Abstract
Some individuals remain inexplicably seronegative and lack evidence for human immunodeficiency virus type 1 (HIV-1) infection by conventional serologic or virologic testing despite repeated high-risk virus exposures. Here, we examined 10 exposed seronegative (ES) individuals exhibiting HIV-1-specific cytotoxicity for the presence of HIV-1. We discovered HIV-1 DNA in resting CD4+ T cells (mean, 0.05 ± 0.01 copies per million cells) at multiple visits spanning 69 to 130 weeks in two ES individuals at levels that were on average 104- to 106-fold lower than those of other HIV-1-infected populations reported. Sequences of HIV-1 envelope and gag genes remained markedly homogeneous, indicating little to undetectable virus replication. These results provide the evidence for HIV-1 infection in ES individuals below the detection limit of standard assays, suggesting that extraordinary control of infection can occur. The two HIV-infected ES individuals remained healthy and were not superinfected with other HIV-1 strains despite continued high-risk sexual exposures to multiple HIV-infected partners. Understanding the mechanisms that confer diminished replicative capacity of HIV-1 in these hosts is paramount to developing strategies for protection against and control of HIV-1 infection.
Rare individuals have been identified since early in the AIDS pandemic who fail to seroconvert and lack evidence for human immunodeficiency virus type 1 (HIV-1) infection despite high-risk and/or multiple exposures to HIV-1. These exposed seronegative (ES) individuals include infants born to HIV-1-infected mothers (11, 42), commercial sex workers in areas where the disease is epidemic (25, 43), hemophiliacs who received tainted factor VIII preparations (16), health care personnel with accidental percutaneous exposure to infected blood (9, 37), and sexual partners of known HIV-1-infected persons (17, 30). Transient infection (2, 6, 40) or silent infection (20) was proposed to explain the case of some ES individuals in whom HIV infection had been detected either by culture or by PCR of peripheral blood mononuclear cells (PBMCs). However, this pattern of HIV infection has not been unambiguously confirmed by genetic analyses (32) and remains highly controversial (15). Nevertheless, one-third to one-half of ES individuals have detectable HIV-1-specific T-cell responses (16, 17, 30, 37, 42, 43) and, in some cases, mucosal immunoglobulin A (IgA) antibodies (30), suggesting that some ES individuals may have acquired HIV-1 infection but that the virus is either cleared or no longer detectable by routine methods (17, 30, 37).
An HIV-1 reservoir in resting CD4+ T cells has been identified as early as 5 days after infection (7, 8, 14, 49) and may persist over the patient's lifetime with highly active antiretroviral therapy (HAART) (13). We hypothesize that during initial active HIV-1 infection, a reservoir is established in quiescent cells of some ES individuals, who then fail to manifest clinical infections or laboratory evidence of infection by standard serologic or virologic testing. To examine this hypothesis, we screened for HIV-1 DNA sequences in purified resting CD4+ T cells from our ES cohort (17). The intense labor involved in these studies precluded assessment of a large number of ES individuals (see below). The present study was not designed to determine the frequency of HIV-1 infection in ES populations. Instead, our investigation focused on 10 ES individuals with detectable HIV-1-specific cytotoxic T lymphocytes (CTL), based on the assumption that memory T-cell responses were more likely to be present in persons with previous HIV-1 exposure or infection (16, 17, 30, 37, 42, 43). We demonstrate here the persistence of HIV-1 DNA at levels below the detection limit of conventional assays in 2 of these 10 persons: ES1 and ES38.
MATERIALS AND METHODS
Subjects studied.
We focused on 10 ES individuals (17) with (i) detectable HIV-1-specific CTL, (ii) higher frequencies of exposure to HIV-1, and (iii) large volumes of samples from multiple time points, allowing us to carry out extensive and repeated PCR analyses. Both ES1 and ES38 reported repeated exposures to HIV-infected sexual partners (Fig. 1). These individuals, both Caucasians, had the wild-type CCR5 coreceptor genotype (17). Class I HLA DNA sequence-specific primer typing showed that ES1 was HLA-A*02, -A*68, -B*44, -B*51, -Bw4, -Cw*05, and -Cw*15, whereas ES38 was HLA-A*02, -A*11, -B*07, -B*35, -Bw6, -Cw*04, and -Cw*07. Over the past 7 years, ES1 and ES38 exhibited no evidence of active HIV-1 infection when evaluated at 1- to 6-month intervals by serologic (enzyme immunoassay and Western blot tests), standard DNA PCR, and PBMC HIV-1 coculture (with or without CD8+-T-cell depletion) assays (17). In addition, no HIV-1 RNA was detected by reverse transcription-PCR (RT-PCR) in seminal plasma, blood plasma, or PBMC from either ES individual. Both ES individuals entered the study with normal CD4+-T-cell counts (ES1, 1,391 cells/μl; ES38, 937 cells/μl), remained healthy without HIV-1-associated opportunistic infections, and maintained strong recall antigen responses by in vitro proliferation (17). ES1, but not ES38, also demonstrated T-cell lymphoproliferative responses to HIV-1 p24, gp120, and gp160 antigens (17). Cytolytic activities recognizing HIV-1 Gag, Env, Pol, and Nef were detected in ES1 and ES38 (17).
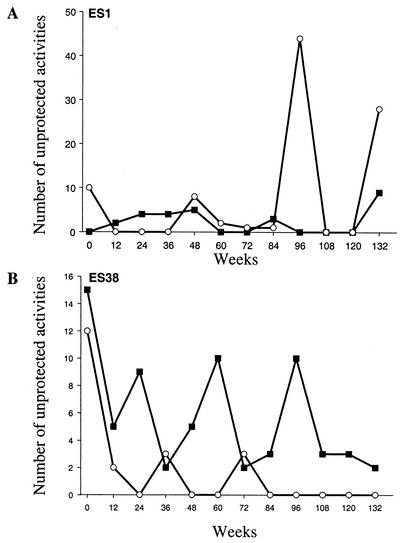
FIG. 1.
Sexual activities, defined as receptive or insertive intercourse, of ES1 (A) and ES38 (B) with their known HIV-1-infected partners (▪) and one or more partners of unknown HIV-1 serostatus (○). Volunteers reported cumulative activities over 3-month intervals.
Cell purification and DNA extraction.
Each sample of fresh blood or cryopreserved PBMC was divided into three portions: one was sent to the University of Washington laboratory, one was sent to the Fred Hutchinson Cancer Research Center laboratory, and one was stored in liquid nitrogen for confirmatory studies. Purification of the three cell populations was performed separately in both laboratories (51). Genomic DNA was extracted separately from purified resting CD4+ T cells, activated CD4+ T cells, and monocytes (51), one sample per day, in a biosafety cabinet housed in a “PCR-clean” room. The room, hood, and equipment were decontaminated after each use. Experiments with ES specimens were separated by intervals of at least 4 weeks to further avoid cross-contamination between specimens.
PCR and sequence analyses.
Limiting-dilution nested PCR was performed to amplify the env and gag of HIV-1 sequences with the following primers: C2-V3-C3 region outer P5-2 and PV3; C2-V3-C3 region inner P5 (50) and Bsu (residues 7341 to 7315 of HIV-1 HXB2 sequence in the Los Alamos Database, 5′-TTACAATTTCTGGGTCCCCTCCTGAGG-3′); V3-C3 region outer Pvu (residues 7061 to 7090, 5′-CAATGCTAAAACCATAATAGTACAGCTGAA-3′) and PV3; V3-C3 inner Pvu and Bsu; gag p17 outer PG3 (residues 763 to 789, 5′-TGACTAGCGGAGGCTAGAAGGAGAGAG-3′) and PG8 (residues 1326 to 1299, 5′-GGCTCCTTCTGATAATGCTGAAAACATG-3′); and gag p17 inner PG1 (residues 790 to 814, 5′-ATGGGTGCGAGAGCGTCAGTATTAA-3′) and PG6 (residues 1262 to 1231, 5′-TCACCTAGAACTTTAAATGCATGGGTAAAAGT-3′). Extend High Fidelity PCR system (Roche Molecular Biochemicals) was used for PCR as described previously (50, 51) with 10 pmol of each primer. Amplification conditions for the first round PCR were 94°C for 2 min, 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s for 32 cycles, followed by a final extension at 72°C for 10 min. Then, 4 μl of the first-round PCR products were used in a second-round PCR at 94°C for 20 s, 55°C for 20 s, and 72°C for 60 s for 35 to 40 cycles, followed by a final extension at 72°C for 10 min. All PCRs were performed in the Perkin-Elmer model 9600 thermocycler. Controls for PCR included genomic DNA containing 1, 4, 25, and 100 copies of HIV-1 DNA, as well as reagent and HIV-1-negative controls. PCR was sensitive to detect one copy per microgram of DNA per reaction and was specific for the detection of HIV-1 (data not shown). HIV-1 copy numbers were calculated based on limiting-dilution PCR and a computer program (QUALITY) described previously (39). PCR products were cloned and sequenced (51). All sequences were aligned by using CLUSTALW (46). Maximum-likelihood estimation (MLE) phylogram was constructed by using PAUP* (Sinauer Associates, Inc., Sunderland, Mass.). The models of evolution (GTR+G) were selected by using the AIC (1) in Modeltest version 3.06 (38). It was necessary to perform 600 or more independent limiting-dilution PCRs to identify the HIV-1 sequences in each sample. Similar intensive PCR assays with positive samples from the same time point but stored initially in another laboratory and or in liquid nitrogen were carried out for confirmatory studies. Typically, billions of PBMC through leukapheresis would be used for cell separation, PCR, and sequence analyses; however, there were not enough cells to perform similar intensive PCR sequence analyses in another laboratory.
GenBank nucleotide sequence accession numbers.
The nucleotide sequences described in the paper have been submitted to GenBank (sequence accession no. AY162341 through AY162373 and AY162474 through AY162825).
RESULTS
Persistence of extraordinarily low levels of HIV-1 DNA in peripheral blood.
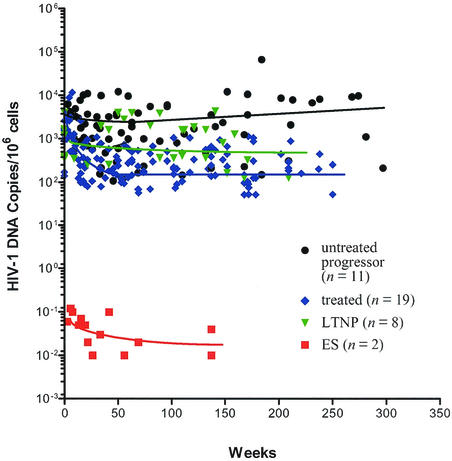
We used a limiting-dilution PCR to detect and quantitate HIV-1 env and gag DNA, and the viral copy numbers shown in Table 1 were derived from the results of hundreds or thousands of independent PCRs performed on each sample (39, 51). HIV-1 DNA was detected at extraordinarily low levels (mean, 0.05 ± 0.01 per million cells) in resting CD4+ T cells isolated at different times and appeared to decline slowly over a period of 137 (ES1) and 69 weeks (ES38) (Table 1). When multiple cell fractions (week 137 of ES1 and week 69 of ES38) were available for study, resting CD4+ T cells had a higher HIV-1 copy number than the total PBMC, monocytes, and activated CD4+ T cells (Table 1). The levels of HIV-1 DNA in these ES individuals were remarkably lower than those typically found in other HIV-1-infected populations (Fig. 2). We detected HIV-1 DNA in resting CD4+ T cells from the sexual partner of ES1 at levels approximately 105 fold higher than in ES individuals (Table 1). HIV-1 DNA levels in ES individuals were, on average, 104-fold lower than those in patients with undetectable plasma viremia during HAART (491.4 ± 123.5 copies per million resting CD4+ T cells; P < 0.0001), approximately 5 × 105-fold lower than those in long-term nonprogressors (LTNP) (1,189 ± 301.10 copies per million PBMC, P < 0.0001), and up to 106-fold lower than in patients with typical HIV-1 infection without therapy (5,704 ± 1,016 copies per million PBMC; P < 0.0001) (Fig. 2) (3-5, 51).
TABLE 1.
Identification of HIV-1 in ES mononuclear cells and comparison of copy numbers to the HIV-1-infected sexual partner of ES1
| Subject | Wk of studya | Cell type | No. of HIV-1 DNA copies/106 cellsb | Genetic markersc |
|---|---|---|---|---|
| ES1 | 0 | PBMC | <0.08 | BB, AB, BB, AA, CC |
| 8 | Resting CD4+ | 0.11 | BB, AB, BB, AA, CC | |
| 44 | Resting CD4+ | 0.12 | BB, AB, BB, AA, CC | |
| 57 | Resting CD4+ | 0.10 | BB, AB, BB, AA, CC | |
| 96 | PBMC | <0.05 | BB, AB, BB, AA, CC | |
| 112 | Resting CD4+ | 0.07 | BB, AB, BB, AA, CC | |
| 112 | PBMC | <0.05 | BB, AB, BB, AA, CC | |
| 132 | Resting CD4+ | 0.06 | BB, AB, BB, AA, CC | |
| 132 | Monocyte | <0.08 | BB, AB, BB, AA, CC | |
| 137 | Resting CD4+ | 0.04 | BB, AB, BB, AA, CC | |
| 137 | Activated CD4+ | 0.01 | BB, AB, BB, AA, CC | |
| 137 | Monocyte | 0.01 | BB, AB, BB, AA, CC | |
| ES38 | 0 | Resting CD4+ | 0.06 | AA, AB, BB, AB, CC |
| 5 | Resting CD4+ | 0.03 | AA, AB, BB, AB, CC | |
| 5 | PBMC | <0.03 | AA, AB, BB, AB, CC | |
| 10 | Activated CD4+ | <0.06 | AA, AB, BB, AB, CC | |
| 10 | Monocyte | <0.05 | AA, AB, BB, AB, CC | |
| 14 | PBMC | <0.05 | AA, AB, BB, AB, CC | |
| 18 | Resting CD4+ | 0.04 | AA, AB, BB, AB, CC | |
| 24 | PBMC | <0.05 | AA, AB, BB, AB, CC | |
| 32 | Resting CD4+ | 0.03 | AA, AB, BB, AB, CC | |
| 34 | PBMC | <0.05 | AA, AB, BB, AB, CC | |
| 39 | PBMC | ND | ND | |
| 51 | PBMC | ND | ND | |
| 69 | Resting CD4+ | 0.02 | AA, AB, BB, AB, CC | |
| 69 | Activated CD4+ | <0.02 | AA, AB, BB, AB, CC | |
| 69 | Monocyte | <0.02 | AA, AB, BB, AB, CC | |
| 69 | PBMC | <0.02 | AA, AB, BB, AB, CC | |
| ES1 partner | 20 | PBMC | ND | BB, AA, AA, AB, AA |
| 27 | Resting CD4+ | 5698 | BB, AA, AA, AB, AA | |
| 96 | Resting CD4+ | 4987 | BB, AA, AA, AB, AA |
Numbers designate the number of weeks after enrollment that the specimens were collected. Fresh cells from ES1 were analyzed on weeks 132 and 137 and from ES38 on week 69. The remainder of the analyses were performed on cryopreserved cells.
HIV-1 copy numbers were calculated based on limiting-dilution PCR and a computer program (QUALITY) described previously (39).
AmpliType PM PCR amplification and typing kits (Perkin-Elmer Cetus) was used to genotype HLA PM molecules in all isolated cells. The types sequentially represent LDLR, GYPA, HBGG, D7S8, and GC. ND, not done.
FIG. 2.
Representative patterns of variation in the level of cell-associated HIV-1 DNA in PBMC during the course of infection. Shown are data from our existing HIV-1 cohorts, including ES individuals (red square, n = 2) (Table 1), untreated, chronically infected patients followed up since acute infection (black circle) (n = 11) (T. Zhu et al., unpublished data) (3), and treated patients from the initiation of HAART (blue diamond) (n = 19) (Zhu et al., unpublished) (3, 51). The data of long-term nonprogressors were from Brostrom et al. (5) (green triangle) (n = 8). Week zero is the time of first analyzed samples from each individual (5). For treated and untreated patients, the plot depicts the mean viral DNA copies per 106 mononuclear cells derived from a minimum of three independent tests on each sample. Nonlinear regression curves (solid lines) depict the trend in viral load for each group by using GraphPad Prison version 3.00 (GraphPad Software, Inc., San Diego, Calif.).
Homogeneity of HIV-1 sequences in longitudinal samples: lack of viral evolution.
We then examined HIV-1 env V3-C3 coding sequences from each time point. Deduced amino acid sequences of HIV-1 V3-C3 regions were markedly similar in ES1 cells obtained from all six time points over the 130 weeks of study (Fig. 3A). To confirm these findings, we retrieved a third portion of frozen cells from weeks 8 and 137 and used different primers to amplify by PCR and sequence the HIV-1 C2-V3-C3 region (Fig. 3A). These sequences were virtually identical to those generated previously by amplification of the shorter envelope region (V3-C3). Moreover, HIV-1 sequences in ES1 found in activated CD4+ T cells and monocytes were extremely homogeneous, as in the resting CD4+ T cells. Homogeneous sequences of HIV-1 V3-C3 region were also observed in the resting CD4+ T cells from ES38 (Fig. 3B). Except for clone 35 of V3-C3 sequence from ES1 activated CD4+ T cells obtained at week 137 (Fig. 3A), no HIV-1 sequence examined contains any stop codons or insertions or deletions. It is unlikely that this homogeneity resulted from repeated sequencing of only a few HIV-1 proviruses, since up to 50 clonal sequences from 6 to 15 independent positive PCRs were used for each ES specimen. Comparison of env sequence changes over time revealed estimated rates of viral evolution not different from zero (−0.012% per site per year in ES1; P = 0.22; 0.00013% in ES38, P = 0.17). These results suggest that HIV-1 sequences in the ES individuals had not evolved or were evolving very slowly over the 69- to 130-week study period. This finding stands in contrast to a higher evolution rate of HIV-1 V3-C3 sequences (mean, 0.437% per site per year; P < 0.001) in patients with typical disease progression (45). The large amounts of cells required to perform these experiments precluded us from carrying out similar extensive studies in another laboratory (see Materials and Methods). However, to prevent contamination, we performed special procedures to process samples throughout the study (see Materials and Methods) (15, 51). To further exclude sample mislabeling or contamination, we performed PCR typing of five genetic markers (Amplitype PM; Perkin-Elmer Cetus, Branchburg, N.J.), which can discriminate the relatedness between two individuals by a power of 0.9954. All samples from a given ES individual had identical individual genotypes for the five markers (Table 1). The genotype of ES1 was also distinct from his HIV-1-infected sexual partner (Table 1).
FIG. 3.
Deduced amino acid sequence alignment of HIV-1 env C2-V3-C3 regions from ES1 (A) and ES38 (B). The majority of sequences (V3-C3 region) were generated from amplified products with primer pair P5 and PV32 (outer) and primer pair PVU and PV32 (inner) (see Materials and Methods). Longer sequences (C2-V3-C3 region) of clones, e.g., clone 21 at week 8 in panel A were generated with primer pair P5-2 and PV3 (outer) and primer pair P5 and Bsu (inner). All sequences are aligned with the consensus sequences from each ES individual. Weeks are numbered after the first sample was collected from each ES individual. The numbers of clones with identical sequences out of the total analyzed are indicated in the right column. Dots indicate identical sequences; asterisks denote the stop codons.
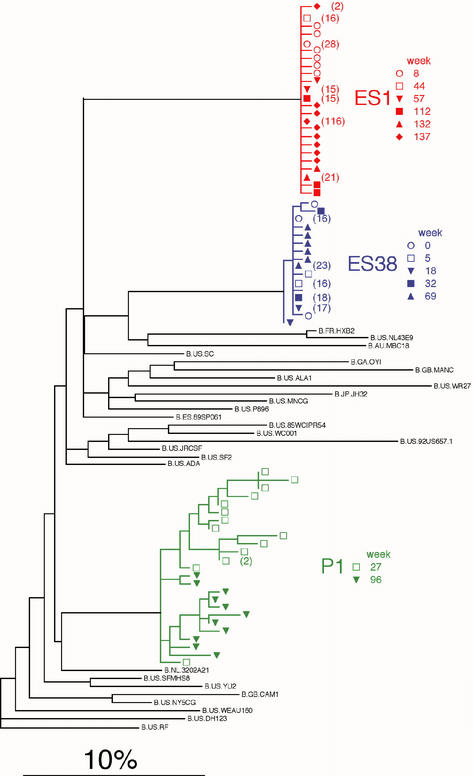
Phylogenetic relationships were estimated between HIV-1 sequences from ES1, ES38, and the contemporaneous, HIV-1-infected, sexual partner of ES1 (Fig. 4). Sequences from ES1 and ES38 were grouped closely in separate monophyletic clusters. In contrast, pronounced sequence variation was observed in the sexual partner of ES1, who was diagnosed with HIV-1 infection 2 years after initial contact with ES1. However, HIV-1 sequences from ES1 and his sexual partner were not closely related, suggesting neither was the source of infection of the other (26). There was also no evidence of superinfection in the ES individuals despite repeated high-risk unprotected sexual activity with multiple partners over the study period (Fig. 1), suggesting a possible protection against new HIV infection. Sequences from ES1 and ES38 were also distinct from those of prototypic laboratory HIV-1 strains, as well as the full catalog of HIV-1 sequences in the Los Alamos National Laboratory database (24) and a database of local sequences by using FASTA (26).
FIG. 4.
Phylogenetic analysis of HIV-1 env V3-C3 sequences of ES1 and ES38. An MLE phylogram (lnL = −269.2349) was constructed by using PAUP* illustrating the inferred evolutionary relationships of sequences obtained from ES1, ES38, and the HIV-1-infected sexual partner of ES1 (P1) and from the Los Alamos Retrovirus Database (24) and a database of local sequences by using FASTA (26). The model of evolution (GTR+G) was selected by using the AIC (1) in Modeltest version 3.06 (38). The parameters of this model were as follows: equilibrium nucleotide frequencies, fA = 0.4632, fC = 0.1299, fG = 0.1971, and fT = 0. 2098; shape parameter (α) of the 71 distributions reflecting the site-to-site rate variability of variable sites, α = 0.7535; R matrix values, RA<->C = 2.531, RA<->G = 2.962, RA<->T = 0.655, RC<->G = 1.571, RC<->T = 4.728, and RG<->T = 1. The scale bar represents the 10% MLE sequence distance. Multiple identical sequences from ES1, ES38, and P1 were represented by a single sequence in the phylogenetic reconstruction analyses; the numbers of sequences represented by a single node are shown in parentheses.
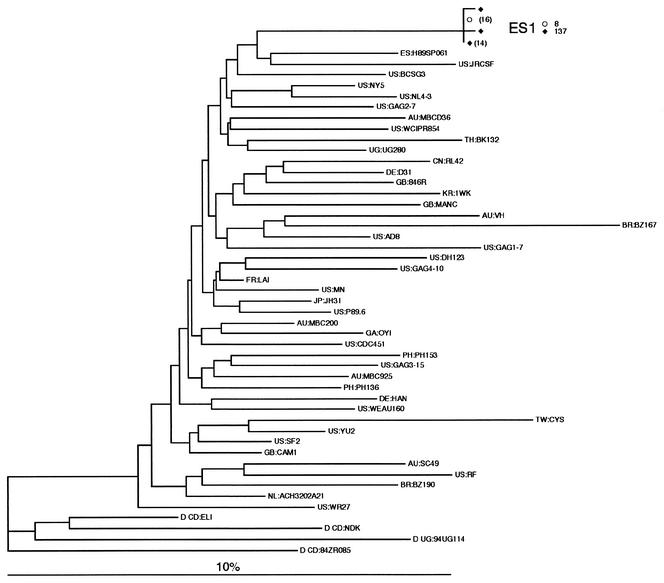
We also amplified gag p17 of HIV-1 sequences from resting CD4+ T cells at weeks 8 and 137 of ES1. The copy numbers of HIV-1 provirus detected by HIV-1 gag PCR were 0.09 and 0.03 per million resting CD4+ T cells at weeks 8 and 137, respectively. The evolution rate of HIV-1 gag p17 sequences was 0.012% per site per year (95% confident interval, −0.051 to 0.028%), which is not significantly different from zero (P = 0.169). Further phylogenetic analysis to determine the evolutionary relationships of sequences obtained from ES1 and other sequences in the databases (24, 26) excluded any possible contamination of known HIV-1 strains (Fig. 5). These results are consistent with findings derived from the analyses on HIV-1 env in ES1, indicating that HIV-1 sequences from ES1 and ES38 are remarkably fixed in evolution and do not represent contaminants from any identified source (24, 26).
FIG. 5.
Neighbor-joining tree of gag p17 region sequences from ES1 and the Los Alamos HIV Sequence Database subtype B reference set (24) rooted by using four sequences from subtype D. The tree was inferred by using a pairwise distance matrix from PAUP* version 4.0b4a. The model of evolution for this estimation (TVM+G) was selected by using Modeltest version 3.06 (38) under the AIC (1). The parameters of the model were as follows: equilibrium nucleotide frequencies, fA = 0.3954, fC = 0.2076, fG = 0.2141, and fT = 0.1829; proportion of invariable sites, 0.2769; shape parameter (α) of the 71 Γ distribution reflecting the site-to-site rate variability of variable sites, α = 0.6701; R matrix values, RA<->C = RG<->T = 1, RA<->G = RC<->T = 2.723, and RA<->T = RC<->G = 0.4653. The numbers of identical sequences represented by a single node are shown in parentheses.
DISCUSSION
These results shed new light on the puzzling observation that some persons can repeatedly engage in unprotected sexual activities with HIV-1-infected partners and yet fail to acquire overt HIV-1 infection. Our study indicates that HIV-1 infection can be demonstrated at extremely low levels in these seronegative persons only by sampling large cell numbers and by performing hundreds or thousands of PCR amplifications and sequence analyses. One of the remarkable findings in the present study is that the HIV-1 DNA levels in ES1 and ES38 are, on average, 104- to 106-fold lower than those of other HIV-1-infected populations studied and also typically 102-fold lower than could have been detected in a previous study of possible transient HIV infection (15). Both ES1 and ES38 have remained healthy and free of superinfection of other new HIV strains despite the fact that they had engaged repeatedly in unprotected sexual intercourse with multiple HIV-1-infected partners over the study period. However, epidemiological data supporting the possibility of recognizable HIV-1 infection from a distant exposure or set of exposures do not exist. This issue may be addressed only by prospective, longitudinal ES studies, which are in progress.
Understanding the mechanisms that account for the control of HIV infection, and the potential protection against new infection in these ES individuals should be of great interest to efforts to develop an effective vaccine. First, the low viral copy numbers may be the result of exposure to a low viral inoculum, reaching a minimum threshold to establish latency but insufficient to result in overt systemic infection. In support of this possibility, transient infection or occult systemic low-level infection was found in macaques challenged with low doses of simian immunodeficiency virus (SIV) (12, 31, 34, 36, 47, 48). In all of these studies, the animals did not seroconvert to SIV antigens by conventional assays, and most of these animals did not develop any signs of SIV-associated disease. Second, ES individuals may have encountered attenuated HIV-1 strains, an idea that in accord with observations of silent infection in monkeys inoculated with attenuated SIVmac strains (29). This may explain our inability to isolate infectious HIV-1 in vitro. Coculture of up to 600 million resting CD4+ T cells and CD8-depleted PBMC of ES individuals with different mitogens, such as phytohemagglutinin, anti-CD3, and tumor necrosis factor alpha alone or in combination, was carried out repeatedly without detection of virus (data not shown). In contrast, infectious virus was consistently isolated from seropositive donor cell controls. However, given that the HIV-1 proviral copy numbers in cells from ES individuals were extremely low (ranging from 1 per 10 to 100 million cells), the sensitivity of this technique might not have been adequate to identify a rare infectious virus. Nevertheless, based upon the extreme homogeneity of HIV-1 sequences over the study period, the repeatedly undetectable HIV-1 RNA in plasma, and the failure to isolate infectious virus in vitro, the HIV-1 in the resting CD4+ T cells from ES1 and ES38 may be attenuated or replication defective. Third, acquired T-cell immunity may play an important role in the control of HIV-1 infection (16, 17, 23, 30, 35, 37, 41-43). This is supported in a recent study (21, 22) by events surrounding the acquisition of HIV-1 infection in Nairobi HIV-1 exposed women who had resisted infection for many years, which the investigators believe may be linked to waning HIV-1-specific cytotoxic responses. Although we observed that the trend of the HIV-1 DNA decline in ES1 paralleled the fall in pCTL frequencies (17), we could not determine whether CTL were involved in controlling viral replication at such low levels. It is possible that T-cell responses in ES individuals only denote previous HIV-1 exposure or perhaps even cross-priming from exposure to their partner's infected cells. The capacity to maintain detectable CTL in the setting of such low levels of antigen exposure is an issue that will require further prospective analysis in order to understand their functional relevance in this population.
In summary, it is likely that more than one mechanism can account for the sustained, extraordinary control of infection and that both viral and host factors, including inheritance of genetic defects and distinct HLA or major histocompatibility complex types may contribute to this outcome (10, 18, 19, 27, 28, 33, 44). Key unresolved issues include (i) the proportion of ES individuals with low levels of HIV-1 infection, (ii) the likelihood that these individuals will subsequently manifest clinical HIV-1 disease, and (iii) the probability that they can transmit HIV-1 infection to their sexual partners. Because the HIV-1 copy number is extremely low and the effort to document low levels of infection is substantial, routine assessment of infection in resting CD4+ T cells is not likely to be feasible by current methodologies. Thus, although our findings provide new insight into protection against active HIV-1 infection, the challenge ahead lies in better understanding the long-term sequelae in these ES individuals and in translating the mechanisms underlying these circumstances to therapeutic and immunization strategies.
Acknowledgments
We thank E. Wakabayashi, F. Feng, J. Kim, H. Liu, K. Diem, C. Alef, and R. Akridge for technical assistance; Y. Wang and A. J. Rossini for advice on statistical analyses; and the study participants for their time and commitment.
This work is supported by NIH grants AI45402, AI41535, AI45206, AI35605, AI26503, RR00166, and AI49109; the Burroughs-Wellcome Clinical Scientist Award; and the University of Washington Center for AIDS Research Young Investigator Award (NIH AI27757).
REFERENCES
- 1.Akaike, H. 1974. A new look at statistical model identification. IEEE Trans. Autom. Contr. 19:716-723. [Google Scholar]
- 2.Bakshi, S. S., S. Tetali, E. J. Abrams, M. O. Paul, and S. G. Pahwa. 1995. Repeatedly positive HIV-1 DNA polymerase chain reaction in HIV-exposed seroreverting infants. Pediatr. Infect. Dis. J. 14:658-662. [DOI] [PubMed] [Google Scholar]
- 3.Berrey, M. M., T. Schacker, A. C. Collier, T. Shea, S. J. Brodie, D. Mayers, R. Coombs, J. Krieger, T. W. Chun, A. Fauci, S. G. Self, and L. Corey. 2001. Treatment of primary human immunodeficiency virus type 1 infection with potent antiretroviral therapy reduces frequency of rapid progression to aids. J. Infect. Dis. 183:1466-1475. [DOI] [PubMed] [Google Scholar]
- 4.Brodie, S. J., D. A. Lewinsohn, B. K. Patterson, D. Jiyamapa, J. Krieger, L. Corey, P. D. Greenberg, and S. R. Riddell. 1999. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat. Med. 5:34-41. [DOI] [PubMed] [Google Scholar]
- 5.Brostrom, C., U. Visco-Comandini, Z. Yun, and A. Sonnerborg. 1999. Longitudinal quantification of human immunodeficiency virus type 1 DNA and RNA in long-term nonprogressors. J. Infect. Dis. 179:1542-1548. [DOI] [PubMed] [Google Scholar]
- 6.Bryson, Y. J., S. Pang, L. S. Wei, R. Dickover, A. Diagne, and I. S. Y. Chen. 1995. Clearance of HIV infection in a perinatally infected infant. N. Engl. J. Med. 332:833-838. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., D. Engel, M. M. Berrey, T. Shea, L. Corey, and A. S. Fauci. 1998. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 95:8869-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerici, M., J. M. Levin, H. A. Kessler, A. Harris, J. A. Berzofsky, A. L. Landay, and G. M. Shearer. 1994. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA 271:42-46. [PubMed] [Google Scholar]
- 10.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 11.De Maria, A., C. Cirillo, and L. Moretta. 1994. Occurrence of human immunodeficiency virus type 1 (HIV-1)-specific cytolytic T-cell activity in apparently uninfected children born to HIV-1-infected mothers. J. Infect. Dis. 170:1296-1299. [DOI] [PubMed] [Google Scholar]
- 12.Dittmar, M. T., K. Cichutek, P. N. Fultz, and R. Kurth. 1995. The U3 promoter region of the acutely lethal SIV clone smmPBj1.9 confers related biological activity on the apathogenic clone agm3mc. Proc. Natl. Acad. Sci. USA 92:1362-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 14.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 15.Frenkel, L. M., J. I. Mullins, G. H. Learn, L. Manns-Arcuino, B. L. Herring, M. L. Kalish, R. W. Steketee, D. M. Thea, J. E. Nichols, S.-L. Liu, A. Harmache, X. He, D. Muthui, A. Madan, L. Hood, A. T. Haase, M. Zupancic, K. Staskus, S. M. Wolinsky, P. Krogstad, J. Zhao, I. Chen, R. Koup, D. D. Ho, B. T. Korber, R. J. Apple, R. W. Coombs, S. Pahwa, and N. J. J. Roberts. 1998. Genetic evaluation of suspected cases of transient HIV-1 infection of infants. Science 280:1073-1077. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons, J., J. M. Cory, I. K. Hewlett, J. S. Epstein, and M. E. Eyster. 1990. Silent infections with human immunodeficiency virus type 1 are highly unlikely in multitransfused seronegative hemophiliacs. Blood 76:1624-1626. [PubMed] [Google Scholar]
- 17.Goh, W. C., J. Markee, R. E. Akridge, M. Meldorf, L. Musey, T. Karchmer, M. Krone, A. Collier, L. Corey, M. Emerman, and M. J. McElrath. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T-cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548-557. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 20.Imagawa, D. T., M. H. Lee, S. M. Wolinsky, K. Sano, F. Morales, S. Kwok, J. J. Sninsky, P. G. Nishanian, J. Giorgi, J. L. Fahey, et al. 1989. Human immunodeficiency virus type 1 infection in homosexual men who remain seronegative for prolonged periods. N. Engl. J. Med. 320:1458-1462. [DOI] [PubMed] [Google Scholar]
- 21.Kaul, R., T. Dong, F. A. Plummer, J. Kimani, T. Rostron, P. Kiama, E. Njagi, E. Irungu, B. Farah, J. Oyugi, R. Chakraborty, K. S. MacDonald, J. J. Bwayo, A. McMichael, and S. L. Rowland-Jones. 2001. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Investig. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaul, R., S. L. Rowland-Jones, J. Kimani, T. Dong, H. B. Yang, P. Kiama, T. Rostron, E. Njagi, J. J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 2001. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J. Clin. Investig. 107:341-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary HIV-1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. T. M. Korber. 1999. Human retroviruses and AIDS. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 25.Langlade-Demoyen, P., N. Ngo-Giang-Huong, F. Ferchal, and E. Oksenhendler. 1994. Human immunodeficiency virus (HIV) nef-specific cytotoxic T lymphocytes in noninfected heterosexual contact of HIV-infected patients. J. Clin. Investig. 93:1293-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Learn, G. H., B. T. M. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of HIV sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, H., D. Chao, E. E. Nakayama, H. Taguchi, M. Goto, X. Xin, J. K. Takamatsu, H. Saito, Y. Ishikawa, T. Akaza, T. Juji, Y. Takebe, T. Ohishi, K. Fukutake, Y. Maruyama, S. Yashiki, S. Sonoda, T. Nakamura, Y. Nagai, A. Iwamoto, and T. Shioda. 1999. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. USA 96:4581-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald, K. S., K. R. Fowke, J. Kimani, V. A. Dunand, N. J. Nagelkerke, T. B. Ball, J. Oyugi, E. Njagi, L. K. Gaur, R. C. Brunham, J. Wade, M. A. Luscher, P. Krausa, S. Rowland-Jones, E. Ngugi, J. J. Bwayo, and F. A. Plummer. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:1581-1589. [DOI] [PubMed] [Google Scholar]
- 29.Marthas, M., R. A. Ramos, B. L. Lohman, K. K. A. Van Rompay, R. E. Unger, J. C. Miller, B. Banapour, N. C. Pedersen, and P. A. Luciw. 1993. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J. Virology 67:6047-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzoli, S., D. Trabattoni, S. Lo Caputo, S. Piconi, C. Ble, F. Meacci, S. Ruzzante, A. Salvi, F. Semplici, R. Longhi, M. Fusi, N. Tofani, M. Biasin, M. Villa, F. Mazzotta, and M. Clerici. 1997. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 3:1250-1257. [DOI] [PubMed] [Google Scholar]
- 31.McChesney, M. B., J. R. Collins, D. Lu, X. Lu, J. Torten, R. L. Ashley, M. W. Cloyd, and C. J. Miller. 1998. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 72:10029-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClure, M. O., P. D. Bleniasz, J. N. Weber, T. S. Tedder, S. O'Shea, J. E. Banatvaia, G. Tudor-Williams, P. Simmonds, and E. C. Holmes. 1995. HIV clearance in an infant? Nature 375:637-638. [DOI] [PubMed] [Google Scholar]
- 33.Michael, N. L., L. G. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C. E. Wang, and H. W. Sheppard. 1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160-1162. [DOI] [PubMed] [Google Scholar]
- 34.Miller, C. J., M. Marthas, J. Torten, N. J. Alexander, J. P. Moore, G. F. Doncel, and A. G. Hendrickx. 1994. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J. Virol. 68:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific T lymphocytes and plasma viral load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 36.Pauza, C. D., P. Emau, M. S. Salvato, P. Trivedi, D. MacKenzie, M. Malkovsky, H. Uno, and K. T. Schultz. 1993. Pathogenesis of SIVmac251 after atraumatic inoculation of the rectal mucosa in rhesus monkeys. J. Med. Primatol. 22:154-161. [PubMed] [Google Scholar]
- 37.Pinto, L. A., J. Sullivan, J. A. Berzofsky, M. Clerici, H. A. Kessler, A. L. Landay, and G. M. Shearer. 1995. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J. Clin. Investig. 96:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737-742. [DOI] [PubMed] [Google Scholar]
- 40.Roques, P. A., G. Gras, F. Parnet-Mathieu, A. M. Mabondzo, C. Dollfus, R. Narwa, D. Marcé, J. Tranchot-Diallo, F. Hervé, G. Lasfargues, C. Courpotin, and D. Dormont. 1995. Clearance of HIV infection in 12 perinatally infected children: clinical, virological and immunological data. AIDS 9:F19-F26. [PubMed] [Google Scholar]
- 41.Rosenberg, E. S., M. Altfeld, S. H. Poon, M. N. Phillips, B. M. Wilkes, R. L. Eldridge, G. K. Robbins, R. T. D'Aquila, P. J. Goulder, and B. D. Walker. 2000. Immune control of HIV-1 after early treatment of acute infection. Nature 407:523-526. [DOI] [PubMed] [Google Scholar]
- 42.Rowland-Jones, S. L., D. F. Nixon, M. C. Aldhous, F. Gotch, K. Ariyoshi, N. Hallam, J. S. Kroll, K. Froebel, and A. McMichael. 1993. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341:860-861. [DOI] [PubMed] [Google Scholar]
- 43.Rowland-Jones, S. L., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, M. Takiguchi, T. Schultz, A. McMichael, and H. Whittle. 1995. HIV-specific cytotoxic T cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 44.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 45.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trivedi, P., D. Horejsh, S. B. Hinds, P. W. Hinds II, M. S. Wu, M. S. Salvato, and C. D. Pauza. 1996. Intrarectal transmission of simian immunodeficiency virus in rhesus macaques: selective amplification and host responses to transient or persistent viremia. J. Virol. 70:6876-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rompay, K. K. A., M. L. Marthas, J. D. Lifson, C. J. Berardi, G. M. Vasquez, E. Agatep, A. Q. Dehqanzada, K. C. Cundy, N. Bischofberger, and N. C. Pedersen. 1998. Administration of 9-[2-(phosphonomethoxy)propyl]adenine (PMPA) for prevention of perinatal simian immunodeficiency virus infection in rhesus macaques. AIDS Res. Hum. Retrovir. 14:761-773. [DOI] [PubMed] [Google Scholar]
- 49.Wong, J. K., M. Hezareh, H. F. Günthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1300. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, T., B. T. Korber, A. J. Nahmias, E. Hooper, P. M. Sharp, and D. D. Ho. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594-597. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]