Abstract
Infection with genital human papillomaviruses (HPVs) is the primary cause of cervical cancer. The infection is widespread, and little is known about the secondary factors associated with progression from subclinical infection to invasive carcinoma. Here we report that HPV genomes are efficiently targeted in vivo by CpG methylation, a well-known mechanism of transcriptional repression. Indeed, it has been shown previously that in vitro-methylated HPV type 16 (HPV-16) DNA is transcriptionally repressed after transfection into cell cultures. By using a scan with the restriction enzyme McrBC, we observed a conserved profile of CpG hyper- and hypomethylation throughout the HPV-16 genomes of the tumor-derived cell lines SiHa and CaSki. Methylation is particularly high in genomic segments overlying the late genes, while the long control region (LCR) and the oncogenes are unmethylated in the single HPV-16 copy in SiHa cells. In 81 patients from two different cohorts, the LCR and the E6 gene of HPV-16 DNA were found to be hypermethylated in 52% of asymptomatic smears, 21.7% of precursor lesions, and 6.1% of invasive carcinomas. This suggests that neoplastic transformation may be suppressed by CpG methylation, while demethylation occurs as the cause of or concomitant with neoplastic progression. These prevalences of hyper- and hypomethylation also indicate that CpG methylation plays an important role in the papillomavirus life cycle, which takes place in asymptomatic infections and precursor lesions but not in carcinomas. Bisulfite modification revealed that in most of the HPV-16 genomes of CaSki cells and of asymptomatic patients, all 11 CpG dinucleotides that overlap with the enhancer and the promoter were methylated, while in SiHa cells and cervical lesions, the same 11 or a subset of CpGs remained unmethylated. Our report introduces papillomaviruses as models to study the mechanism of CpG methylation, opens research on the importance of this mechanism during the viral life cycle, and provides a marker relevant for the etiology and diagnosis of cervical cancer.
Human papillomavirus type 16 (HPV-16) and related HPV types are carcinogenic, and persistent HPV, infection is a prerequisite in the etiology of most or even all cervical cancers (22, 38, 52). Most women become infected by HPVs, and while some of these infections progress malignantly, most remain subclinical or lead only to precursor lesions. The factors that determine these outcomes are poorly understood. Transformation by HPVs depends on the oncoproteins E6 and E7, whose transcription is modulated by numerous transcription factors and epigenetic mechanisms (5, 18). Tumor progression may result from stimulated oncoprotein expression through transcriptional induction by steroids (10, 17), by deletion of transcriptional silencers (25), or by integration of HPV genomes into cellular DNA (3, 40, 43).
cis-responsive elements that regulate E6 and E7 oncogene transcription are spread throughout the long control region (LCR) of HPV-16, an 850-bp segment between the L1 and E6 genes. Transcription starts at the E6 promoter P97, which is regulated by one binding site for Sp1 and two for the HPV-encoded factor E2 (11, 47). The activity of P97 is stimulated by an enhancer with binding sites for several cellular factors, including AP1, NF1, and the progesterone receptor (1, 10, 16-18). Two specifically positioned nucleosomes can form over the enhancer and promoter (42) and repress transcription when they are modified by histone deacetylases (HDACs). HDACs are associated with CDP, which binds a silencer between the enhancer and promoter (27, 30). When HPV genomes integrate into cellular DNA during progression to malignancy, a nuclear matrix attachment region located downstream of P97 becomes a strong transcriptional stimulator (43). Here we report that a well-known and potent epigenetic mechanism, the repression of transcription by methylation of DNA at CpG dinucleotides, targets HPV-16 genomes, with likely consequences for viral oncogene expression, the viral life cycle, and carcinogenic progression of HPV-16 lesions.
In mammals, DNA methylation mediated by a family of DNA-[cytosine-5] methyltransferases (DNMT1, DNMT3a, and DNMT3b) (28, 34) occurs at 2 to 4% of all cytosines located 5′ to guanosine (CpG) (7). CpGs are present in mammalian genomes at only 20% of the statistically expected frequency (39), suggesting adverse functional consequences of methylation as a cause for their loss during evolution. CpG methylation leading to gene silencing is involved in X-chromosome inactivation and genomic imprinting (31). The rare case of tissue-specific gene expression mediated by CpG methylation has recently been confirmed (15). Furthermore, CpG methylation is required for stabilization of the genome against recombination, which is implicated by the preferential methylation of repeated sequences (51). During carcinogenesis, global hypomethylation and regional (CpG islands) hypermethylation (21) are frequently observed. CpG methylation represses gene expression either by preventing transcription factors from recognizing their cognate binding sites (36, 48) or by attracting proteins that bind methylated DNA (MeCPs) (20), which recruit HDAC1 to condense adjacent chromatin.
Methylation of the genomes of the carcinogenic HPV types in situ has never been studied, but there is evidence for its potential involvement in the regulation of HPV transcription. First, CpGs are underrepresented in HPVs, similar to the situation in their human host. Second, when in vitro-methylated HPV-16 DNA was transfected into cells, it failed to express (37), similar to the EcoRI-resistant HPV-16 DNA studied in one specific cell line (24). Third, methylation of the DNAs of HPV-1, which causes flat warts, and of cottontail rabbit papillomavirus was reported 20 years ago (9, 44, 50), but this report was never followed up by systematic investigation of medically more important papillomaviruses. Here, we report findings from a study of CpG methylation of HPV-16 in cell lines and in clinical samples. We found that HPV-16 DNA is an efficient target for DNA methylation, that preferentially methylated regions are asymmetrically distributed over the genome, that methylation occurs at CpGs overlapping with the enhancer and promoter, and that the frequency of CpG methylation decreases during progressive stages of cancer. These observations suggest that CpG methylation is involved in the biology of HPV-16 as well as in the etiology of cervical cancer, where it may be as a diagnostic marker.
MATERIALS AND METHODS
Analysis of the DNA of cell lines.
CaSki and SiHa cells originated from the laboratory of H. zur Hausen at the German Cancer Research Center, Heidelberg, Germany, where they were frozen in 1986 and freshly thawed for this study. Cellular DNA was purified with genomic tips (Qiagen). Fifteen and 150 ng of each of these DNAs were digested with HpaII and MspI (New England Biolabs), respectively, and, after heat inactivation of the enzymes, were amplified by PCR. For digestions with McrBC (New England Biolabs), 250 ng of SiHa or CaSki DNA was digested with 3 U of enzyme for 1 h at 37°C in 25 μl of NE buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol [pH 7.9]).
Clinical specimens.
Brazilian samples (obtained by L. L.Villa) originated from cross-sectional studies in two cities in the northeastern part of the country and consisted of cervical scrapings and tumor biopsies. Cytological and histopathological analyses were done according to the Bethesda system. For smears classified as being “asymptomatic,” ectocervical and endocervical cells were collected with a cytobrush. Tissue biopsies were classified as cervical intraepithelial lesions (cervical intraepithelial neoplasia I [CIN I] and CIN III) or invasive carcinoma and were digested with proteinase K prior to DNA isolation. DNA samples were purified by spin column chromatography and tested for the presence of HPV DNA by the MY09/11 PCR protocol for specific HPV types (6). All specimens were number coded for the privacy of the subjects. Asymptomatic and precursor samples from New Mexico were obtained and characterized during published epidemiological research (32). Invasive cervical cancer specimens were obtained from ongoing case-control studies (conducted by C. M. Wheeler) of the same population diagnosed between 1980 and 1999. In both countries, the original collection of clinical samples followed the respective guidelines for the protection of human subjects.
PCR.
Table 1 lists the primers used for dissection of the HPV-16 genome and mapping of the LCR-E6 segment. PCR was carried out in a 25-μl volume containing 0.2 mM concentrations of each of the four deoxynucleoside triphosphates (dNTPs), 10 pmol of the primers, 2.5 μl of buffer B (20 mM Tris-HCl [pH 8], 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 50% glycerol, 1% Triton X-100) (Promega), 2 mM MgCl2, and 0.75 U of Taq (Promega) with 25 or 0.25 μl of SiHa or CaSki DNA, respectively, uncleaved or cleaved by McrBC. The PCR started at 94°C for 1 min, followed by 35 amplification cycles (denaturation at 94°C for 10 s, annealing at 58°C for 30 s, and extension at 68°C for 45 s, increasing by 10 s per cycle), with final extension at 68°C for 7 min. PCR with Taq Gold was carried out in a 25-μl volume containing template DNA, 5 mM concentrations of dNTPs, 10 pmol of primers, 2.5 μl of magnesium ion-free buffer supplied by the manufacturer (Roche), 2 mM MgCl2, and 1.25 U of AmpliTaq Gold (Applied Biosystems) under the following conditions: 94°C for 9 min, followed by 40 amplification cycles (denaturation at 94°C for 10 s, annealing at 58°C for 30 s, and extension at 68°C for 45 s, increasing by 10 s per cycle), with final extension at 68°C for 7 min.
TABLE 1.
Oligonucleotides used for amplification of segments of the HPV-16 genome
| Amplicona | Primer | Position | Sequence |
|---|---|---|---|
| G1 | G1F | 515-534 | GTCTTGTTGCAGATCATCAAGA |
| G1R | 1528-1509 | ATTCTGAAAAACTCACCCCG | |
| G2 | G2F | 1501-1520 | GAGTTATACGGGGTGAGTTT |
| G2R | 2526-2505 | CAATGGTCTATGCTTTACATCC | |
| G3 | G3F | 2501-2522 | CTATGGATGTAAAGCATAGACC |
| G3R | 3501-3481 | TTTCCGGTGTCTGGCTCTGAT | |
| G4 | G4F | 3471-3490 | AGCGACCAAGATCAGAGCCA |
| G4R | 4472-4453 | GGCCTTGTTCCCAATGGAAT | |
| G5 | G5F | 4402-4421 | GGTGGGTTAGGAATTGGAAC |
| G5R | 5402-5383 | GATGTAGAGGGTACAGATGG | |
| G6 | G6F | 5367-5386 | TTCTACAACCCCGGTACCAT |
| G6R | 6370-6351 | GTCGCCATATGGTTCTGACA | |
| G7 | G7F | 6348-6367 | TGGTGTCAGAACCATATGGC |
| G7R | 7173-7150 | CAACATACATACAATACTTACAGC | |
| G8 | G8F | 7145-7184 | CGTAAGCTGTAAGTATTGTATGTATGTTGAATTAGTGTTG |
| G8R | 559-528 | TTACAGCTGGGTTTCTCTACGTGTTCTTGATG | |
| P2 | P1F | 7145-7184 | CGTAAGCTGTAAGTATTGTATGTATGTTGAATTAGTGTTG |
| P2R | 122-94 | CCTGTGGGTCCTGAAACATTGCAGTTCT | |
| P4 | P4F | 7851-7886 | TGTGTGCAAACCGTTTTGGGTTACACATTTACAAG |
| P1R | 559-528 | TTACAGCTGGGTTTCTCTACGTGTTCTTGATG | |
| P5 | P5F | 99-132 | ATGCACCAAAAGAGAACTGCAATGTTTCAGGAC |
| P5R | 559-528 | TTACAGCTGGGTTTCTCTACGTGTTCTTGATG | |
| P11 | P11F | 1-38 | ACTACAATAATTCATGTATAAAACTAAGGGCGTAACCG |
| P2R | 122-94 | CCTGTGGGTCCTGAAACATTGCAGTTCT |
Amplicons G1 to G8 overlap and cover comprehensively the whole HPV-16 genome. Amplicons P2, P4, P5, and P11 were used to study specifically the enhancer-promoter-E6 segment.
Reverse transcription and PCR.
RNA was prepared by using a Qiagen RNA kit. One microgram of RNA was primed with 5 pmol of oligo(dT) primer, topped up to 11 μl with H2O, heated at 70°C for 10 min, and placed on ice for 5 min. A master mix containing 2 μl of 10× Moloney murine leukemia virus reverse transcriptase buffer (New England Biolabs), 100 mM dithiothreitol, 0.5 mM concentrations of each of the four dNTPs, 100 U of reverse transcriptase enzyme (New England Biolabs), and 20 U of RNase inhibitor (Roche) was added to each sample, followed by incubation for 50 min at 42°C. The enzyme was heat inactivated at 70°C for 15 min. PCR was carried out in a 25-μl volume containing 0.2 mM concentrations of each of the four dNTPs, 10 pmol of primers, 2.5 μl of buffer B (Promega), 2 mM MgCl2, 0.75 U of Taq polymerase (Promega), and 1.5 μl of the cDNA preparation. The PCR started at 94°C for 2 min, followed by 25 amplification cycles (denaturation at 94°C for 10 s, annealing at 55°C for 30 s, and extension at 68°C for 45 s), with final extension at 68°C for 7 min.
Bisulfite sequencing.
For mapping of methylated cytosine residues (14), DNAs were modified by a CpGenomeTm DNA modification kit (Intergen Inc.) and the reaction products were amplified with primers specific for modified HPV-16 DNA. Msp3F (genomic positions 4322 to 4348; ATTTGATATTATATTTAAGGTTGAA) and msp3R (4970 to 4946; AATAATTACAAAAACAAAATCTACA) spanned part of the L2 gene. Msp4F (7498 to 7522; TAGTTTTATGTTAGTAATTATGGTT) and msp4R (161 to 140; ACAACTCTATACATAACTATAATA) amplified a segment with the enhancer-promoter. The amplification products were directly sequenced with the same primers.
RESULTS
Most HPV-16 genomes of the cancer cell line CaSki, but not the single HPV-16 genome of SiHa cells, contain meCpGs.
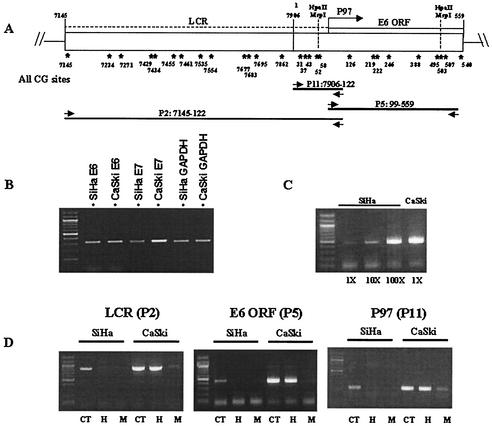
With genome sizes of 7,900 bp and G+C contents close to 40%, HPV genomes may be expected to contain about 400 CpG sites. However, the common papillomavirus types, HPV-6, -11, -16, -18, -31, and -45 (26), have 160, 155, 112, 172, 122, and 154 CpGs, respectively. This lower-than-expected frequency, similar to what is found in the human host, suggests a function of CpGs in the biology of genital HPVs. Methylated CpGs (meCpGs) can be detected with the restriction enzymes HpaII and MspI. Both enzymes cleave the sequence CCGG, but CpG methylation inhibits cleavage by HpaII. Figure 1A shows the positions of two HpaII/MspI sites in the LCR and E6 gene of HPV-16.
FIG. 1.
HpaII/MspI cleavage identifies CpG methylation of SiHa and CaSki DNA as a likely source of transcriptional repression. (A) Distribution of CpGs in the LCR (positions 7154 to 7906 and 1 to 96) and in the E6 gene of HPV-16 (positions 97 to 559) and positions of two HpaII/MspI sites, one (position 57) overlapping with elements of the E6 promoter P97 and the other located in the 3′ part of the E6 gene (position 502). (B) Reverse transcription-PCR confirms that similar amounts of E6 and E7 transcripts are generated by SiHa and CaSki cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase, served as the cellular control transcript. (C) Genomic PCR confirms the large excess of HPV-16 DNA in CaSki cells. (D) Chromosomal DNA of SiHa and CaSki cells was not cut (CT, control) or was cleaved with one of the two enzymes (H, HpaII; M, MspI) and PCR amplified to generate the amplicons P2, P5, and P11. None of the three amplicons could be generated after cleavage with either of the two enzymes in the case of SiHa cells, indicating lack of any methylated CCGG sequences. In the case of CaSki cells, most of the DNA was resistant to HpaII digestion but was readily cleaved by MspI, indicating methylation of these two sites in most of the 500 HPV-16 copies in CaSki cells.
The cervical carcinoma cell lines CaSki and SiHa contain about 500 genomes and a single genome, respectively, of HPV-16 integrated into the chromosomal DNA. The HPV-16 genome in SiHa cells is integrated by interruption of its E2 gene at positions 3132 and 3384, accompanied by deletion of 251 nucleotides of viral sequence (3) and 7.8 kb of the cellular DNA flanking the site of recombination (4). The HPV-16 genomes in CaSki cells are inserted in numerous loci. In most of these, HPV-16 is transcriptionally silent, while E6 and E7 transcripts stem from a single locus (49), possibly due to an epigenetic repression mechanism. We confirmed that SiHa and CaSki cells express similar levels of E6 and E7 transcripts, in spite of the large excess of HPV-16 genomes in CaSki cells (3) (Fig. 1B and C).
To determine whether CpG methylation may explain this observation, we analyzed the methylation statuses of the two HpaII/MspI sites in the LCR and the E6 gene, which are present in the PCR amplicons P5 (corresponding to the E6 gene), P2 (containing the LCR), and P11 (containing just E6 promoter sequences) in both DNAs. In the case of SiHa DNA, these amplicons could not be generated after cleavage with either of the two enzymes, while in the case of CaSki cells, most of the DNA was resistant to HpaII but was readily cleaved by MspI (Fig. 1D). These findings indicate that the LCR and the E6 gene contain CmeCGG in most HPV-16 genomes of CaSki cells but not in the HPV-16 genome of SiHa cells.
Conservation of preferential CpG methylation throughout the HPV-16 genomes of SiHa and CaSki cells and a cervical carcinoma.
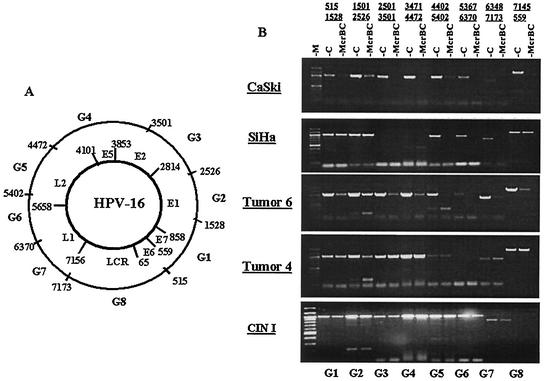
Screening with the restriction enzyme McrBC is more powerful than analysis by HpaII/MspI, as McrBC cuts close to the sequence purine-meC (PumeC) in the context of a second, arbitrarily spaced PumeC (41, 45). As a consequence, McrBC recognizes pairs of PumeCpG residues and, on average, cleaves every other meCpG, while HpaII/MspI analysis resolves the methylation status of only 1 in 16 CpGs.
The HPV-16 genome contains 112 CpGs, and 81 of these are part of the sequence purine-CpG and are potential targets of methylation as well as for cleavage by McrBC. To establish a crude map of the distribution of methylation throughout the HPV-16 genomes of SiHa and CaSki cells, we used McrBC to digest the chromosomal DNA and PCR amplified segments with sizes of about 1 kb (Fig. 2). As expected from the HpaII/MspI data, the LCR-E6 segment from positions 7145 to 559 could not be digested in SiHa DNA, indicating a lack of methylated targets. Most of this amplicon was digested in CaSki DNA and therefore methylated, while a reproducible weak undigested band indicated lack of methylation of some copies. Also, the segments 515-1528 and 1501-2526 were not cleaved in SiHa cells and were partially cleaved in CaSki cells, suggesting lack of and partial methylation, respectively, of the genomic region spanning the E7 and E1 genes. The amplicon between positions 2501 and 3501 could not be amplified with SiHa DNA, as expected from the recombination of this segment. Most of this amplicon was digestible with CaSki DNA. Three of the remaining segments (3471-4472, 5367-6370, and 6348-7143) were almost completely cleaved with both SiHa and CaSki DNA, while a fourth segment (4402-5402) was partially cleaved. This part of the genome, with the late genes L2 and L1, is apparently well recognized by the cellular methylation machinery. In SiHa, this segment is—due to the recombination—upstream of viral promoters and may be transcriptionally silent.
FIG. 2.
McrBC cleavage of segments of the HPV-16 genomes in SiHa and CaSki cells and three clinical samples detects patterns of hyper- and hypomethylation. (A) Genomic map of HPV-16 (7,906 bp), with genes E6, E7, E1, E2, E5, L2, and L1, the LCR, and the relative locations of amplicons G1 to G8 indicated. (B) Cleavage of amplicons G1 to G8 by McrBC (right lane of each pair of samples) indicates methylation, compared with uncleaved controls (left lane of each pair). The cleavage pattern indicates hypermethylation throughout most HPV-16 genomes in DNA from CaSki cells and tumor 6, hypermethylation of the late genes in the single HPV-16 genome of SiHa cells, and hypo- or no methylation in tumor 4 and a CIN I lesion.
To examine whether one would find a similar distribution of McrBC cleavage in situ, we analyzed the DNA from a cervical carcinoma (tumor 6), our only DNA preparation among 33 tumor-derived DNAs which contained a strongly methylated LCR-E6 segment (see below). This tumor DNA was preferentially cleaved in an uneven manner that was similar to what was done with the HPV-16 genomes in SiHa and CaSki cells, with strong methylation of all segments between positions 3471 and 7173. The 7145-557 and 2501-3501 segments were moderately methylated, while there was low methylation in the segment from positions 515 to 2526 (Fig. 2). Differential cleavage by McrBC was not biased by unequal distribution of McrBC sites, since amplicons G1 to G8 all have numerous potential McrBC sites (11, 8, 15, 13, 5, 7, 10, and 13 sites, respectively). For example, G6, with seven sites, is efficiently cleaved in CaSki, SiHa, and T6 DNA, in contrast to G2, which has eight sites.
We also examined the DNA from a carcinoma, whose LCR-E6 segment, like those of nearly all of the tumors we studied, had no detectable DNA methylation (see below). The DNA of tumor 4 was not cleaved by McrBC throughout the HPV-16 genome (Fig. 2B). The segment 5367-6370 led only to a weak amplicon. We also analyzed the complete HPV-16 genome of a precursor lesion (CIN I). None of the amplicons of this sample, initially found to have a hypomethylated LCR-E6 region (see below), was cleaved by McrBC.
We conclude that HPV-16 DNA can be extensively modified by DNA methylation, that methylation varies quantitatively in different parts of the genome, and that hypermethylation in three DNA isolates correlates with the positions of the late genes. Methylation of HPV-16 DNA is not necessary but depends on as-yet-unknown circumstances, as suggested by the lack of detectable methylation in two lesions, with the tumor and CIN I likely containing integrated and episomal DNA, respectively.
HPV-16 methylation is frequent in asymptomatic smears, uncommon in precursors lesions, and rare in carcinomas.
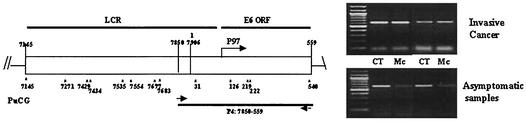
Since we asked whether CpG methylation occurs only sporadically in cell lines and invasive carcinomas or is frequent in clinical specimens, we analyzed DNA from cervical smears classified as being normal (asymptomatic smears), biopsies of low-grade and high-grade precursor lesions (CIN I and CIN III), and invasive carcinomas by McrBC digestion and PCR amplification. The strategy and selected data are shown in Fig. 3.
FIG. 3.
McrBC cleavage identifies hypomethylation of HPV-16 DNA in cervical cancers and hypermethylation in asymptomatic smears. Amplicon P4 includes the genomic segment from positions 7850 to 559 with the E6 promoter elements, the E6 oncogene, and cis-responsive elements within E6 (left side of figure). The upper panel at right shows two typical McrBC-resistant, i.e., unmethylated, amplicons detected in DNA from invasive cancers, and the lower panel shows two typical McrBC-sensitive and methylated amplicons from asymptomatic cervical smears.
We examined DNA from 48 clinical specimens from New Mexico and 33 from Brazil. Among the latter group of specimens, 8 of 14 asymptomatic smears, 0 of 3 CIN I lesions, and 2 of 16 samples from carcinomas contained hypermethylated viral DNA (Table 2). One of these two carcinoma DNAs was lightly methylated, while the other, from tumor 6, was strongly methylated and was described in detail above. Among the samples from New Mexico, DNA of 5 of 11 asymptomatic smears, 3 of 10 CIN I lesions, 2 of 10 CIN III lesions, and 0 of 17 cancers contained methylated HPV-16 genomes. Taking both cohorts together, 52% of the HPV-16 genomes from asymptomatic smears, 21.7% of those from precursor lesions, and only 6.1% of those from the carcinomas contained methylated DNA, indicating that, as judged by McrBC cleavage, methylation of the LCR and the E6 gene decreases with progression of the infection (Table 2). A scan of the methylation pattern of the whole HPV-16 genome from cervical smears of asymptomatic patients could not be attempted because of the small amounts of DNA available.
TABLE 2.
Methylation of the LCR and E6 gene of HPV-16 in clinical samples from Brazil and New Mexicoa
| Site(s) | Sample type (total no.) | No. of samples showing:
|
% of amplifiable samples with methylation | |
|---|---|---|---|---|
| Methylation | No methylation | |||
| Brazil | Asymptomatic (14) | 8 | 6 | 57.1 |
| CIN I (3) | 0 | 3 | 0 | |
| Invasive cancer (16) | 2 | 14 | 12.5 | |
| New Mexico | Asymptomatic (11) | 5 | 6 | 45.4 |
| CIN I (10) | 3 | 7 | 30 | |
| CIN III (10) | 2 | 8 | 20 | |
| Invasive cancer (17) | 0 | 17 | 0 | |
| Bothb | Asymptomatic (25) | 13 | 12 | 52 |
| CIN I and III (23) | 5 | 18 | 21.7 | |
| Invasive cancer (33) | 2 | 31 | 6.1 | |
CpG methylation was determined by cleavage of the DNA preparations with the restriction enzyme McrBC and subsequent PCR amplification of a genomic segment of HPV-16 corresponding to the promoter and E6 gene as shown in Fig. 3.
Data from both cohorts were combined, results with CIN I and CIN III were united into one group of precursor lesions, and DNA preparations that could not be amplified were omitted.
Mapping of meCpGs by bisulfite modification.
meCpGs can be mapped precisely by sequencing after bisulfite modification (14). As it is very laborious to completely analyze 7.9-kb genomes in numerous samples, we used this technique only for selected samples and an analysis of the enhancer-promoter region between genomic positions 7498 and 161. This segment contains 11 CpGs, at positions 7535, 7554, 7677, 7683, 7695, 7862, 31, 37, 43, 52, and 58 (Fig. 1). The first five of these CpGs are in close proximity to important AP1 and NFI sites of the HPV-16 enhancer (1, 5, 18). The last five CpGs overlap with an Sp1 and two E2 binding sites, which regulate the transcription start at position 97 (11, 47). In SiHa cells, all 11 of these CpGs were modified, i.e., unmethylated. In contrast, the same 11 CpGs were methylated in most of the 500 HPV-16 DNA copies in CaSki cells. An unmodified and transcriptionally active background of one or a few HPV-16 genomes would not have been detectable in this experiment. A segment of the L2 gene (4322-4946) served as a control and did not differ between SiHa and CaSki cells, as all four CpGs of this amplicon were methylated.
We also studied 15 patient samples by bisulfite modification (Table 3). In five asymptomatic smears, all 11 CpGs were methylated. In two separate CIN I samples, three and four of the five CpGs at the promoter were not methylated, while all CpGs of the enhancer were methylated in one sample and two were methylated in the other. In the five invasive tumors, we observed heterogeneous methylation patterns, while in a CIN III sample and two tumor samples, all CpGs of the enhancer and promoter were methylated. Notably, however, the only CpG residue between the enhancer and promoter (position 7862) was unmethylated in these three samples and all other CIN and invasive cancer lesions. This site is the only CpG dinucleotide between the two specifically positioned nucleosomes that overlap the enhancer and promoter elements and is close to CDP and AP-1 sites that are essential for transcriptional repression and activation during epithelial differentiation (5, 30, 42, 43). We conclude that bisulfite modification detected the same trend of hypomethylation, when lesions were compared with asymptomatic smears, as that observed with McrBC cleavage, although the latter technique is less sensitive and underreported CpG methylation.
TABLE 3.
Methylation of 11 CpGs in the LCR of HPV-16 in two cell lines and 15 clinical samples
| Cell line or samplea | Methylation of CpG at positionb:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7535 | 7554 | 7677 | 7683 | 7695 | 7862 | 31 | 37 | 43 | 52 | 58 | |
| SiHa | − | − | − | − | − | − | − | − | − | − | − |
| CaSki | + | + | + | + | + | + | + | + | + | + | + |
| As16 | + | + | + | + | + | + | + | + | + | + | + |
| As22 | + | + | + | + | + | + | + | + | + | + | + |
| As23 | + | + | + | + | + | + | + | + | + | + | + |
| As24 | + | + | + | + | + | + | + | + | + | + | + |
| As33 | + | + | + | + | + | + | + | + | + | + | + |
| CIN I A1 | + | + | + | + | + | − | + | − | − | − | − |
| CIN I A6 | + | + | − | − | − | − | + | + | − | − | − |
| CIN III A20 | + | + | + | + | + | − | + | + | + | + | + |
| T6 | + | + | + | + | + | − | + | + | + | + | + |
| T8 | + | ± | − | + | − | − | − | − | + | + | + |
| T9 | ? | + | + | ± | ± | ± | + | + | + | + | + |
| T10 | + | + | + | + | + | − | + | + | + | + | + |
| T20 | ? | ? | + | ± | ± | ± | + | + | + | + | + |
| T37 | − | − | − | − | − | − | + | + | + | + | + |
| T39 | + | + | + | + | + | − | − | − | − | − | − |
The five CpGs between positions 7535 and 7695 are close to cis-responsive elements of the viral enhancer; the CpGs between positions 31 and 58 overlap with promoter elements; and the CpG at position 7862 is located between the borders of two specifically positioned nucleosomes, one of which overlaps the enhancer while the other overlaps the promoter.
+, methylation; −, lack of methylation; ±, both methylated and unmethylated residues in the same sample; ?, unknown.
DISCUSSION
Our data provide compelling evidence that the HPV-16 genome is efficiently modified by cellular DNA methylation and raise questions about how HPV-16 genomes are recognized, which role DNA methylation has in the HPV-16 life cycle, and whether analyzing methylation helps us to understand the etiology of and to diagnose cervical cancer.
Our data suggest that the recognition of HPV-16 DNA is not based on a single physical property. HPV-16 DNA is efficiently targeted as an episome, since we observed methylation very frequently in asymptomatic smears, which generally contain only episomally replicating HPV-16. However, since we also detected methylation in a few invasive tumors and in cell lines, which typically contain HPV-16 genomes integrated into chromosomal DNA (40, 52), one may consider that either arrangement into tandem repeats or chromosomal recombination also provides a methylation signal. Repeated DNA is known to be a preferred target for CpG methylation (51), and in the case of cottontail rabbit papillomavirus, tandem repeats of the viral DNA that were generated during progression of cutaneous lesions correlated with increased methylation (44, 50). Even in the absence of tandem arrangements, nontranscribed parts of the single intrachromosomal HPV-16 genome are strongly methylated in the SiHa cell line, suggesting that recombination per se is a de novo methylation signal, as extensively studied in the adenovirus system, which has been the paradigm for DNA methylation studies for more than 20 years (33, 46).
We showed by meCpG mapping that the methylation of the HPV-16 LCR does not follow a singular and simple pattern but that groups of CpGs in alternative locations and with alternative extents can become modified. Methylation of numerous CpGs of the enhancer may indicate repression of its function. It is known that CpG methylation does not interfere with binding of the E6 promoter factor Sp1 (19), while it displaces E2 protein (48), which, however, is a repressor of the P97 promoter. Therefore, a methylated promoter may be transcriptionally active independent of the epithelial specific enhancer, particularly as it is under the stimulatory influence of the nuclear matrix attachment region in the E6 gene (43).
The normal HPV-16 life cycle is restricted to asymptomatic infections and CIN lesions, while carcinogenic progression is an accident, as there is no further virus production. Consequently, the high prevalence of methylated HPV-16 genomes in asymptomatic epithelia may indicate that methylation is part of normal HPV-16 biology. Two speculative and opposing views might explain this observation. The cell may have an antiviral defense that senses viral DNA as foreign and targets it for transcriptional repression. Alternatively, DNA methylation may be yet another example of the numerous strategies developed by HPVs that favor a subclinical, long-term maintenance of the viral infection (5). Such a model would be reminiscent of the life cycle of Epstein-Barr virus, which includes DNA methylation-dependent silencing of a specific promoter during one form of latency (35, 36). Likewise, the HPV genome may contain molecular properties that induce methylation-dependent latency under some conditions, while under different conditions this mechanism may be overruled. This could be achieved by the conserved nuclear matrix attachment regions (43) and Sp1 sites at the E6 promoters of all genital HPVs (47). Both elements have been reported to antagonize DNA methylation in the context of certain cellular genes (8, 12, 23). Preferential methylation of late genes may suggest an additional role in the early-late switch, which is a multifaceted event involving differential promoter usage, splicing, elongation, and mRNA stability (2, 29). This possibility has been discussed based on studies of the HPV-1 genome with the limited HpaII/MspI technology (9).
Our data document the power of molecular diagnoses by revealing the epigenetic diversity of pathologically similar samples. The analysis is limited by the problem that one may study DNA populations that may be heterogeneous due to inclusion of surgical margins, different progression stages within one biopsy, and molecularly divergent HPV-16 genomes even in single cells. McrBC is an excellent tool for a crude scan of whole viral genomes but may be prone to underreporting of meCpGs, as suggested by the detection of meCpGs in bisulfite-modified tumor DNAs, which were not cleaved by McrBC. HPV DNA methylation may become important for the diagnosis of cervical smears if further research confirms that asymptomatic infections and precursor lesions fall into classes that either express or repress the viral oncogenes, leading to different prognoses, as E6/E7 expression is required for continued transformation and viability of HPV-containing tumors and tumor cell lines (13).
Acknowledgments
B.F.L.L. and H.-U.B. contributed equally to this work.
This research was supported, in part, by funds provided by A-Star (Singapore) through the Institute of Molecular and Cell Biology and by NIH grant ROICA91964 to H.-U.B.
REFERENCES
- 1.Apt, D., T. Chong, Y. Liu, and H.-U. Bernard. 1993. Nuclear factor I and epithelial cell-specific transcription of human papillomavirus type 16. J. Virol. 67:4455-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. C., and C. Calef. 1995. Maps of papillomavirus transcripts, part III-A, p. 3-19. In G. Myers, H.-U. Bernard, H. Delius, C. Baker, J. Icenogle, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses 1995 compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 3.Baker, C. C., W. C. Phelps, V. Lindgren, M. J. Braun, M. J. Gonda, and P. M. Howley. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer-Hofmann, R., C. Borghouts, E. Auvinen, E. Bourda, F. Rosl, and A. Alonso. 1996. Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa. Virology 217:33-41. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, H.-U. 2000. Regulation of human papillomavirus gene expression by the factor CDP, nucleosomes and the nuclear matrix. Papillomavirus Rep. 11:73-80. [Google Scholar]
- 6.Bernard, H.-U., S. Y. Chan, M. M. Manos, C. K. Ong, L. L. Villa, H. Delius, H. M. Bauer, C. Peyton, and C. M. Wheeler. 1994. Assessment of known and novel human papillomaviruses by polymerase chain reaction, restriction digest, nucleotide sequence, and phylogenetic algorithms. J. Infect. Dis. 170:1077-1085. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. P. 1992. The essentials of DNA methylation. Cell 70:5-8. [DOI] [PubMed] [Google Scholar]
- 8.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 9.Burnett, T. S., and J. P. Sleeman. 1984. Uneven distribution of methylation sites within the human papillomavirus la genome: possible relevance to viral gene expression. Nucleic Acids Res. 12:8847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, W. K., G. Klock, and H.-U. Bernard. 1989. Progesterone and glucocorticoid response elements occur in the long control regions of several human papillomaviruses involved in anogenital neoplasia. J. Virol. 63:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeret, C., C. Desaintes, M. Yaniv, and F. Thierry. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 71:9343-9349. [DOI] [PMC free article] [PubMed]
- 12.Forrester, W. C., L. A. Fernandez, and R. Grosschedl. 1999. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 13:3003-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futscher, B. W., M. M. Oshiro, R. J. Wozniak, N. Holtan, C. L. Hanigan, H. Duan, and F. E. Domann. 2002. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 31:175-179. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Carranca, A., F. Thierry, and M. Yaniv. 1988. Interplay of viral and cellular proteins along the long control region of human papillomavirus type 18. J. Virol. 62:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloss, B., H.-U. Bernard, K. Seedorf, and G. Klock. 1987. The upstream regulatory region of the human papillomavirus-16 contains an E2 protein independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 6:3735-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloss, B., T. Chong, and H.-U. Bernard. 1989. Numerous nuclear proteins bind the long control region of human papillomavirus type 16: a subset of 6 of 23 DNaseI-protected segments coincides with the location of the cell-type-specific enhancer. J. Virol. 63:1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington, M. A., P. A. Jones, M. Imagawa, and M. Karin. 1988. Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. USA 85:2066-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich, B., and A. Bird. 2000. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation, p. 55-74. In P. A. Jones and P. K. Vogt (ed.), DNA methylation and cancer. Springer, Berlin, Germany. [DOI] [PubMed]
- 21.Herman, J. G., and S. B. Baylin. 2000. Promoter-region hypermethylation and gene silencing in human cancer, p. 35-54. In P. A. Jones and P. K. Vogt (ed.), DNA methylation and cancer. Springer, Berlin, Germany. [DOI] [PubMed]
- 22.International Agency for Research on Cancer. 1995. Monograph on the evaluation of carcinogenic risks to humans, vol. 64. Human papillomaviruses. International Agency for Research on Cancer, Lyon, France.
- 23.Jenuwein, T., W. C. Forrester, L. A. Fernandez-Herrero, G. Laible, M. Dull, and R. Grosschedl. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385:269-272. [DOI] [PubMed] [Google Scholar]
- 24.List, H. J., V. Patzel, U. Zeidler, A. Schopen, G. Ruhl, J. Stollwerk, and G. Klock. 1994. Methylation sensitivity of the enhancer from the human papillomavirus type 16. J. Biol. Chem. 269:11902-11911. [PubMed] [Google Scholar]
- 25.May, M., X. P. Dong, E. Beyer-Finkler, F. Stubenrauch, P. G. Fuchs, and H. Pfister. 1994. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 13:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers, G., H.-U. Bernard, H. Delius, M. Favre, J. Iconogle, M. van Ranst, and C. Wheeler (ed.). 1994. Human papillomaviruses 1994 compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 27.O'Connor, M. J., W. Stünkel, C. H. Koh, H. Zimmermann, and H.-U. Bernard. 2000. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses. J. Virol. 74:401-410. [PMC free article] [PubMed] [Google Scholar]
- 28.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 29.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattison, S., D. G. Skalnik, and A. Roman. 1997. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 71:2013-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulsen, M., and A. C. Ferguson-Smith. 2001. DNA methylation in genomic imprinting, development, and disease. J. Pathol. 195:97-110. [DOI] [PubMed] [Google Scholar]
- 32.Peyton, C. L., P. E. Gravitt, W. C. Hunt, R. S. Hundley, M. Zhao, R. J. Apple, and C. M. Wheeler. 2001. Determinants of genital human papillomavirus detection in a US population. J. Infect. Dis. 183:1554-1564. [DOI] [PubMed] [Google Scholar]
- 33.Remus, R., C. Kämmer, H. Heller, B. Schmitz, G. Schell, and W. Doerfler. 1999. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J. Virol. 73:1010-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 35.Robertson, K. D. 2000. The role of DNA methylation in modulating Epstein-Barr virus gene expression, p. 21-34. In P. A. Jones and P. K. Vogt (ed.), DNA methylation and cancer. Springer Verlag, Berlin, Germany. [DOI] [PubMed]
- 36.Robertson, K. D., and P. A. Jones. 2000. DNA methylation: past, present and future directions. Carcinogenesis 21:461-467. [DOI] [PubMed] [Google Scholar]
- 37.Rosl, F., A. Arab, B. Klevenz, and H. zur Hausen. 1993. The effect of DNA methylation on gene regulation of human papillomaviruses. J. Gen. Virol. 74:791-801. [DOI] [PubMed] [Google Scholar]
- 38.Schiffman, M. H., and L. A. Brinton. 1995. The epidemiology of cervical carcinogenesis. Cancer 76:1888-1901. [DOI] [PubMed] [Google Scholar]
- 39.Schorderet, D. F., and S. M. Gartler. 1992. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA 89:957-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 41.Stewart, F. J., and E. A. Raleigh. 1998. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 379:611-616. [PubMed] [Google Scholar]
- 42.Stünkel, W., and H.-U. Bernard. 1999. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J. Virol. 73:1918-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stünkel, W., Z. Huang, S. H. Tan, M. O'Connor, and H.-U. Bernard. 2000. Nuclear matrix attachment regions of human papillomavirus-16 repress or activate the E6 promoter depending on the physical state of the viral DNA. J. Virol. 74:2489-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara, K., K. Fujinaga, T. Yamashita, and Y. Ito. 1983. Integration and methylation of shope papilloma virus DNA in the transplantable Vx2 and Vx7 rabbit carcinomas. Virology 131:88-99. [DOI] [PubMed] [Google Scholar]
- 45.Sutherland, E., L. Coe, and E. A. Raleigh. 1992. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225:327-348. [DOI] [PubMed] [Google Scholar]
- 46.Sutter, D, and W. Doerfler. 1980. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl. Acad. Sci. USA 77:253-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, S. H., L. E. C. Leong, P. A. Walker, and H.-U. Bernard. 1994. The human papillomavirus type 16 transcription factor E2 binds with low cooperativity to two flanking binding sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 68:6411-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thain, A., O. Jenkins, A. R. Clarke, and K. Gaston. 1996. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 70:7233-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Tine, B. A., J. Knops, T. R. Broker, L. T. Chow, and P. T. Moen. 2001. In situ analysis of the transcriptional activity of integrated viral DNA using tyramide-FISH. Dev. Biol. (Basel) 106:381-385. [PubMed] [Google Scholar]
- 50.Wettstein, F. O., and J. G. Stevens. 1983. Shope papilloma virus DNA is extensively methylated in non-virus-producing neoplasms. Virology 126:493-504. [DOI] [PubMed] [Google Scholar]
- 51.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Gen. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 52.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]