated IAP Gag polyproteins which were still associated with the ribosomes due to the missing stop codons. These ribosome-nascent-chain complexes were subsequently sedimented by centrifugation at 7,500 rpm in a TLA 100.2 rotor in the Beckman Optima TLX Ultracentrifuge.
(ii) Chemical cross-linking. Sedimented ribosome-nascent-chain complexes were resuspended in XL buffer (50 mM triethanolamine, pH 7.5; 50 mM potassium acetate, 5 mM magnesium acetate, 200 mM sucrose). Cross-linking reactions were performed for 2 h at 0°C by addition of disuccinimidyl suberate (DSS) (final concentration, 0.4 mM; Pierce) from a 27.1 mM (10-mg/ml) stock in dimethyl sulfoxide. Quenching of the cross-linking reagent was carried out by adding 50 mM Tris-HCl buffer, pH 7.5, and incubation on ice for 10 min.
(iii) Immunoprecipitation. Quenched cross-linking products were incubated in 1% SDS for 5 min at 85°C and subsequently diluted with 4 volumes of water followed by a further incubation for 5 min at the same temperature. The samples were then adjusted to a final concentration of 20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100; 0.5% sodium deoxycholate; and 0.1% SDS. Immunoprecipitations were carried out with a suspension of protein A-Sepharose (Pharmacia) containing antiserum against the 54-kDa subunit of SRP (25) for 16 h at 4°C under vigorous agitation. Protein A-Sepharose beads were collected by low-speed centrifugation and washed extensively in RIPA buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS). Bound antigen was eluted by addition of SDS loading buffer and heating at 100°C for 10 min.
Sample analysis and quantitation.
Samples from the translation reactions were analyzed on SDS-polyacrylamide gels containing 17.5% polyacrylamide (200:1 ratio of acrylamide to N,N-methylenebisacrylamide). The products of cross-linking and immunoprecipitation experiments were electrophoresed on SDS-19.4% polyacrylamide gels containing 6 M urea (60:0.8 ratio of acrylamide to N,N-methylenebisacrylamide). Following electrophoresis, the gels were stained, dried, and autoradiographed or analyzed using a Fuji BAS 2000 Bioimager and TINA 2.09 software.
RESULTS
In a previous series of experiments, we had analyzed the intracellular localization of the wild-type polyprotein of the murine IAP element MIA14 and of derivatives thereof, following transient transfection of expression plasmids (70). These studies revealed that ER targeting of IAP Gag required the N-terminal 28 amino acids of Gag. Deleting this sequence abolished ER transport, and substituting the targeting signal of the Src protein for this region redirected the polyprotein to the plasma membrane, leading to release of extracellular particles and activation of proteolysis (70). Here, we study the molecular mechanism of IAP Gag transport and assembly in an in vitro system. To this end, coupled in vitro transcription and translation reactions were performed in the presence and absence of microsomal membranes or synthetic proteoliposomes, respectively, and the resulting products were analyzed for membrane association.
The N terminus of MIA14 Gag mediates ER transport in vitro.
Initially, we analyzed whether MIA14 Gag polyproteins associate with ER membranes in vitro and whether this association requires the N-terminal hydrophobic region of Gag. To this end, different variants of the Gag polyprotein were compared (Fig. 1A): MIA2 corresponds to the wild-type sequence of the IAP element MIA14, which contains the coding regions for Gag, PR, and part of the pol gene products (70). MIA5 contains a deletion of the first 28 codons of gag, while this region was replaced by the Src membrane localization signal in MIA4. To characterize the association of newly synthesized IAP polyproteins with ER membranes, sedimentation experiments were performed following in vitro transcription and translation in the presence or absence of microsomal membranes (Fig. 1B). In the case of MIA2, ca. 75% of the Gag polyprotein was observed in the pellet fraction when synthesized in the presence of microsomal membranes, while less than 10% was pelletable in the absence of membranes (Fig. 1B, compare lanes 2 and 8). MIA5 polyproteins, on the other hand, remained largely soluble independent of the presence or absence of microsomal membranes (Fig. 1B, compare lanes 6 and 12). MIA4 polyproteins synthesized in the presence of microsomal membranes were detected mostly (ca. 80%) in the pellet fraction, similar to MIA2 (Fig. 1B, lane 10). However, in this case there was also a significant amount of Gag in the pellet fraction even in the absence of microsomal membranes (30 to 40%; Fig. 1B, lane 4). This may be caused by protein aggregation of the MIA4 translation product.
FIG. 1.
(A) Map of the IAP-specific sequence in plasmids pTM1-MIA2, pTM1-MIA4, and pTM1-MIA5. MIA2 corresponds to the wild-type sequence of the IAP MIA14 (38). In MIA4, the first 10 codons of src were substituted for the first 28 codons of the gag gene, and in MIA5 the first 28 codons of gag were deleted. Nucleotide numbering is according to the published sequence (38). Proteins derived from different reading frames are depicted in different lanes. (B) Analysis of in vitro-translated and membrane-bound IAP polyproteins. Coupled transcription-translation reactions were programmed with pTM1-MIA2, pTM1-MIA4, or pTM1-MIA5 in the absence or presence of microsomal membranes as indicated and subsequently centrifuged for 15 min at 12,000 × g. Supernatants (S) and resuspended pellets (P) were applied in equivalent amounts and analyzed by SDS-PAGE. Translation products were labeled with [35S]methionine and detected by autoradiography. The positions of the Gag, Gag-PR, and Gag-PR-Pol polyproteins are indicated on the left. (C) Analysis of membrane-bound IAP polyproteins by sucrose flotation. Coupled transcription-translation reactions were programmed with pTM1-MIA2 or pTM1-MIA5 in the absence or presence of microsomal membranes as indicated. After translation, the samples were adjusted to 85% (wt/vol) sucrose and overlaid with 65 and 10% sucrose. The step gradient was centrifuged at 43,000 rpm in a Beckman Optima TLX ultracentrifuge for 18 h at 4°C. Fractions were collected from top (fraction 1) to bottom (fraction 8), and proteins were resolved by SDS-PAGE.
To confirm that the sedimentation behavior of in vitro-synthesized Gag polyproteins was indeed due to membrane association, MIA2 and MIA5 translation products were analyzed by membrane flotation using a sucrose step gradient. In contrast to sedimentation assays, where unspecific aggregates are pelleted as well, membrane flotation through a dense sucrose cushion is a more stringent assay for membrane association. As shown in Fig. 1C, 50 to 60% of the MIA2 translation products were detected in the membrane fractions (fractions 1 to 4) when synthesized in the presence (lower panel), but not in the absence, of microsomal membranes (upper panel). Reflotation of these fractions led to a recovery of more than 80% of MIA2 Gag in the membrane fractions (data not shown). In contrast, there was no flotation of the MIA5 translation products into the membrane fractions, independent of the presence or absence of microsomal membranes (Fig. 1C, right panels). Taken together, these findings indicate that the IAP polyproteins associate with ER membranes in vitro and that this association requires the N-terminal 28 amino acids of Gag.
IAP Gag polyprotein transport to the ER membrane occurs cotranslationally.
To differentiate between cotranslational (suggesting SRP-mediated transport) and posttranslational membrane binding, in vitro synthesis of IAP polyproteins was performed in the presence or absence of microsomal membranes (Fig. 2). For analysis of posttranslational binding, the translation reaction was stopped with puromycin before microsomal membranes were added. As shown above, <10% of wild-type IAP polyproteins were sedimented in the absence of membranes, while 80% of polyproteins were sedimented in the presence of microsomal membranes (Fig. 2A, lanes 1 to 4). If microsomal membranes were added after the translation had been stopped, membrane association of IAP polyproteins was significantly lower (ca. 30%; Fig. 2A, lanes 5 to 6), indicating that efficient ER transport occurs mostly during translation. A different picture was observed for MIA4, which carries the Src membrane transport signal substituted for the IAP N terminus. In this case, membrane binding was approximately equal (ca. 80%) when membranes were present cotranslationally (Fig. 2B, lanes 3 and 4) or added posttranslationally (Fig. 2B, lanes 5 and 6). These results indicate that the IAP-specific sequence at the N terminus of Gag directs the viral polyproteins preferentially into a cotranslational transport pathway to the ER membrane, while the Src N terminus mediates efficient posttranslational transport, presumably due to its N-terminal myristoylation.
FIG. 2.
Analysis of co- and posttranslational binding of IAP polyproteins. Coupled transcription-translation reactions were programmed with pTM1-MIA2 (A) or pTM1-MIA4 (B) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of microsomal membranes for 1 h. For analysis of posttranslational transport, reactions without membranes were adjusted to 1.25 mM puromycin and microsomal membranes were added for an additional hour (lanes 5 and 6). Subsequently, translation mixtures were centrifuged for 15 min at 12,000 × g and supernatants (odd lanes), and resuspended pellets (even lanes) were applied in equivalent amounts and analyzed by SDS-PAGE. The relative amounts of soluble and pelleted products were calculated by quantifying radioactively labeled proteins using a Fuji BAS 2000 Bioimager and correcting for the background in each lane. The lower panels depict quantitation of the results from three independent experiments.
To characterize membrane factors which may be important for membrane-association of the polyproteins, the experiments were repeated with microsomes that had been pretreated with high concentrations of salt. Salt extraction (1 M NaCl) is conventionally used to release peripheral but not integral membrane proteins. Sedimentation of wild-type IAP polyproteins in vitro translated in the presence of salt-extracted microsomes revealed that their ability to associate with microsomal membranes was retained (data not shown), indicating that membrane binding is independent of peripheral ER-associated proteins.
IAP polyprotein transport requires only the minimal translocation apparatus in the target membrane.
The result that IAP polyprotein transport occurs mostly cotranslationally suggests that components of the SRP pathway are involved (66, 67). The minimal functional translocation apparatus for SRP-mediated protein transport is composed of two membrane protein complexes: the SRP receptor complex and the Sec61p complex forming the translocation channel (21). To analyze whether IAP polyproteins require specific membrane components for their membrane association, flotation experiments were performed after translation of IAP proteins in the presence of synthetic liposomes or proteoliposomes selectively reconstituted with different ER proteins. The liposomes were prepared from synthetic phospholipids and were reconstituted with purified ER proteins as described previously (21).
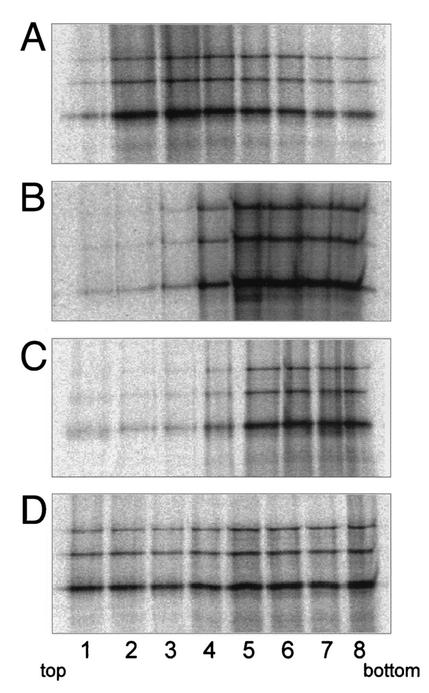
Figure 3 shows the protein distribution after flotation of MIA2 polyproteins, in vitro-synthesized in the presence of microsomal membranes (Fig. 3A); synthetic liposomes lacking any protein component (Fig. 3B); or proteoliposomes reconstituted with either signal peptidase (Fig. 3C) or SRP receptor and Sec61p complex (Fig. 3D). Clearly, IAP polyproteins did not associate with synthetic liposomes (Fig. 3B, lanes 1 to 4) or proteoliposomes containing signal peptidase, a transmembrane protein which is not involved in ER targeting of proteins (Fig. 3C, lanes 1 to 4), while ca. 70% of IAP polyproteins associated with microsomal membranes (Fig. 3A, lanes 1 to 4). If synthesized in the presence of synthetic proteoliposomes containing SRP receptor and Sec61p complex, ca. 40% of IAP polyproteins were found associated with the membrane fraction (Fig. 3D, fraction 1 to 4). These results indicate that there is very little nonspecific lipid binding of the IAP polyproteins and the specific membrane association requires the SRP docking and translocation apparatus. The decreased binding to SRP receptor/Sec61p proteoliposomes compared to native microsomes may be due to the lower concentration of Sec61p complex in the proteoliposomes versus microsomes and to nonfunctional membrane-insertion of SRP receptor and Sec61p complexes during preparation of proteoliposomes.
FIG. 3.
Analysis of IAP polyprotein association with liposomes and reconstituted proteoliposomes. Coupled transcription-translation reactions were programmed with pTM1-MIA2 in the presence of microsomal membranes (A), liposomes prepared from synthetic phospholipids without any protein (B), proteoliposomes reconstituted with purified signal peptidase complex (C), or proteoliposomes reconstituted with SRP receptor and Sec61p complex (D). After translation, all mixtures were subjected to sucrose flotation as described in the legend to Fig. 1C. Fractions were collected from top (fraction 1) to bottom (fraction 8), and proteins were resolved by SDS-PAGE and analyzed by autoradiography.
The N-terminal region of IAP Gag polyproteins interacts with SRP.
The process of protein translocation across or integration into the ER membrane can be divided into the targeting and translocation phases. Transport of a nascent, growing polypeptide chain to the ER membrane occurs after initial binding of its signal sequence to the 54-kDa polypeptide subunit of the cytosolic SRP (SRP54) (27). The subsequent interaction of SRP with its receptor at the ER membrane causes immediate SRP displacement and concomitant delivery of the ribosome-nascent-chain complex to the Sec61p complex, initiating the translocation phase. In order to analyze whether targeting of IAP polyproteins is SRP mediated, we performed chemical cross-linking experiments and determined whether there is a direct interaction between SRP54 and IAP polyproteins. Nascent-chain-ribosome complexes were generated by cleaving pTM1-MIA2 with different restriction endonucleases before adding the DNA to coupled in vitro transcription-translation reactions. Because the resulting truncated mRNAs lack stop codons, translation of nascent peptide chains is not terminated and nascent-chain-ribosome complexes are generated. Synthesis of truncated IAP polyproteins was performed in the absence or presence of microsomal membranes. Subsequently, nascent-chain-ribosome complexes were sedimented by centrifugation and subjected to cross-linking experiments using the homobifunctional cross-linker DSS.
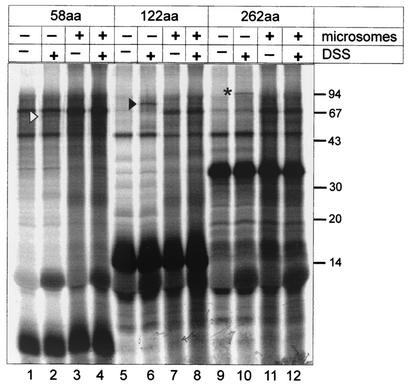
Figure 4 shows the results of cross-linking experiments for nascent chains of different lengths (58 amino acids, lanes 1 to 4; 122 amino acids, lanes 5 to 8; 262 amino acids, lanes 9 to 12). In each case, the first two lanes represent translations in the absence of microsomes (with or without DSS), while the next two lanes correspond to translations in the presence of microsomes. In the case of all three nascent chains, the predominant translation products exhibited the expected migration pattern (Fig. 4). Additional products observed in all lanes probably correspond to nonspecific labeling of components of the translation mix or of microsomal membranes and were disregarded. Specific cross-linking products, on the other hand, were expected to give rise to products of different migration depending on the length of the nascent chain and should be found specifically in the reactions without addition of membranes (since SRP54 dissociates upon ER association of the complex). Such specific products were indeed detected for all three nascent-chain complexes and are marked as described in the legend to Fig. 4. The apparent molecular mass of the cross-linking product for the 58-amino-acid peptide corresponded to 60 kDa (Fig. 4, lane 2). This product was absent when DSS was omitted (Fig. 4, lane 1) and when the translation was performed in the presence of membranes (Fig. 4, lanes 3 and 4). Subtracting the molecular mass of the 58-amino-acid peptide from the 60-kDa product indicated a cross-linked protein with a molecular mass of ca. 54 kDa. The same calculations for the cross-linking products of the 122-amino-acid peptide (ca. 70 kDa) and the 262-amino-acid peptide (ca. 90 kDa) reveal cross-linked proteins of the same molecular mass of ca. 55 kDa. Taken together, these results strongly suggest that nascent-chain complexes of IAP polyproteins bind to SRP54 and use the SRP transport pathway for ER targeting.
FIG. 4.
Cross-linking of ribosome-nascent-chain complexes to SRP components. Coupled transcription-translation reactions were programmed with pTM1-MIA2 linearized with either StuI (lanes 1 to 4), EcoRI (lanes 5 to 8), or NgoMI (lanes 9 to 12) to produce short mRNAs lacking a stop codon. Reactions were performed for 30 min in the absence or presence of microsomal membranes as indicated. Subsequently, ribosome-nascent-chain complexes were centrifuged at 75,000 rpm in a Beckman Optima TLX ultracentrifuge for 30 min at 2°C. Aliquots from the resuspended pellets were subjected to a cross-linking reaction (2 h, 0°C) with the homobifunctional cross-linking reagent DSS as indicated. Molecular mass markers are indicated on the right. The open arrowhead marks the cross-linked product (58aa × unknown protein) of about 60 kDa in lane 2; a solid arrowhead and an asterisk identify the cross-linked products in lanes 6 and 10, respectively. Note that no specific cross-linking products were detected in lanes 4, 8, and 12, where the corresponding reactions translated in the presence of microsomal membranes were applied.
To verify that IAP polyproteins interact with SRP54, immunoprecipitation of cross-linking products was performed. Nascent-chain complexes of the 122-amino-acid IAP peptide were synthesized in the absence or presence of microsomal membranes as described above. Subsequently, cross-linking with DSS was performed and cross-linking products were immunoprecipitated with a rabbit polyclonal antiserum specific for the 54-kDa subunit of SRP (αSRP54) (25). Figure 5 shows that a specific cross-linking product was observed when translation was performed in the absence of microsomes (lane 2) and this product of ca. 70 kDa was specifically immunoprecipitated by αSRP54 (lane 3). Cross-linking after translation in the presence of membranes did not yield this product (lane 5), but a small amount of the 70-kDa cross-linking product was also detected after immunoprecipitation (lane 6), indicating that part of the nascent chains were not dissociated from SRP despite the presence of microsomal membranes. These immunoprecipitation experiments prove that the N-terminal region of IAP Gag functions as a signal sequence, which is recognized and bound by SRP.
FIG. 5.
Immunoprecipitation of cross-linking products derived from the 122-amino-acid MIA2 polyprotein fragment. Coupled transcription-translation and cross-linking reactions were performed as described in the legend to Fig. 4. Labeled products cross-linked to the 54-kDa subunit of SRP were immunoprecipitated using a specific antiserum directed against SRP54 (lanes 3 and 6). The positions of molecular mass markers are indicated on the right.
In vitro-translated IAP Gag is not translocated into the ER lumen.
To determine whether Gag polyproteins are translocated into the ER lumen, we performed protease protection experiments. To this end, proteins synthesized in the presence or absence of microsomal membranes were subjected to proteinase K treatment. As a positive control, ppl was synthesized in parallel, which is translocated into microsomal membranes and subsequently cleaved by signal peptidase. Figure 6B shows that ca. 40% of ppl was translocated into the ER in the presence of microsomal membranes as indicated by its cleavage to prolactin (pl; compare lanes 1 and 2). The translocated pl was protected from proteinase K digestion, while the remaining ppl was degraded (Fig. 6B, lane 3). Treating the samples with detergent prior to proteinase K digestion led to complete degradation of the translation products (Fig. 6B, lane 4). Analysis of the MIA2 translation products, on the other hand, revealed no protease protected polyprotein even in the absence of detergent (Fig. 6B, lanes 5 to 7), indicating that IAP-specific products are not translocated into the ER. The residual 25-kDa product observed in lanes 7 and 8 probably corresponds to a protease-resistant fragment of the polyprotein, since it is also present in the detergent-treated sample (lane 8).
FIG. 6.
(A) Amino acid sequence at the N termini of ppl, MIA2, and the fusion protein SPpplMIA, in which the signal sequence of MIA2 was replaced by the ppl signal peptide. The amino acid sequences are given in single-letter code, and positively (+) and negatively (−) charged amino acid side chains as well as the cleavage site for signal peptidase (gap) are indicated. The domain representing the ppl signal peptide is shadowed, and the domain representing the IAP signal peptide is boxed. (B) ER transport of ppl, wt IAP Gag and the SPpplMIA fusion protein. Proteins were synthesized by coupled in vitro transcription-translation in the presence or absence of microsomal membranes as indicated. ER import of translation products was analyzed by incubation with proteinase K (50 μg/ml) for 1 h at 0°C in the absence or presence of 0.1% Triton X-100 as indicated. Aliquots of each reaction mixture were analyzed by SDS-PAGE (ppl, lanes 1 to 4; MIA2, lanes 5 to 8, SPpplMIA, lanes 9 to 12). The positions of ppl and the cleaved prolactin (pl) are indicated on the left.
Both, the IAP signal sequence and the downstream segments of Gag are responsible for the lack of translocation.
The lack of ER translocation of the IAP polyproteins may be due to a translocation-incompetent signal sequence or to the downstream part of Gag blocking ER transport. To analyze these two possibilities, we constructed fusion proteins containing either the first 28 amino acids of MIA14 Gag in the position of the ppl signal sequence or the ppl signal sequence in the position of the IAP N-terminal region. Figure 6A shows the N-terminal sequence of the fusion protein SPpplMIA containing the ppl signal sequence, including the signal peptidase cleavage site at the N terminus of the IAP polyprotein. In vitro synthesis of this fusion protein yielded amounts of Gag-specific proteins similar to those observed for the wild-type construct. However, the translation products carrying the ppl signal sequence were also not translocated into the ER (Fig. 6B, lanes 9 to 12). No cleavage of the heterologous signal sequence was observed (Fig. 6B, compare lanes 9 and 10), and proteinase K treatment in the presence and absence of detergent led to complete degradation of the fusion protein with the protease-resistant 25-kDa product remaining (Fig. 6B, lanes 11 and 12).
To determine whether the MIA14 signal sequence can function in a heterologous context to translocate an ER protein through the Sec61p channel, we constructed two fusion proteins containing either the first 23 or the first 28 amino acids of the MIA14 polyprotein at the N terminus of pl (Fig. 7A, plMIA28 and plMIA23). The shorter fusion protein was made to avoid the positively charged amino acids in positions 24 and 27 of the IAP polyprotein, which could have an inhibitory effect on translocation (61). In both constructs, the signal peptidase cleavage site of ppl was maintained (Fig. 7A). In vitro transcription and translation in the presence of microsomal membranes revealed that both fusion proteins were produced at levels similar to those of wild-type ppl (Fig. 7B, lanes 1, 4, and 7). However, both fusion proteins failed to be processed by signal peptidase, while ca. 40% of ppl was cleaved. Furthermore, no protease-protected proteins were observed for the two fusion proteins, while pl was largely protected unless treated in the presence of detergent (Fig. 7B). These results show that the MIA14 signal sequence is not capable of mediating luminal translocation of a bona fide transport competent protein.
FIG. 7.
(A) Amino acid sequence at the N termini of ppl, MIA2, and the fusion proteins plMIA28 and plMIA23, in which the signal sequence of ppl was replaced by the first 28 or 23 amino acids of MIA2, respectively. The amino acid sequences are given in single letter code and positively (+) and negatively (−) charged amino acid side chains as well as the cleavage site for signal peptidase (gap) are indicated. The domain representing the IAP signal peptide is boxed. (B) ER transport of ppl, plMIA28, and plMIA23. Proteins were synthesized and protease treated as described in the legend to Fig. 6 (ppl: lanes 1 to 3; plMIA23:lanes 4 to 6; plMIA28: lane 7 to 9).
IAP Gag polyproteins are strongly associated with but not inserted into the ER membrane.
The previous experiments showed that IAP polyproteins are transported to the ER membrane in an SRP-dependent manner but are not translocated into the ER lumen. To analyze whether these proteins remain peripherally associated with the cytoplasmic side of the membrane or are inserted as transmembrane proteins, we performed membrane extraction experiments following in vitro translation of MIA2 in the presence of microsomal membranes. Extraction with high-concentration salt (1 M NaCl), chelating divalent cations with EDTA, and treatment with high-pH carbonate buffer is expected to release trapped soluble and peripherally membrane-associated proteins, while not affecting integral membrane proteins. Detergent extraction, on the other hand, should solubilize all membrane proteins. Figure 8 shows that extraction of membrane-associated IAP-specific polyproteins with 1 M salt released ca. 35% of the protein (lanes 3 and 4), and extraction with 50 mM EDTA released ca. 50% (lanes 5 and 6). Treatment with high-pH buffer, which is a more stringent extraction method, resulted in the release of ca. 70% of the polyprotein (lanes 7 and 8). As a control, dissolving the membranes with 0.1% Triton X-100 resulted in 85% soluble IAP Gag polyproteins (lanes 9 and 10), while extraction with buffer alone solubilized only ca. 20% of the protein (lanes 1 and 2). These results indicate that membrane association of the IAP polyproteins is very tight but that they do not behave as integral membrane proteins.
FIG. 8.
Analysis of IAP Gag polyprotein membrane-association. Coupled in vitro transcription-translation reactions were programmed with pTM-MIA2 in the presence of microsomal membranes, and membrane-bound products were collected by centrifugation. Subsequently, membranes were extracted for 1 h at 25°C with buffer only (lanes 1 and 2); with 1 M NaCl (lanes 3 and 4), 50 mM EDTA (lanes 5 and 6), or 0.1% Triton X-100 (lanes 9 and 10); or with 0.1 M carbonate buffer (pH 11.5) for 30 min at 0°C (lanes 7 and 8). Treated samples were centrifuged for 15 min at 12,000 × g and supernatants (odd lanes) and resuspended pellets (even lanes) were applied in equivalent amounts and analyzed by SDS-PAGE and autoradiography. Relative amounts of soluble and membrane-associated products were calculated as described before, and the lower panel depicts quantitation of the results obtained in three independent experiments.
DISCUSSION
Our current knowledge regarding the intracellular transport, assembly, and budding of enveloped viruses is mostly derived from infection and transfection experiments in tissue culture. It appears likely, however, that a detailed understanding of the underlying molecular principles will require the development of cell-free in vitro systems, as has been the case for nuclear (1), ER (4), and vesicular transport (18). IAPs are a particularly suitable viral system for such experiments, because virus budding occurs at the ER (with microsomal membranes being readily available for in vitro studies) and requires only the viral Gag polyprotein. This is in contrast to, e.g., alphaviruses (35, 75), hepadnaviruses (5), and foamy viruses (51), which also bud into the ER but require the viral glycoproteins for budding and/or ER transport. Therefore, we attempted to reconstitute ER transport of IAP Gag polyproteins in an in vitro system, using in vitro translation in the presence of microsomal membranes or reconstituted (proteo-)liposomes. Here we show that ER transport of the MIA14 polyproteins is mediated by an N-terminal signal sequence which directs the polyproteins into the SRP-dependent transport pathway. The viral polyproteins associate with the ER membrane via SRP receptor and Sec61p complex but are not translocated into the ER lumen. Protease digestion experiments detected no protected IAP polyproteins, indicating that budding and release of virus-like particles into the ER lumen did not take place in this in vitro system. This may be due to the low concentration of translation products or to the lack of essential cofactors.
The first indication that SRP may be involved in IAP polyprotein transport came from the observation that transport occurred mostly cotranslationally. This was in contrast to a Gag derivative which contained the Src protein N-terminal sequence instead of its own N terminus. In the latter case, adding membranes to the completed translation reaction yielded the same amount of membrane-associated proteins as for translation in the presence of microsomal membranes, while the wild-type polyproteins showed much more efficient ER transport when membranes were present during translation. Cotranslational transport is dependent on the well-characterized SRP pathway (reviewed in reference 68) and needs a specific signal sequence in the nascent peptide (61). Typically, eukaryotic signal sequences are composed of a short, positively charged hydrophilic amino-terminal segment and a central hydrophobic part (7 to 15 residues), followed by a more polar carboxy-terminal region (61). This signal sequence is recognized by SRP, a ribonucleoprotein complex consisting of a 7S RNA and six different polypeptide subunits. SRP interacts through its 54-kDa subunit with the signal sequence of nascent polypeptide chains (27) and directs the entire complex, which consists of the ribosome, nascent chain, and SRP to the ER membrane. Continued elongation of the polypeptide chain is delayed or even arrested (65) until SRP is bound to the SRP receptor (also called docking protein), an integral protein complex of the ER membrane (37). As a result, SRP is released from the signal sequence in a GTP-dependent manner (10), and the ribosome together with the nascent chain is subsequently passed onto the heterotrimeric Sec61p complex which forms a protein-conducting channel through the ER membrane (11, 53). The continuing translation inserts the polypeptide into the Sec61p channel. Cleavable signal sequences are cotranslationally processed by signal peptidase at the luminal side of the ER membrane.
Several lines of evidence show that IAP polyprotein transport occurs via the SRP pathway and depends on an N-terminal signal sequence in the Gag polyprotein. Deletion of the first 28 predominantly hydrophobic amino acids of Gag led to complete loss of membrane transport as shown by sedimentation and flotation experiments, and this result corresponds to the requirement for this region to mediate ER transport in transfection experiments (70). This N-terminal region can be specifically cross-linked to the 54-kDa subunit of SRP, indicating that it is a functional signal sequence. Furthermore, translation in the presence of reconstituted proteoliposomes identified SRP receptor and Sec61p complex as IAP Gag receptors in the ER membrane. These two complexes form the minimal translocation apparatus which is necessary and sufficient for SRP-dependent transport and insertion into reconstituted proteoliposomes (21). A cross-link between IAP Gag nascent chains and subunits of the Sec61 complex could not be detected. This may be explained by the lack of lysine residues in suitable positions and/or a very brief interaction between the two peptides.
SRP-mediated ER transport of cellular proteins normally leads either to luminal transport through the Sec61p channel or to membrane integration of their hydrophobic transmembrane domains. Some proteins insert with their N- or the C-terminal region serving as membrane anchor. Retroviral budding, on the other hand, requires that the viral polyproteins remain on the cytoplasmic face of the membrane. Furthermore, cryo-electron microscopy analysis of immature HIV particles revealed that the globular MA domain of Gag is closely apposed to but separable from the inner leaflet of the viral membrane (19, 72), indicating that MA is not inserted into the membrane. Protease digestion of in vitro translation products produced in the presence of microsomal membranes showed that IAP polyproteins were not protected, while the bona fide secretory protein ppl was translocated into the ER and cleaved by signal peptidase, and the resulting pl was protected from digestion. Experiments with chimeric proteins containing segments of IAP Gag and ppl indicated not only that the signal sequence from IAP Gag is not translocation competent but also that the downstream segments of Gag block translocation, even when the translocation-competent signal sequence from ppl is attached to the protein. Analysis of the IAP N-terminal sequence showed a cluster of positively charged residues (Fig. 6A). Statistically, positive charges are enriched on the cytosolic side and depleted from the exoplasmic side of noncleavable signal sequences (the “positive inside” rule) (63, 64). In bacterial signal sequences, a detrimental effect of downstream positive charges on translocation has been documented in a number of cases, both in vivo and in vitro (34, 73). The topogenic role of the flanking charges has also been experimentally confirmed by site-directed mutagenesis (39, 49, 50, 60). Thus, the cluster of basic residues may prevent translocation of Gag through the Sec61p channel, even in the heterologous context of the chimeric protein SPpplMIA.
Membrane extraction experiments revealed that IAP Gag polyproteins are tightly associated with the ER membrane but do not behave as integral membrane proteins. The IAP signal sequence is not cleaved by signal peptidase and could function as a type I signal anchor. Signal-anchor sequences mainly differ from cleavable signal sequences by long hydrophobic cores of 19 to 27 hydrophobic residues forming an α-helix across the lipid bilayer (15, 43, 62). Mutational analysis of the influenza A M2 protein signal-anchor showed that at least 16 hydrophobic amino acids are required to integrate the protein in the membrane (29). The N terminus of IAP Gag contains two stretches of 5 and 12 uncharged amino acid residues, respectively (Fig. 6A). Both of these regions are preceded by a negatively charged amino acid, and the second stretch contains four polar amino acids, features that are incompatible with a functional signal anchor. Conceivably, the N-terminal segment of Gag may extend into the lipid bilayer or is tightly apposed to the membrane, rendering it relatively resistant to conditions that characteristically remove peripherally associated membrane proteins. A similarly tight membrane association was observed for the MA proteins of influenza A virus (31) and vesicular stomatitis virus (VSV) (8, 9). However, mutants of influenza A MA associated with membranes in the absence of the hydrophobic regions (31) and the MA proteins of influenza A (7, 23) and VSV (44, 74) associated with artificial phospholipid vesicles, in contrast to the IAP polyproteins. In the case of VSV, the MA protein may form an amphipathic helix which is partially embedded into the lipid bilayer (33).
Comparing IAP sequences with those of other retroviruses revealed homologies extending throughout most of gag and pol with a conspicuous absence of homologies in the 5′ terminal part of gag corresponding to the MA coding region (3, 14, 38, 55). Conceivably, IAPs acquired cellular targeting signals through retrotransposition to fulfill the specific requirements of ER transport and budding. Interestingly, however, these transport signals share functional properties of the targeting signals of exogenous retroviruses. In the case of HIV, plasma membrane association depends on a bipartite signal formed by N-terminal myristoylation of Gag and a cluster of basic residues within the MA domain (76). We speculate that stable membrane association of IAP Gag is mediated in a similar way by (i) partial insertion of the hydrophobic N terminus into the lipid bilayer, functionally analogous to myristoylated Gag polyproteins, and (ii) the cluster of basic residues following the signal peptide, thereby mimicking the bipartite transport signals of other polyproteins. Taken together, our results suggest that this IAP has acquired an N-terminal targeting signal which serves at least two functions. First, it mediates SRP-dependent specific transport of viral polyproteins to the ER membrane. Second, it serves to stably attach the polyproteins to the membrane in a tight manner to prepare for budding, while preventing translocation into the ER and both of these features may depend on the basic amino acids following the signal sequence. By analogy, structural polyproteins of other retroviruses may also use (yet-unknown) cellular transport pathways to reach the viral budding site.
Acknowledgments
We are grateful to B. Dobberstein for antiserum against SRP54 and to B. Müller for help in preparation of the figures.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft to H.-G.K. (Kr906/2) and R.Z. (SFB530).
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. J., K. E. Mostov, and G. Blobel. 1983. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc. Natl. Acad. Sci. USA 80:7249-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aota, S., T. Gojobori, K. Shigesada, H. Ozeki, and T. Ikemura. 1987. Nucleotide sequence and molecular evolution of mouse retrovirus-like IAP elements. Gene 56:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, G., and B. Dobberstein. 1975. Transfer to proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67:852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucher, D. J., I. G. Kharitonenkov, J. A. Zakomirdin, V. B. Grigoriev, S. M. Klimenko, and J. F. Davis. 1980. Incorporation of influenza virus M-protein into liposomes. J. Virol. 36:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong, L. D., and J. K. Rose. 1994. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J. Virol. 68:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong, L. D., and J. K. Rose. 1993. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J. Virol. 67:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly, T., and R. Gilmore. 1989. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell 57:599-610. [DOI] [PubMed] [Google Scholar]
- 11.Corsi, A. K., and R. Schekman. 1996. Mechanism of polypeptide translocation into the endoplasmic reticulum. J. Biol. Chem. 271:30299-30302. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies, R. J., S. L. Sanders, D. A. Feldheim, and R. Schekman. 1991. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349:806-808. [DOI] [PubMed] [Google Scholar]
- 13.Deshaies, R. J., and R. Schekman. 1987. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 105:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorner, A. J., F. Bonneville, R. Kriz, K. Kelleher, K. Bean, and R. J. Kaufman. 1991. Molecular cloning and characterization of a complete Chinese hamster provirus related to intracisternal A particle genomes. J. Virol. 65:4713-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg, D. 1984. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 53:595-623. [DOI] [PubMed] [Google Scholar]
- 16.Facke, M., A. Janetzko, R. L. Shoeman, and H. G. Krausslich. 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 67:4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehrmann, F., R. Welker, and H. G. Krausslich. 1997. Intracisternal A-type particles express their proteinase in a separate reading frame by translational frameshifting, similar to D-type retroviruses. Virology 235:352-359. [DOI] [PubMed] [Google Scholar]
- 18.Fries, E., and J. E. Rothman. 1980. Transport of vesicular stomatitis virus glycoprotein in a cell-free extract. Proc. Natl. Acad. Sci. USA 77:3870-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Krausslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich, D., E. Hartmann, S. Prehn, and T. A. Rapoport. 1992. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357:47-52. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich, D., and T. A. Rapoport. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75:615-630. [DOI] [PubMed] [Google Scholar]
- 22.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregoriades, A., and B. Frangione. 1981. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J. Virol. 40:323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, M., L. Jelinek, S. Whiting, and E. Barklis. 1990. Transport and assembly of gag proteins into Moloney murine leukemia virus. J. Virol. 64:5306-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser, S., G. Bacher, B. Dobberstein, and H. Lutcke. 1995. A complex of the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J. 14:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, L. E., H. C. Krutzsch, and S. Oroszlan. 1983. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational proteins modification. Proc. Natl. Acad. Sci. USA 80:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.High, S., and B. Dobberstein. 1991. The signal sequence interacts with the methionine-rich domain of the 54-kD protein of signal recognition particle. J. Cell Biol. 113:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hull, J. D., R. Gilmore, and R. A. Lamb. 1988. Integration of a small integral membrane protein, M2, of influenza virus into the endoplasmic reticulum: analysis of the internal signal-anchor domain of a protein with an ectoplasmic NH2 terminus. J. Cell Biol. 106:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krausslich, H. G., and R. Welker. 1996. Intracellular transport of retroviral capsid components. Curr. Top. Microbiol. Immunol. 214:25-63. [DOI] [PubMed] [Google Scholar]
- 31.Kretzschmar, E., M. Bui, and J. K. Rose. 1996. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology 220:37-45. [DOI] [PubMed] [Google Scholar]
- 32.Kuff, E. L., and K. K. Lueders. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183-276. [DOI] [PubMed] [Google Scholar]
- 33.Lenard, J., and R. Vanderoef. 1990. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J. Virol. 64:3486-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, P., J. Beckwith, and H. Inouye. 1988. Alteration of the amino terminus of the mature sequence of a periplasmic protein can severely affect protein export in Escherichia coli. Proc. Natl. Acad. Sci. USA 85:7685-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez, S., J. S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luftig, R. B., and L. D. Lupo. 1994. Viral interactions with the host-cell cytoskeleton: the role of retroviral proteases. Trends Microbiol. 2:178-182. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, D. I., E. Krause, and B. Dobberstein. 1982. Secretory protein translocation across membranes-the role of the “docking protein'. Nature 297:647-650. [DOI] [PubMed] [Google Scholar]
- 38.Mietz, J. A., Z. Grossman, K. K. Lueders, and E. L. Kuff. 1987. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J. Virol. 61:3020-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monier, S., P. Van Luc, G. Kreibich, D. D. Sabatini, and M. Adesnik. 1988. Signals for the incorporation and orientation of cytochrome P450 in the endoplasmic reticulum membrane. J. Cell Biol. 107:457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 41.Muller, G., and R. Zimmermann. 1987. Import of honeybee prepromelittin into the endoplasmic reticulum: structural basis for independence of SRP and docking protein. EMBO J. 6:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson, I., P. Whitley, and G. von Heijne. 1994. The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J. Cell Biol. 126:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogden, J. R., R. Pal, and R. R. Wagner. 1986. Mapping regions of the matrix protein of vesicular stomatitis virus which bind to ribonucleocapsids, liposomes, and monoclonal antibodies. J. Virol. 58:860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono, M., H. Toh, T. Miyata, and T. Awaya. 1985. Nucleotide sequence of the Syrian hamster intracisternal A-particle gene: close evolutionary relationship of type A particle gene to types B and D oncovirus genes. J. Virol. 55:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panzner, S., L. Dreier, E. Hartmann, S. Kostka, and T. A. Rapoport. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81:561-570. [DOI] [PubMed] [Google Scholar]
- 48.Parent, L. J., C. B. Wilson, M. D. Resh, and J. W. Wills. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks, G. D., and R. A. Lamb. 1993. Role of NH2-terminal positively charged residues in establishing membrane protein topology. J. Biol. Chem. 268:19101-19109. [PubMed] [Google Scholar]
- 50.Parks, G. D., and R. A. Lamb. 1991. Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64:777-787. [DOI] [PubMed] [Google Scholar]
- 51.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 53.Rapoport, T. A., M. M. Rolls, and B. Jungnickel. 1996. Approaching the mechanism of protein transport across the ER membrane. Curr. Opin. Cell Biol. 8:499-504. [DOI] [PubMed] [Google Scholar]
- 54.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reuss, F. U., and H. C. Schaller. 1991. cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J. Virol. 65:5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual., 2 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Schatz, P. J., and J. Beckwith. 1990. Genetic analysis of protein export in Escherichia coli. Annu. Rev. Genet. 24:215-248. [DOI] [PubMed] [Google Scholar]
- 58.Schlenstedt, G., G. H. Gudmundsson, H. G. Boman, and R. Zimmermann. 1990. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Biol. Chem. 265:13960-13968. [PubMed] [Google Scholar]
- 59.Schlenstedt, G., G. H. Gudmundsson, H. G. Boman, and R. Zimmermann. 1992. Structural requirements for transport of preprocecropinA and related presecretory proteins into mammalian microsomes. J. Biol. Chem. 267:24328-24332. [PubMed] [Google Scholar]
- 60.Szczesna-Skorupa, E., N. Browne, D. Mead, and B. Kemper. 1988. Positive charges at the NH2 terminus convert the membrane-anchor signal peptide of cytochrome P-450 to a secretory signal peptide. Proc. Natl. Acad. Sci. USA 85:738-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 62.von Heijne, G. 1985. Signal sequences. The limits of variation. J. Mol. Biol. 184:99-105. [DOI] [PubMed] [Google Scholar]
- 63.von Heijne, G. 1986. Towards a comparative anatomy of N-terminal topogenic protein sequences. J. Mol. Biol. 189:239-242. [DOI] [PubMed] [Google Scholar]
- 64.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 65.Walter, P., and G. Blobel. 1981. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter, P., and G. Blobel. 1981. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walter, P., I. Ibrahimi, and G. Blobel. 1981. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91:545-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walter, P., and A. E. Johnson. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87-119. [DOI] [PubMed] [Google Scholar]
- 69.Watts, C., W. Wickner, and R. Zimmermann. 1983. M13 procoat and a pre-immunoglobulin share processing specificity but use different membrane receptor mechanisms. Proc. Natl. Acad. Sci. USA 80:2809-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welker, R., A. Janetzko, and H. G. Krausslich. 1997. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J. Virol. 71:5209-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickner, W., and M. R. Leonard. 1996. Escherichia coli preprotein translocase. J. Biol. Chem. 271:29514-29516. [DOI] [PubMed] [Google Scholar]
- 72.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamane, K., and S. Mizushima. 1988. Introduction of basic amino acid residues after the signal peptide inhibits protein translocation across the cytoplasmic membrane of Escherichia coli. Relation to the orientation of membrane proteins. J. Biol. Chem. 263:19690-19696. [PubMed] [Google Scholar]
- 74.Zakowski, J. J., W. A. Petri, Jr., and R. R. Wagner. 1981. Role of matrix protein in assembling the membrane of vesicular stomatitis virus: reconstitution of matrix protein with negatively charged phospholipid vesicles. Biochemistry 20:3902-3907. [DOI] [PubMed] [Google Scholar]
- 75.Zhao, H., B. Lindqvist, H. Garoff, C. H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]