Abstract
Japanese encephalitis virus (JEV) is a common agent of viral encephalitis that causes high mortality and morbidity among children. Molecular genetic studies of JEV are hampered by the lack of a genetically stable full-length infectious JEV cDNA clone. We describe here the development of such a clone. A JEV isolate was fully sequenced, and then its full-length cDNA was cloned into a bacterial artificial chromosome. This was then further engineered so that transcription of the cDNA in vitro would generate synthetic RNAs with authentic 5′ and 3′ ends. The synthetic RNAs thus produced were highly infectious in susceptible cells (>106 PFU/μg), and these cells rapidly generated a high titer of synthetic viruses (>5 × 106 PFU/ml). The recovered viruses were indistinguishable from the parental virus in terms of plaque morphology, growth kinetics, RNA accumulation, protein expression, and cytopathogenicity. Significantly, the structural and functional integrity of the cDNA was maintained even after 180 generations of growth in Escherichia coli. A single point mutation acting as a genetic marker was introduced into the cDNA and was found in the genome of the recovered virus, indicating that the cDNA can be manipulated. Furthermore, we showed that JEV is an attractive vector for the expression of heterologous genes in a wide variety of cell types. This novel reverse genetics system for JEV will greatly facilitate research into JEV biology. It will also be useful as a heterologous gene expression vector and will aid the development of a vaccine against JEV.
Research investigating positive-sense RNA viruses has been considerably advanced by the development of the reverse genetics system. Here, infectious cDNA clones of the viral genome in question are constructed and become the templates for infectious RNA synthesis that generates synthetic viruses. In the classical RNA-launched approach, cells are transfected with RNA transcripts made from the infectious cDNA clones, and the synthetic viruses are then recovered from these cells (18, 29, 30, 32, 43). However, an alternative DNA-launched approach also exists. This approach was first reported for poliovirus (27) and has been adapted for alphaviruses (34). Here, synthetic viruses are generated by directly transfecting infectious cDNA clones into susceptible cells. Both of these approaches have been used to construct infectious cDNA clones for many positive-sense RNA virus families, including coronaviruses, which have the largest RNA genomes (2). These clones have been invaluable in addressing many questions regarding the positive-sense RNA viruses. However, the construction of a full-length infectious cDNA clone for Japanese encephalitis virus (JEV) has been hampered, largely because of the genetic instability of the cloned cDNA. Despite extensive efforts (23, 38, 39, 51), a genetically stable full-length infectious cDNA molecular clone for JEV does not exist.
JEV is a member of the Flaviviridae family and is transmitted by mosquitoes. It is an important human pathogen that causes permanent neuropsychiatric sequelae and even fatal disease, especially in children (37, 40, 41). Transmission of the virus has recently been observed in the southern hemisphere, indicating that this virus could become a worldwide public health threat (12, 13, 20). From its genome structure, which is similar to that of other flaviviruses, JEV is a small-enveloped virus with a single-stranded, positive-sense RNA genome approximately 11 kb in length. The genome contains a single long open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTRs) that are important cis-acting elements for viral replication. The RNA genome has a type I cap structure at its 5′ terminus but lacks a poly(A) tail at its 3′ terminus. The ORF is translated into a large polyprotein that is co- or posttranslationally processed into three structural and seven nonstructural proteins whose genes are arranged in the genome as follows: C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 (7, 19, 45). Further information, for example, on the function of the majority of the JEV gene products and the molecular mechanisms involved in JEV replication, neurovirulence, and pathogenesis, is limited largely because of the lack of a reliable reverse genetics system.
Here we report the JEV reverse genetics system that we have developed. We showed that in vitro transcription of the full-length cDNA produces synthetic RNAs that, when transfected into susceptible BHK-21 cells, have a specific infectivity exceeding 106 PFU/μg. Synthetic JEVs recovered from the culture supernatant of the transfected cells 24 h posttransfection exceeded 5 × 106 PFU/ml. The recovered viruses were similar to the parent in terms of plaque morphology, growth kinetics, RNA accumulation, protein expression, and cytopathogenicity. A genetic marker, a single point mutation that had been introduced into the infectious cDNA, was observed in the genome of the recovered virus. Most significantly, the infectious bacterial artificial chromosome (BAC) remained genetically stable even after 180 generations of serial growth in Escherichia coli. Furthermore, foreign genes engineered into the JEV cDNA were expressed as genomic RNA transcripts, indicating that JEV could be exploited as a eukaryotic expression vector. Thus, we have established a reverse genetics system for JEV that will be a useful tool for many areas of biological research.
MATERIALS AND METHODS
Cell lines and viruses.
BHK-21 cells were maintained in alpha minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, vitamins, and antibiotics. All reagents used in cell culture were purchased from Gibco-BRL Life Technologies, Inc., Gaithersburg, Md. The Korean JEV strain K87P39 (8) was obtained from the Korean National Institute of Health. This isolate was isolated from wild mosquitoes in Korea in 1987 and underwent five passages in suckling mouse brains. The YF17D yellow fever virus strain was generated from the infectious cDNA pACNR/YF17D by SP6 polymerase runoff transcription as described below.
Isolation of viruses by plaque purification.
Cells infected with the JEV K87P39 strain were overlaid with MEM containing 10% fetal bovine serum and 0.5% SeaKem LE agarose (FMC BioProducts, Rockland, Maine) and incubated for 3 to 4 days. Individual plaques were picked with sterile Pasteur pipettes and resuspended in 1 ml of medium. Viruses were eluted from the agarose at 4°C for 2 h. The eluate was amplified only once in BHK-21 cells and stored at −80°C until use.
Complete nucleotide sequence analysis of JEV genomic RNA.
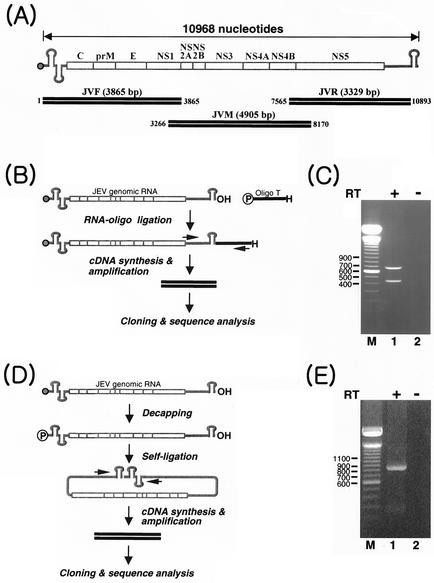
Viral genomic RNA was extracted from 100 μl of virus-containing culture fluid with 300 μl of Trizol LS reagent as recommended by the manufacturer (Gibco-BRL) and then resuspended in 20 μl of RNase-free water. To analyze the complete nucleotide sequence of the viral genomic RNA, three overlapping cDNAs (JVF, JVM, and JVR) representing the entire viral RNA genome apart from the 3′-terminal sequences were amplified by long reverse transcription (RT)-PCR as illustrated in Fig. 2. Oligonucleotides used for cDNA synthesis and amplification were designed according to the consensus sequence of all 16 fully sequenced JEV RNA genomes available from the GenBank database [CH2195LA, CH2195SA, FU, GP78, HVI, JaOAr01, JaGArS982, K94P05, Vellore P20778, p3, SA(A), SA(V), SA14, SA14-14-2, TC, and TL strains] (Table 1).
FIG. 2.
Strategies used to sequence genomic RNA of CNU/LP2. (A) Scheme for RT-PCR amplification of three overlapping cDNA amplicons representing the entire JEV genomic RNA apart from the 5′ and 3′ termini. RNA is indicated in gray, and cDNA is indicated by solid parallel lines. The top panel schematically depicts the CNU/LP2 JEV genomic RNA (10,968 bp in length). The bottom panels portray the three overlapping cDNAs, JVF (nt 1 to 3865), JVM (nt 3266 to 8170), and JVR (nt 7565 to 10893). (B) Scheme used to sequence the 3′ end of CNU/LP2 genomic RNA. The 5′-phosphorylated and 3′-blocked oligonucleotide T (Oligo T) was ligated to the 3′ end of JEV genomic RNA by T4 RNA ligase, and the resulting RNA was then used for cDNA synthesis and amplification with the primers indicated by an arrow. The resulting products were cloned and sequenced. (C) JEV-specific amplicons synthesized from the oligonucleotide T-ligated JEV genomic RNA described in B. First-strand cDNA was synthesized with oligonucleotide TR, complementary to oligonucleotide T, and the RT reaction was carried out in the presence (lane 1) or absence (lane 2) of Superscript II RT. The cDNA was amplified with oligonucleotide TR and primer J35, which is complementary to nt 10259 to 10276. The expected size of the PCR product is 727 bp. The products were separated on a 1.2% agarose gel and visualized by staining with ethidium bromide. (D) Scheme used to sequence the 5′ end of CNU/LP2 genomic RNA. The cap structure of viral genomic RNA was removed with tobacco acid pyrophosphatase, and the decapped viral RNA was then self-ligated with T4 RNA ligase and used for cDNA synthesis and amplification. The resulting amplified products were cloned and sequenced. (E) JEV-specific amplicons synthesized from the self-ligated JEV genomic RNA described in D. First-strand cDNA synthesis was carried out with primer J40, which is complementary to nt 215 to 232. The RT reaction was performed in the presence (lane 1) or absence (lane 2) of Superscript II RT. The cDNA was amplified with primer J35 and primer J39, which is complementary to nt 164 to 181. The expected size of the PCR product is 890 bp. The products were analyzed on a 1.2% agarose gel as described above. Lane M, 100-bp DNA size ladder (in base pairs).
TABLE 1.
Oligonucleotides used for ligation, cDNA synthesis, and PCR amplification
| Oligonucleotide | Sequencea | Positionb | Polarity |
|---|---|---|---|
| J1 | 5′-GATCCTGTGTTCTTCCTCACC | 10947-10967 | Antisense |
| J2 | 5′-AGAAGATCTCCCAGTCTATTCCCA | 10870-10893 | Antisense |
| J3 | 5′-GATTTAATTAACACCTCCTCTACAGCTTCG | 10795-10813 | Antisense |
| J4 | 5′-AGGACGCGTAGTGTGCGTTGT | 8150-8170 | Antisense |
| J6 | 5′-GCCCCTAGGACCAGAACCACG | 3845-3865 | Antisense |
| J7 | 5′-AGCGCTAAGACTGGCATG | 3986-4003 | Antisense |
| J8 | 5′-GATCGGACCGAGAAGTTTATCTGTGTGA | 1-18 | Sense |
| J12 | 5′-GATCGGACCGAATTCCACCACAGCCAC | 7565-7582 | Sense |
| J20 | 5′-AAACCAGGGACCTTGGGA | 3266-3283 | Sense |
| J31 | 5′-GGCTGTGGGCCCACTTGT | 8821-8838 | Sense |
| J35 | 5′-AGCAACCTGGGCTGAGAA | 10259-10276 | Sense |
| J39 | 5′-CCCACTAGTGGGAATACG | 164-181 | Antisense |
| J40 | 5′-AAACGTACTGGTCCTCTG | 215-232 | Antisense |
| J41 | 5′-TCCGTGGAATGAACAATG | 17911-17928 | Sense |
| J42 | 5′-GAGAAGTTTATCTGTGTG | 1-17 | Sense |
| J43 | 5′-ACAGATAAACTTCTCTATAGTGTCCCCTAA | 1-14 | Antisense |
| J45 | 5′-AGTACTAGTCGGTCCGCGGCCGCTCGAGATCCTGTGTTCTT | 10954-10968 | Antisense |
| J46 | 5′-AGTACTAGTCGGTCCGCGGCCGCTCTAGAGATCCTGTGTTCTT | 10954-10968 | Antisense |
| J47 | 5′-CCAAAGCTTCAAACTCAAGATACC | 9127-9150 | Antisense |
| J48 | 5′-ACTGAGCTCACGCGTCCTCGAGATGAC | 8162-8179 | Sense |
| J72 | 5′-GAAGGTACCCCATTGTATGG | ||
| J73 | 5′-TTCTCCTTTACCCATGGTTGTGGCAAGCTT | ||
| J74 | 5′-ATGGGTAAAGGAGAAGAA | ||
| J75 | 5′-AAGATGCATTCATTAACCGTCGACTGCAGA | ||
| J76 | 5′-TTTGGCGTCTTCCATGGTTGTGGCAAGCTT | ||
| J77 | 5′-ATGGAAGACGCCAAAAAC | ||
| J78 | 5′-CTTAAGATGCATTCATTACACGGCGATCTT | ||
| J80 | 5′-ACAGATAAACTTCTCTATAGTGAGTCGTAT | 1-14 | Antisense |
| J81 | 5′-TCTTGCCCGCCTGATGAA | ||
| J82 | 5′-GCCCATGGTAAGCTTAGG | 636-653 | Antisense |
| J89 | 5′-TGCTTTGGCCTTCTTGGCCAC | 2417-2437 | Sense |
| J90 | 5′-TTGAGGCCCCCACGGCCCAA | 10687-10706 | Sense |
| J91 | 5′-ACCCGCATATTCTGTGATCCGTGGTTCCAG | 5566-5575, 5659-5678 | Antisense |
| J92 | 5′-ACAGAATATGCGGGTAAA | 5664-5681 | Sense |
| J93 | 5′-AGCTAACGGCCGATCTCTTTC | 6457-6477 | Antisense |
| T | 5′-CCAGTGTTGTGGCCTGCAGGGCGAATT | ||
| TR | 5′-GATGAATTCGCCCTGCAGGCCACAACA |
JEV-specific sequences are shown in boldface type. Restriction enzyme recognition sites used for cDNA cloning are underlined.
Nucleotide position refers to the complete nucleotide sequence of pBACSP6/JVFLx/XbaI.
For JVF amplicons (nucleotides [nt] 1 to 3865), primer J7, complementary to nt 3986 to 4003 of the JEV genome, was used for cDNA synthesis. The primers for PCR amplification were primer J8, complementary to nt 1 to 18, and primer J6, complementary to nt 3845 to 3865. For JVM amplicons (nt 3266 to 8170), primer J4, complementary to nt 8150 to 8170, was used for cDNA synthesis. The primers for PCR amplification were primer J20, complementary to nt 3266 to 3283, and primer J4. For JVR amplicons (nt 7565 to 10893), primer J1, complementary to nt 10947 to 10967, was used for cDNA synthesis. The primers for PCR amplification were primer J12, complementary to nt 7565 to 7582, and primer J2, complementary to nt 10870 to 10893.
The standard RT reaction was conducted in a 20-μl reaction mixture containing 10 μl of extracted viral RNA, 5 pmol of the appropriate primer, 100 U of Superscript II RT (Gibco-BRL), 40 U of RNaseOUT (Gibco-BRL), 0.1 mM dithiothreitol (DTT), 10 mM deoxynucleoside triphosphate mix, and the RT buffer supplied by the manufacturer (Gibco-BRL). The reaction mixture was incubated at 37°C for 1 h and then heated at 70°C for 15 min. A 5-μl aliquot of the RT mixture was subsequently used for PCR amplification with Pyrobest DNA polymerase (Takara Bio Inc., Shiga, Japan) and the appropriate primer pair. The PCRs were performed with 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 5 min, followed by a final extension step at 72°C for 10 min.
To sequence the 3′-terminal sequences, a synthetic oligodeoxyribonucleotide T was ligated to the 3′ end of the viral genomic RNA to provide a primer-binding site for cDNA synthesis and PCR amplification as previously described (17). The 3′ end of oligonucleotide T was first modified by incorporating ddATP with terminal deoxynucleotidyltransferase (Takara), which blocks the intramolecular and intermolecular ligation of oligonucleotide T. The 5′ end of oligonucleotide T was also phosphorylated with T4 polynucleotide kinase (Takara). Thereafter, the modified oligonucleotide T was ligated to the 3′ end of the viral genomic RNA by T4 RNA ligase (New England Biolabs, Inc., Beverly, Mass.). The ligation reaction mixture (20 μl) contained 10 U of T4 RNA ligase, 40 U of RNaseOUT, 10 pmol of oligonucleotide T, viral genomic RNA, and the buffer supplied by the manufacturer (NEB). After incubation at 16°C for 12 h, the ligated viral RNA was phenol extracted, precipitated with ethanol, and resuspended with 20 μl of RNase-free water.
Subsequently, half of the oligonucleotide-ligated viral RNA was used for cDNA synthesis with oligonucleotide TR, which is complementary to oligonucleotide T, as previously described. First-strand cDNA was amplified with primer J35, complementary to nt 10259 to 10276, and primer TR. For PCR, one quarter of the RT reaction mix was amplified with Pyrobest DNA polymerase and 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, followed by a final extension of 10 min at 72°C. The PCR mixtures were as described above. cDNA amplicons were cloned into the pRS2 vector with HindIII and EcoRI sites incorporated in the positive-sense and negative-sense primers, respectively.
The 5′-terminal sequence was determined by self-ligation of viral RNA (5). The cap structure of viral genomic RNA was first cleaved off with tobacco acid pyrophosphatase. The cleavage reaction mixture (20 μl) contained 10 U of tobacco acid pyrophosphatase (Epicentre Technology Co., Madison, Wis.), 10 μl of viral RNA, and the buffer supplied by the manufacturer (Epicentre Technology Co.). After incubation at 37°C for 1 h, the tobacco acid pyrophosphatase-treated viral RNA was subjected to phenol extraction and ethanol precipitation and resuspended with 20 μl of RNase-free water. Half of the decapped viral RNA was self-ligated in a 20-μl reaction mixture with T4 RNA ligase as described above. A quarter of the self-ligated viral RNA was used for cDNA synthesis with primer J40, complementary to nt 215 to 232. First-strand cDNA was PCR amplified with primer J39, which is complementary to nt 164 to 181, and primer J35. cDNA amplicons were digested with ApoI and SpeI and ligated into the pRS2 vector which had been digested with ApoI and XbaI, leading to construct pRS2/JV3′5′.
Complete nucleotide sequence analysis was carried out by directly sequencing the cDNA amplicons representing the entire viral genome of JEV and the individual clones inserted into pRS2 vectors with an automatic 3700 DNA sequencer.
Construction of full-length infectious cDNAs for JEV.
Recombinant DNA techniques were used according to standard procedures (31). Three overlapping cDNA amplicons (JVF, JVM, and JVR) originally used for complete nucleotide sequence analysis were first subcloned into pBAC/SV, a derivative of the pBeloBAC11 plasmid (S.-I. Yun and Y.-M. Lee, unpublished data). The pBAC/SV plasmid contains the 491-bp NotI-AatII (T4 DNA polymerase-treated) fragment of pACNR/NADL (21), the 9,215-bp SacI (T4 DNA polymerase-treated)-SspI (T4 DNA polymerase-treated) fragment of pSINrep 19 (10), and the 6,875-bp SfiI (T4 DNA polymerase-treated)-NotI fragment of pBeloBAC11. Thus, the 3,863-bp RsrII-AvrII fragment of the JVF amplicons, the 4,717-bp BspE I-MluI fragment of the JVM amplicons, and the 3,326-bp RsrII-BglII fragment of the JVR amplicons were inserted into the pBAC/SV plasmid which had been digested with the same enzymes. This led to the pBAC/JVF, pBAC/JVM, and pBAC/JVR subclone constructs, respectively. These BAC plasmids were grown in E. coli DH10B cells and sequenced. The nucleotide sequences of the cloned cDNAs were identical to that of CNU/LP2 with the exception of a point mutation, T8906→ C (silent), within the NS5 gene in pBAC/JVR. The T8906→ C substitution was corrected by recloning a 315-bp ApaI-HindIII fragment corresponding to nt 8827 to 9142, leading to the construct pBAC/JVRR.
To facilitate the precise adjoining of the bacteriophage SP6 promoter transcription start to the 5′ end of the full-length JEV cDNA, pBAC/JVF was modified. Two fragments were first isolated by PCR of pBAC/SV with primer J41 and primer J43, which incorporates the negative-sense sequence of the SP6 promoter and PCR of pBAC/JVF with primers J42 and J40. These two fragments were fused by a second round of PCR with primers J41 and J40. The resulting amplicons were digested with PacI and PmeI and ligated with pBAC/JVF which had been digested with the same two enzymes. This produced pBACSP6/JVF.
To generate an authentic or nearly authentic 3′ terminus during runoff transcription of plasmid linearized at the 3′ end of the viral genome, we modified pBAC/JVRR so that the nucleotide sequence of the authentic 3′ terminus was followed by a unique restriction endonuclease recognition site, either XhoI or XbaI. To create the pBAC/JVRR/XhoI subclone containing a unique XhoI site at the end of the viral genome, fragment I was synthesized by PCR amplification of pRS2/JV3′5′ with primer J90 and primer J45, which incorporates an XhoI site (Table 1; underlined). The 298-bp SfiI-SpeI portion of fragment I amplicons was ligated with pBAC/JVRR which had been digested with SfiI and NheI. To create pBAC/JVRRx/XbaI, which has an XbaI site at the end of the viral genome, the existing XbaI site at nt 9131 to 9136 within the NS5 gene was first inactivated by introducing a silent point mutation (A9134 → T) by PCR. Here, pBAC/JVRR was amplified with primer J31 and primer J47, which incorporated the A9134 → T substitution. The 315-bp ApaI-HindIII portion of the cDNA amplicons, corresponding to nt 8828 to 9143, was cloned into pBAC/JVRR, leading to the construct pBAC/JVRRx. Subsequently, pBAC/JVRRx/XbaI was constructed in the same manner as described for pBAC/JVRR/XhoI. Thus, fragment II was obtained by PCR amplification of pRS2/JV3′5′ with primer J90 and primer J46, which incorporated an XbaI site (Table 1; underlined). The 298-bp SfiI-SpeI portion of the fragment II amplicons was then ligated into pBAC/JVRRx which had been digested with SfiI and NheI. To create pBAC/JVRRx/XhoI containing a unique XhoI site and the A9134→ T substitution, the 298-bp SfiI-SpeI portion of fragment I amplicons was ligated into pBAC/JVRRx which had been digested with SfiI and NheI.
Thus, we constructed five plasmids, pBACSP6/JVF, pBAC/JVM, pBAC/JVRR/XhoI, pBAC/JVRRx/XbaI, and pBAC/JVRRx/XhoI. These contained contiguous regions of the JEV genome and could now be used to assemble three different full-length JEV cDNAs, as illustrated in Fig. 3. First, the pBACSP6/JVFM subclone was constructed by ligating together the 4,717-bp BspEI-MluI fragment of pBAC/JVM, the 8,970-bp BspEI-XbaI fragment of pBACSP6/JVF, and the 3,670-bp XbaI-MluI fragment of pBAC/SV. Subsequently, two fragments of pBACSP6/JVFM (the 8,142-bp PacI-SapI fragment and the 4,801-bp PacI-BsrGI fragment) were ligated with either (i) the 5,620-bp SapI-BsrGI fragment of pBAC/JVRR/XhoI to generate pBACSP6/JVFL/XhoI, (ii) the 5,622-bp SapI-BsrGI fragment of pBAC/JVRRx/XbaI to generate pBACSP6/JVFLx/XbaI, or (iii) the 5,620-bp SapI-BsrGI fragment of pBAC/JVRRx/XhoI to generate pBACSP6/JVFLx/XhoI.
FIG. 3.
Construction of full-length JEV cDNA clones in bacterial artificial chromosome pBeloBAC11. (A) Schematic diagram of the full-length JEV cDNAs constructed in this study. Viral proteins are shown with thick solid lines at both termini representing the 5′ and 3′ NTRs of the viral genome. The SP6 and T7 promoter transcription start sites and the unique restriction endonuclease recognition site ensuring runoff transcription are shown at the 5′ and 3′ ends, respectively. (B and C) 5′ and 3′ termini of full-length JEV cDNA clones. Nucleotide sequences of JEV genomic RNA are shown as bold italic lowercase letters. Illustrated are the 5′ termini of four SP6-driven (B) and four T7-driven (C) full-length JEV cDNA templates. To produce SP6 and T7 RNA polymerase runoff products, the 3′ termini of two SP6-driven (B, pBACSP6/JVFL/XhoI and pBACSP6/JVFLx/XhoI) and two T7-driven (C, pBACT7/JVFL/XhoI and pBACT7/JVFLx/XhoI) JEV cDNA templates were linearized by XhoI digestion, resulting in three nucleotides (CGA) of virus-unrelated sequence at the 3′ ends. Similarly, the cutting of the 3′ termini of an SP6-driven (B, pBACSP6/JVFLx/XbaI) and a T7-driven (C, pBACT7/JVFLx/XbaI) JEV cDNA template with XbaI resulted in four nucleotides (CTAG) of virus-unrelated sequence at the 3′ ends. In contrast, the authentic 3′ end of JEV genomic RNA was present when SP6-driven (B, pBACSP6/JVFLx/XbaIMBN) and T7-driven (C, pBACT7/JVFLx/XbaIMBN) JEV cDNA templates were linearized by XbaI digestion and then treated with mung bean nuclease to remove the unrelated single-stranded sequences. Underlined is the restriction endonuclease recognition site introduced at the 3′ end of the viral genome. An arrowhead indicates a cleavage site.
In addition to the SP6-driven JEV cDNAs, we also constructed a set of three T7-driven full-length cDNAs. A fragment from pBAC/NADLcIns−/PAC (Y.-M. Lee and C. M. Rice, unpublished data) was first synthesized by PCR with the primers J81 and J80. A fragment from pBACSP6/JVFLx/XbaI was also synthesized with the primers J42 and J82. These two fragments were fused by a second round of PCR with the primers J81 and J82. The 793-bp EcoRI-SpeI fragment of the resulting amplicons was inserted into the pRS2 vector digested with EcoRI and XbaI, leading to the construct pRS2T7/5′JV. The 675-bp PvuI-PmeI fragment of pRS2T7/5′JV was ligated with either (i) the 18,364-bp PacI-PmeI fragment of pBACSP6/JVFL/XhoI to create pBACT7/JVFL/XhoI, (ii) the 18,364-bp PacI-PmeI fragment of pBACSP6/JVFLx/XhoI to create pBACT7/JVFLx/XhoI, or (iii) the 18,366-bp PacI-PmeI fragment of pBACSP6/JVFLx/XbaI to create pBACT7/JVFLx/XbaI.
Mutagenesis of full-length infectious JEV cDNA and insertion of foreign genes.
The point mutation A8171→ C (silent) was placed inside the NS5 gene in pBACSP6/JVFLx/XbaI by PCR-based site-directed mutagenesis to generate pBACSP6/JVFLx/gm/XbaI (see Fig. 6A). The point mutation resulted in the acquisition of a unique XhoI restriction endonuclease recognition site. A fragment from pBACSP6/JVFLx/XbaI was first generated by PCR with primer J48, in which the XhoI site (Table 1; underlined) was created by the A8171→ C substitution (Table 1; bold), and primer J3. The 665-bp MluI-ApaI fragment of the resulting amplicons was then ligated with the 4,802-bp ApaI-BsrGI and the 5,874-bp BsrGI-MluI fragments of pBACSP6/JVFLx/XbaI, resulting in the pBACSP6/JVFLx/gm/XbaI construct.
FIG. 6.
Presence of XhoI genetic marker in recombinant JEVs derived from pBACSP6/JVFLx/gm/XbaI. (A) Schematic diagram of the RT-PCR fragments of JVFLx/XbaIMBN and JVFLx/gm/XbaIMBN expected after XhoI digestion. Indicated are the primers used for RT-PCR (arrows), the introduced XhoI site (asterisk), and the sizes of the RT-PCT products (2,580 bp) and the two XhoI digestion products (1,506 bp and 1,074 bp) expected after digestion of JVFLx/gm/XbaIMBN with XhoI. (B) BHK-21 cells were transfected with synthetic RNAs transcribed from either pBACSP6/JVFLx/XbaIMBN or pBACSP6/JVFLx/gm/XbaIMBN. Viruses were recovered 24 h later and serially passaged in BHK-21 cells at a multiplicity of infection of 0.1. At each passage prior to the next round of infection, viruses were incubated with DNase I and RNase A (21). At passages 1 and 3, viral RNA was extracted from the culture supernatant containing the released viruses and used for RT-PCR. The PCR products were incubated in the presence (+) or absence (−) of XhoI, separated on a 1% agarose gel, and stained with ethidium bromide. The expected sizes of the undigested and digested PCR products are shown on the left. Lane M, 1-kb DNA ladder.
To generate the pBACSP6/JVFLx/LUC/XbaI construct (Fig. 8A), a fragment of pBACSP6/JVFLx/XbaI was amplified with primers J72 and J76. A fragment was also amplified from pACNR/NADLcIn−/luciferase (Y.-M. Lee and C. M. Rice, unpublished data) with primers J77 and J78. These two fragments were then fused by a second round of PCR with primers J72 and J78. The 1,801-bp KpnI-NsiI fragment of the resulting amplicons was then ligated with the 8,011-bp NsiI-PacI and 11,021-bp PacI-KpnI fragments of pBACSP6/JVFLx/XbaI, leading to pBACSP6/JVFLx/LUC/XbaI.
FIG. 8.
Expression of foreign genes with JEV cDNA as the vector. (A) Schematic diagram of the pBACSP6/JVFLx/GFP/XbaIMBN, pBACSP6/JVFLx/LUC/XbaIMBN, and pBACSP6/JVFLx/LUCREP−/XbaIMBN cDNA templates used for runoff transcription with SP6 RNA polymerase. Indicated are the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES)-driven GFP and luciferase genes that were inserted at the beginning of the 3′NTR of the viral genome, the SP6 promoter transcription start, and the runoff site generated by XbaI digestion and mung bean nuclease treatment (XbaI/MBN). In pBACSP6/JVFLx/LUCREP−/XbaIMBN, a solid vertical bar indicates an 83-nucleotide deletion (nt 5581 to 5663) in the middle of the NS3 gene that determines viral translation at nt 5596 (asterisk). (B) Expression of GFP protein. BHK-21 cells were mock transfected or transfected with 2 μg of synthetic RNAs transcribed from the pBACSP6/JVFLx/GFP/XbaIMBN template (JVFLx/GFP/XbaIMBN), incubated for 30 h, and then fixed and examined by confocal microscopy. (C) Induction of luciferase protein. BHK-21 cells (8 × 106) were mock transfected or transfected with 2 μg of synthetic RNAs transcribed from the pBACSP6/JVFLx/LUC/XbaIMBN (solid circles) and pBACSP6/JVFLx/LUCREP−/XbaIMBN (open circles) templates and seeded in a six-well plate at a density of 6 × 105 cells per well. Cells were lysed at the indicated time points, and luciferase activity was determined. The standard deviations obtained from three independent experiments are indicated by error bars.
To create the pBACSP6/JVFLx/GFP/XbaI construct (see Fig. 8A), a fragment from pBACSP6/JVFLx/LUC/XbaI was amplified by PCR with the primers J72 and J73. A fragment was also amplified from pRSGFP-C1 with the primers J74 and J75. These two fragments were fused by a second round of PCR with the primers J72 and J75. The 913-bp KpnI-NsiI fragment of the resulting amplicons was then ligated with the 8,011-bp NsiI-PacI and 11,021-bp PacI-KpnI fragments of pBACSP6/JVFLx/LUC/XbaI, resulting in the pBACSP6/JVFLx/GFP/XbaI construct.
To generate pBACSP6/JVFLx/LUCREP−/XbaI (see Fig. 8A), which contains an 83-nucleotide deletion (nt 5581 to 5663) in the middle of the NS3 gene that results in premature termination of viral translation at nt 5596, a fragment of pBACSP6/JVFLx/LUC/XbaI was amplified with primers J89 and J91. A fragment was also amplified from pBACSP6/JVFLx/LUC/XbaI with primers J92 and J93. These two fragments were then fused by a second round of PCR with primers J89 and J93. The 3,960-bp SfiI-EagI fragment of the resulting amplicons was then ligated with the 6,493-bp EagI-SfiI and 10,297-bp SfiI-SfiI fragments of pBACSP6/JVFLx/LUC/XbaI, leading to pBACSP6/JVFLx/LUCREP−/XbaI.
Transcriptions and transfections.
RNA transcripts were synthesized by in vitro transcription. Here, 100 to 200 ng of the template DNA linearized with XhoI or XbaI digestion and in some cases modified with mung bean nuclease was added to a 25-μl reaction mixture consisting of the buffer supplied by the manufacturer (Gibco-BRL) plus 0.6 mM cap analog [m7G(5′)ppp(5′)A or m7G(5′)ppp(5′)G; NEB Inc.], 0.5 μM [3H]UTP (1.0 mCi/ml, 50 Ci/mmol; New England Nuclear Corp., Boston, Mass.), 10 mM DTT, 1 mM each UTP, GTP, CTP, and ATP, 40 U of RNaseOUT, and 15 U of SP6 RNA polymerase (Gibco-BRL). The reaction mixtures were incubated at 37°C for 1 h. RNAs were quantified on the basis of [3H]UTP incorporation as measured by RNA adsorption to DE-81 (Whatman, Maidstone, United Kingdom) filter paper (31). A 1- to 1.5-μl aliquot of reaction mixture was examined by agarose gel electrophoresis, and aliquots were stored at −80°C until use.
For RNA transfection, cells were electroporated with synthetic RNAs with a model ECM 830 electroporator (BTX Inc., San Diego, Calif.) as recommended by the manufacturer. Briefly, subconfluent cells were trypsinized, washed three times with ice-cold RNase-free phosphate-buffered saline (PBS), and resuspended at a density of 2 × 107 cells/ml in PBS. A 400-μl aliquot of the suspension was mixed with 2 μg of synthetic RNA, and the cells were immediately electroporated under the conditions determined previously to be optimal (980 V, 99-μs pulse length, and five pulses). The electroporated mixture was then transferred to 10 ml of fresh medium.
An infectious center assay was used to quantify the specific infectivity of the synthetic RNA. The electroporated cells were serially diluted 10-fold and plated on monolayers of untransfected cells (5 × 105) in a six-well plate. Cells were allowed to attach to the plate for 6 h, after which they were overlaid with 0.5% SeaKem LE agarose-containing MEM as described above. The plates were incubated for 3 to 4 days at 37°C with 5% CO2, and infectious plaque centers were visualized by crystal violet staining.
Western blot analysis.
Cells (3 × 105) were lysed with 200 μl of sample loading buffer [80 mM Tri-HCl (pH 6.8), 2.0% sodium dodecyl sulfate (SDS), 10% glycerol, 0.1 M DTT, 0.2% bromophenol blue], and one-tenth of the lysate was boiled for 5 min and fractionated on an SDS-polyacrylamide gel. Proteins were transferred electrophoretically onto a methanol-activated polyvinylidene difluoride membrane with a Trans-Blot SD electrophoretic transfer cell machine (Bio-Rad Laboratories Inc., Hercules, Calif.), and the membrane was blocked at room temperature for 1 h with 5% nonfat dried milk in washing solution (0.2% Tween 20 in PBS). After three washes with washing solution, membranes were incubated at room temperature for 2 h with either a monoclonal antiactin antibody (A4700) that recognizes the epitope conserved in the C terminus of all actin isoforms (Sigma, St. Louis, Mo.) or mouse hyperimmune ascites fluid specific for JEV (ATCC VR-1259AF; American Type Culture Collection). The membranes were then washed three times with washing solution and incubated at room temperature for 2 h with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Labs Inc., West Grove, Pa.). The membranes were washed three times with washing solution and once with PBS. Actin and JEV protein bands were visualized by incubation with the substrates 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Northern blot analysis.
Total RNA was extracted from infected BHK-21 cells (3 × 105) with 1 ml of Trizol reagent (Gibco-BRL). One-third of the RNA was analyzed for JEV-specific RNA by Northern blot analysis as described elsewhere (31). Briefly, the RNA was electrophoresed in denaturing 2.2 M formaldehyde-1% agarose gels and transferred onto nylon membranes (Amersham Biosciences Inc., Piscataway, N.J.). The RNA on the membranes was cross-linked by irradiation with a 254-nm light source (Stratalinker UV cross-linker; Stratagene, La Jolla, Calif.), and the JEV-specific RNAs were detected by hybridization with a [32P]CTP-labeled antisense riboprobe that binds to nt 9143 to 9351 of the JEV genome. [This probe had been synthesized by in vitro transcription from the BamHI-linearized cDNA clone pGEM3Zf(+)/JV9143, which was constructed by ligating the 209-bp HindIII-SacI fragment of pBACSP6/JVFLx/XbaI with pGEM3Zf(+) digested with the same enzymes. This clone was transcribed with the T7-MEGAscript kit (Ambion, Austin, Tex.) as recommended by the manufacturer with a 20-μl reaction mixture containing 3.12 μM [α-32P]CTP (800 Ci/mmol; Amersham). After being treated with DNase I, the reaction mixture was spun in a Quick Spin G-50 Sephadex column (Boehringer Mannheim) to remove unincorporated ribonucleoside triphosphates.
The membrane was prehybridized at 55°C for 6 h in hybridization solution [5× SSPE (0.9 M NaCl, 50 mM NaH2PO4, and 5 mM EDTA, pH 7.7), 5× Denhardt's reagent, 0.5% SDS, 100 μg of denatured salmon sperm DNA per ml, 50% formamide] and then incubated at 55°C overnight in the hybridization solution containing 107 cpm of the labeled riboprobe. The membrane was washed three times at 55°C for 10 min with 1× SSPE-0.5% SDS and once for 10 min with 0.1× SSPE-0.5% SDS. Viral RNA bands were visualized by autoradiography and quantified with a Molecular Imager (Bio-Rad).
Direct immunofluorescence.
To examine JEV expression in infected BHK-21 cells by confocal microscopy, cells (2 × 105) were seeded in a four-well chamber slide, incubated for 12 h, and then mock-infected or infected at a multiplicity of infection of 1 for 18 h with either the original JEV K87P39 strain, the JEV CNU/LP2 isolate, or the YF17D strain. Immunostaining for JEV viral proteins was accomplished by first fixing the cells by incubation in PBS containing 0.37% (vol/vol) formaldehyde for 30 min at 25°C. The cells were then washed three times with PBS and permeabilized for 10 min at 37°C with PBS containing 0.2% (vol/vol) Triton X-100. Thereafter, the cells were washed four times with PBS, rehydrated in PBS for 15 min, and blocked for 1 h at 37°C with PBS containing 5% (wt/vol) bovine serum albumin. The cells were then incubated for 2 h at 25°C with 1:500-diluted mouse hyperimmune ascites fluid specific for JEV, washed three times with PBS, incubated for 2 h at 25°C with 1:500-diluted fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Labs Inc.), and washed again three times with PBS. Thereafter, the cells were incubated for 30 min at 37°C in PBS containing 5 μg of propidium iodide and 5 μg of RNase A per ml to localize the nuclei and mounted with 0.2 ml of 80% glycerol. Images were acquired on a Zeiss Axioskop confocal microscope equipped with a 63× objective with a Bio-Rad MRC 1024 and LaserSharp software.
To examine green fluorescent protein (GFP) expression, BHK-21 cells were mock-transfected or transfected with 2 μg of JVFLx/GFP/XbaIMBN RNA. Transfected cells (105) were incubated for 30 h in a four-well chamber slide. Cells were washed twice with PBS, fixed by incubation for 30 min at 25°C in PBS containing 0.37% (vol/vol) formaldehyde, and mounted with 0.2 ml of 80% glycerol. Cells were viewed by confocal microscopy and analyzed as described above.
Luciferase assay.
For the luciferase assay, BHK-21 cells (8 × 106) were mock-transfected or transfected with 2 μg of JVFLx/LUC/XbaIMBN RNA or JVFLx/LUCREP−/XbaIMBN RNA. Cells were seeded at a concentration of 6 × 105 cells/well in a six-well plate and cultivated. At the given time points, the cells were washed with Ca2+- and Mg2+-free PBS solution and then lysed by adding 0.2 ml of lysis buffer [25 mM Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100 (vol/vol)] to each well. Cell lysates were incubated for 10 min at room temperature, and cellular debris was then removed by centrifugation. The supernatants were quickly placed at −80°C for storage until use. To determine the luciferase activity, 20 μl of the cell lysates was placed in a luminometer tube containing 100 μl of luciferase assay reagent [20 mM Tricine, 1.07 mM (MgCO3)4Mg(OH)2 · 5H2O, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 270 μM coenzyme A, 470 μM luciferin (Promega), 530 μM ATP]. The activity was usually measured for 10 s. Each data point represents the results of three independent experiments.
Cloned full-length cDNA stability.
The genetic structure and functional integrity of the infectious JEV cDNAs were analyzed as follows. E. coli strain DH10B was transformed with pBACSP6/JVFLx/XbaI, and two independently derived clones were grown at 37°C overnight in 10 ml of 2xYT containing 12.5 μg of chloramphenicol per ml. Cells from these primary cultures were maintained for 9 days by diluting them 106-fold every day. After the third, sixth, and ninth passages, large-scale preparation of the infectious cDNA plasmid was made by the SDS-alkaline method and purified further by cesium chloride density gradient centrifugation (31). The genetic structure of the plasmid DNA was monitored by its restriction endonuclease pattern, and its functional integrity was assessed by measuring the specific infectivities of the synthetic RNAs transcribed from the cDNA template, which was linearized by XbaI digestion and mung bean nuclease treatment.
RESULTS
Isolation and complete nucleotide sequence analysis of JEV isolate CNU/LP2.
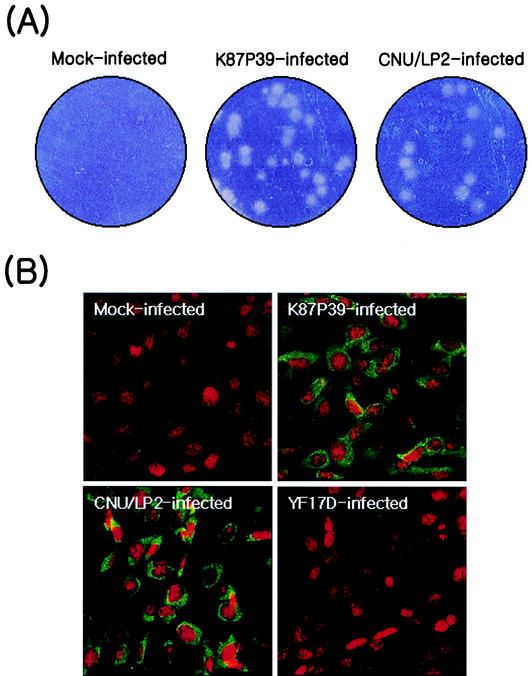
JEV strain K87P39, which was isolated from a wild mosquito in Korea and underwent five passages in suckling mouse brains, was obtained from the Korean National Institute of Health (8). The viral plaque sizes of susceptible BHK-21 cells infected with K87P39 and grown as a monolayer varied, indicating that the viral population is heterogeneous (Fig. 1A, K87P39-infected). Consequently, we employed the plaque purification assay with BHK-21 cells to isolate homogeneous populations of three large-plaque-forming variants that we named CNU/LP1, -2, and -3 (data not shown). Of these three variants, CNU/LP2 consistently maintained its large-plaque phenotype on BHK-21 cells (Fig. 1A, CNU/LP2-infected) or Vero cells (data not shown).
FIG. 1.
Comparison of large-plaque-forming JEV isolate CNU/LP2 and original K87P39 strain. (A) Plaque morphology. BHK-21 cells were mock infected or infected with the original JEV K87P39 strain, which formed a heterogeneous mixture of viral plaque sizes. The CNU/LP2 isolate purified in this study formed a homogeneous population of large plaques (CNU/LP2-infected). (B) Levels and patterns of JEV protein expression. Naïve BHK-21 cells were mock infected or infected with K87P39, CNU/LP2, or the yellow fever virus strain YF17D; 18 h later they were fixed and stained with JEV-specific mouse hyperimmune ascites followed by fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (green fluorescence) and confocal microscopy. Nuclei were visualized by staining with propidium iodide (red fluorescence) in the presence of RNase A.
Confocal microscopy with anti-JEV hyperimmune ascites revealed that CNU/LP2-infected BHK-21 cells expressed JEV viral proteins around the perinuclear membranes (Fig. 1B, CNU/LP2-infected), similar to K87P39-infected cells (Fig. 1B, K87P39-infected). This fluorescence staining was not observed in mock-infected BHK-21 cells (Fig. 1B, mock-infected). As a negative control, BHK-21 cells infected with yellow fever virus 17D, a flavivirus closely related to JEV, did not stain with anti-JEV hyperimmune ascites, as expected (Fig. 1B, YF17D-infected). CNU/LP2 infection of a variety of animal cell lines, including the neuronal SHSY-5Y (human) and B103 (mouse) cell lines and the nonneuronal Vero (monkey) and MDCK (dog) cell lines, resulted in high virus titers (106 to 107 PFU/ml) in the culture supernatants (data not shown). We thus decided to use CNU/LP2 as the parental strain for developing a reverse genetics system for JEV.
Prior to establishing the JEV reverse genetics system, we first had to determine the complete nucleotide sequence of CNU/LP2. Long RT-PCR was used to amplify the entire viral RNA genome apart from the 5′ and 3′ termini (Fig. 2A) and yielded three overlapping cDNA products denoted JVF (nt 1 to 3865), JVM (nt 3266 to 8170), and JVR (nt 7565 to 10893) (about 3.9, 4.9, and 3.3 kb in length, respectively). To ensure faithful cDNA synthesis and amplification, we used the RNase H-negative RT together with the low-error-rate Pfu DNA polymerase. To avoid the selection bias that can occur due to cloning, the uncloned materials of the amplified products were directly sequenced in both directions in all cases. Sequencing analysis with two independently isolated preparations of viral RNA resulted in identical sequences.
The 3′-terminal sequence of CNU/LP2 viral RNA was analyzed after synthetic oligonucleotide T was ligated to it. Oligonucleotide T serves as a specific priming site for cDNA synthesis and PCR amplification (Fig. 2B) and has been used successfully to identify the highly conserved 3′ terminus of the hepatitis C virus RNA genome (17). Thus, synthetic oligonucleotide T that had been modified by adding ddATP at its 3′ end to prevent intramolecular and intermolecular ligation was ligated to the 3′ end of the viral RNA, and RT-PCR was then performed with a negative-sense primer complementary to oligonucleotide T and a positive-sense primer corresponding to a sequence near the 3′ end of the viral genome (nt 10259 to 10276) (Fig. 2B). Agarose gel electrophoresis revealed that the amplified products migrated as two bands, a larger band of approximately 700 bp and a smaller band of about 450 bp (Fig. 2C). Both bands were purified and cloned, and 20 and 10 randomly picked clones containing the larger and the smaller bands, respectively, were sequenced. As has been documented for most of the fully sequenced JEV isolates, we found that all the clones with the larger insert terminated the viral genome with GGA TCT10968. In contrast, all the clones with the smaller insert showed the viral genome truncated at nt 10684, resulting in a band 284 bp shorter. During assembly of the full-length JEV cDNA, we used the nucleotide sequences of the larger insert because the smaller insert did not contain 284 nucleotides at the 3′ end of the viral genome.
The 5′-terminal sequence of CNU/LP2 viral RNA was examined after the cap structure at its 5′ end had been removed by incubation with tobacco acid pyrophosphatase (5). The resulting viral RNA was then self-ligated, and the 3′-5′ junction was subjected to cDNA synthesis and PCR amplification with a positive-sense primer for RT-PCR complementary to a sequence near the viral 3′ end (nt 10259 to nt 10276) and a negative-sense primer corresponding to a sequence near the viral 5′ end (nt 164 to nt 181) (Fig. 2D). Agarose gel electrophoresis revealed the amplified products as a single band of about 850 bp (Fig. 2E). The amplicons were cloned, and 12 randomly picked clones were sequenced. In all 12 clones, the GGA TCT10968 of the viral 3′-terminal sequence (see Fig. 2B and C) was followed by the 5′-terminal sequence1AGA AGT. Identical results were also obtained by direct cycle sequencing of uncloned material.
Thus, we have determined the complete nucleotide sequence of the CNU/LP2 isolate. The nucleotide sequence of CNU/LP2 was identical to that of the consensus primers used with the exception of a substitution, T3851→ C (Table 1, oligonucleotide J6). This sequence information will soon be available in the GenBank database.
Construction of cDNA encoding the JEV RNA genome as a bacterial artificial chromosome.
During our initial attempts to clone the cDNA of the CNU/LP2 RNA genome, it became apparent that a particular region of the viral genome was not compatible with cloning in high-copy-number plasmids in E. coli because the cloned DNA underwent genetic rearrangements (data not shown). Attempts to clone this region into a low-copy-number bacterial plasmid were also unsuccessful due to genetic instability together with a low DNA yield (data not shown). These difficulties have also been reported for other flaviviruses (5, 11, 26, 29, 38, 39). We finally succeeded in cloning this cDNA by employing the bacterial artificial chromosome (BAC) plasmid pBeloBAC11 (22, 36, 46). This vector also later proved itself suitable for housing the full-length JEV cDNA.
Each of the three overlapping amplicons JVF, JVM, and JVR that were originally obtained for sequence analysis was subcloned into the pBeloBAC11 plasmid. The nucleotide sequences of the cloned cDNA in all three subclones were identical to that of the parental virus with the exception of a base substitution (T8906→C) in the NS5 gene contained within JVR. This substitution was translationally silent and must have arisen during the cloning because sequencing of eight randomly picked individual clones revealed a T residue at nt 8906. Although the T8906→C substitution does not alter the corresponding amino acid, it is possible that this change could affect viral replication (43), and thus we corrected this substitution back to a T residue. During their manipulation and propagation in the E. coli strain DH10B, all three subcloned cDNAs remained genetically stable (data not shown).
By sequentially linking the overlapping JEV cDNA fragments at natural restriction sites (see Materials and Methods), we assembled six full-length cDNA clones of CNU/LP2 (Fig. 3). Three were designated pBACSP6/JVFL/XhoI, pBACSP6/ JVFLx/XhoI, and pBACSP6/JVFLx/XbaI (Fig. 3B). These cDNA clones all had the SP6 promoter transcription start at the beginning of the viral genome so that synthetic RNA transcripts with an authentic 5′ end would be generated. To ensure that the 3′ end of the viral genome after runoff transcription would be close to authentic, we placed a unique restriction endonuclease site, either XhoI or XbaI, at the end of the viral genome (Fig. 3B, underlined). Thus, pBACSP6/JVFL/XhoI bears an XhoI site at the end of the viral genome. For the construct with an XbaI site immediately at the end of viral genome, as the viral genome already contains an XbaI site in the NS5 gene, this site had to be destroyed by introducing a silent point mutation (A9134→T). This construct was designated pBACSP6/JVFLx/XbaI, where the x denotes the presence of the silent point mutation that destroyed the original XbaI site. The third clone, pBACSP6/JVFLx/XhoI, contains both the XhoI site at the end of viral genome and the A9134→T substitution. Three other full-length cDNA clones under the control of the T7 RNA promoter were assembled in a similar manner and led to the constructs designated pBACT7/JVFL/XhoI, pBACT7/JVFLx/XhoI, and pBACT7/JVFLx/XbaI (Fig. 3C). At every cloning step during the assembly process, the structural integrity of the cloned cDNAs was assessed by extensive restriction and nucleotide sequence analyses. Structural instability of the inserts leading to deletions or rearrangements was never observed (data not shown).
In vitro generation of highly infectious synthetic JEV RNAs transcribed from BAC constructs.
We first examined the specific infectivity of the synthetic RNAs transcribed from the three SP6-driven constructs. For runoff transcription, the constructs were linearized by digestion with XhoI or XbaI (Fig. 3). SP6 polymerase runoff transcription of the two XhoI-linearized plasmids in the presence of the m7G(5′)ppp(5′)A cap structure analog yielded capped synthetic RNAs containing three nucleotides (CGA) of virus-unrelated sequence at their 3′ ends. This is the result of copying the 5′ overhang left by the XhoI digestion (Fig. 3B). Similarly, SP6 polymerase runoff transcription of the XbaI-linearized pBACSP6/JVFLx/XbaI plasmid produced capped synthetic RNAs with four nucleotides (CTAG) of virus-unrelated sequence at their 3′ ends (Fig. 3B). When susceptible BHK-21 cells were transfected with the synthetic RNAs from these constructs, all were highly infectious (Table 2). That is, the synthetic RNAs obtained from pBACSP6/JVFL/XhoI, pBACSP6/JVFLx/XhoI, and pBACSP6/JVFLx/XbaI transfected under optimal electroporation conditions had specific infectivities of 3.5 × 105, 4.3 × 105, and 3.4 × 105 PFU/μg, respectively (Table 2, infectivity). Similar results were also obtained with synthetic RNAs transcribed from the T7-driven cDNA constructs by T7 polymerase runoff transcription (Table 2, infectivity).
TABLE 2.
Specific infectivity of in vitro RNA transcripts generated from full-length JEV cDNAs and virus titer
| Template used for transcriptiona | Infectivityb (PFU/μg of RNA) | Virus titerc (PFU/ml)
|
|
|---|---|---|---|
| 24 h | 48 h | ||
| pBACSP6/JVFL/XhoI | 3.5 × 105 | 4.4 × 105 | 3.6 × 106 |
| pBACT7/JVFL/XhoI | 2.9 × 105 | 2.0 × 105 | 2.3 × 106 |
| pBACSP6/JVFLx/XhoI | 4.3 × 105 | 2.1 × 105 | 5.2 × 106 |
| pBACT7/JVFLx/XhoI | 3.8 × 105 | 3.3 × 105 | 4.1 × 106 |
| pBACSP6/JVFLx/XbaI | 3.4 × 105 | 3.5 × 105 | 3.2 × 106 |
| pBACT7/JVFLx/XbaI | 3.0 × 105 | 2.4 × 105 | 2.7 × 106 |
| pBACSP6/JVFLx/XbaIMBN | 3.1 × 106 | 6.2 × 106 | 1.4 × 106 |
| pBACT7/JVFLx/XbaIMBN | 2.7 × 106 | 5.6 × 106 | 2.4 × 106 |
All full-length JEV cDNAs were linearized with an appropriate restriction endonuclease (XhoI or XbaI) for runoff transcription as indicated in the names of the cDNAs. For pBACSP6/JVFLx/XbaIMBN and pBACT7/JVFLx/XbaIMBN, these cDNA templates were prepared by linearization with XbaI digestion, which was followed by treatment with MBN.
After in vitro transcription with SP6 or T7 RNA polymerase, as indicated, samples were used to electroporate BHK-21 cells, and infectious plaque centers were determined as described in Materials and Methods.
Virus titers at 24 and 48 h postelectroporation.
It has been reported that for some flaviviruses, the presence of unrelated sequences at the 3′ end of synthetic RNAs transcribed from infectious cDNA diminishes or abrogates their specific infectivity (48). This motivated us to generate synthetic RNAs lacking the unrelated sequences by treating the XbaI-linearized pBACSP6/JVFLx/XbaI plasmid with mung bean nuclease (MBN) prior to the transcription reaction, which removed the four excess nucleotides of CTAG. To verify mung bean nuclease activity, XbaI-linearized and mung bean nuclease-treated pBACSP6/JVFLx/XbaI plasmid was self-ligated, and its viral 3′ end was sequenced, demonstrating removal of the four excess nucleotides of CTAG (data not shown). RNA transcripts from XbaI-linearized and mung bean nuclease-treated pBACSP6/JVFLx/XbaI and pBACT7/JVFLx/XbaI (Fig. 3B, pBACSP6/JVFLx/XbaIMBN, and Fig. 3C, pBACT7/JVFLx/XbaIMBN) both had increased specific infectivities compared to the untreated transcripts (Table 2, infectivity). That is, the infectivity of RNAs transcribed from pBACSP6/JVFLx/XbaIMBN was estimated to be 3.1 × 106 PFU/μg, approximately 10-fold higher than the infectivity (3.4 × 105 PFU/μg) of the unmodified template (Table 2, infectivity). The RNAs derived from pBACT7/JVFLx/XbaI (3.0 × 105 PFU/μg) also had increased infectivity after mung bean nuclease modification (2.7 × 106 PFU/μg) (Table 2, infectivity). Thus, the authentic 3′ end of the JEV genome should be present to ensure highly infectious synthetic JEV RNA transcripts are generated.
The altered specific infectivity of the RNA transcripts due to the presence of three or four virus-unrelated nucleotides at the 3′ end also influences the virus titers harvested from culture supernatants of the transfected BHK-21 cells. Virus titers released from BHK-21 cells transfected with RNA transcripts from mung bean nuclease-untreated pBACSP6/JVFL/XhoI, pBACSP6/JVFLx/XhoI, and pBACSP6/JVFLx/XbaI ranged from 2.1 × 105 to 4.4 × 105 PFU/ml at 24 h posttransfection (Table 2, virus titer 24 h), at which time half of the transfected cells were still attached to culture dishes showing virus-induced strong cytopathic effect. These titers increased about 10-fold to the range of 3.2 × 106 to 5.2 × 106 PFU/ml at 48 h posttransfection (Table 2, virus titer 48 h), at which point most of the cells had died and detached from the bottom of the culture dishes. In contrast, the virus titer released from BHK-21 cells transfected with RNA transcripts from mung bean nuclease-treated pBACSP6/JVFLx/XbaIMBN had already reached 6.2 × 106 PFU/ml at 24 h posttransfection, at which time the majority of the transfected cells had died (Table 2, virus titer 24 h). This titer decreased slightly to 1.4 × 106 PFU/ml at 48 h posttransfection (Table 2, virus titer 48 h). Similar patterns of virus production were seen with the T7 polymerase-driven RNA transcripts (Table 2).
We confirmed that specific infectivity requires the transcription of RNA from the full-length JEV cDNA template by using the full-length cDNA clone pBACSP6/JVFLx/XbaIMBN (Fig. 4). The cDNA template alone was not infectious (Fig. 4A, lane 5, and B, without SP6 Pol), but the intact cDNA template was needed during the transcription reaction because DNase I treatment abolished infectivity (Fig. 4A, lane 2, and B, DNaseI during). Addition of DNase I after the transcription reaction had no effect (Fig. 4A, lane 3, and B, DNaseI after) relative to the intact reaction mixture (Fig. 4A, lane 1, and B, without treatment), but RNase A treatment abolished the infectivity of the transcribed synthetic RNAs (Fig. 4A, lane 4, and B, RNase A after).
FIG. 4.
Full-length JEV cDNA template alone is not infectious but is required for generation of infectious synthetic RNAs during in vitro transcription. pBACSP6/JVFLx/XbaI (100 to 200 ng) linearized with XbaI and treated with mung bean nuclease was used for SP6 polymerase transcription in the absence (A, lane 1; B, without treatment) or presence (A, lane 2; B, DNase I during) of DNase I. After synthesis, the transcription reaction mix was treated for 30 min at 37°C with DNase I (A, lane 3; B, Dnase I after) or RNase A (A, lane 4; B, RNase A after). As a control, the reaction was carried out in the absence of SP6 RNA polymerase (A, lane 5; B, without SP6 Pol). (A) Following treatment, 5% of the reaction mixture was separated on a 0.6% agarose gel, and the cDNA template and RNA transcripts were visualized by staining with ethidium bromide. (B) The reaction mixtures were used to transfect BHK-21 cells, and infectious centers of plaques were estimated.
Synthetic JEVs recovered from full-length infectious cDNAs are indistinguishable from the CNU/LP2 parental virus.
We compared the synthetic JEVs recovered from full-length infectious cDNAs with the parental virus CNU/LP2 originally used for cDNA construction. As shown in Fig. 5A, BHK-21 cells infected with synthetic viruses recovered from pBACSP6/JVFL/XhoI (dish 1), pBACSP6/JVFLx/XhoI (dish 2), pBACSP6/ JVFLx/XbaI (dish 3), and pBACSP6/JVFLx/XbaIMBN (dish 4) formed homogeneous large plaques, similar to the cells infected with CNU/LP2 (dish 5). The growth properties were also similar. Here, BHK-21 cells were infected with low (0.01 PFU/cell), medium (1 PFU/cell), and high (10 PFU/cell) multiplicities of infection, after which the cell culture fluids were harvested periodically and used to determine the kinetics of infectious virus release over time. As shown in Fig. 5B, the multiplicity of infection-dependent virus titers accumulating over time were similar for the four recovered viruses and the parental virus.
FIG. 5.
Comparison of synthetic JEVs with parental virus CNU/LP2. (A) Representative plaque assays of synthetic JEVs and parent CNU/LP2. BHK-21 cells were infected with parent or synthetic viruses, overlaid with agarose, and stained 3 days later with crystal violet. (B) Growth kinetics in BHK-21 cells of synthetic JEVs and parent CNU/LP2 infected at multiplicities of infection of 0.01, 1, and 10. Viruses were harvested at the hour postinfection (h.p.i.) indicated, and titers were determined by plaque assays. The data shown represent one of two independent experiments yielding similar results. (C and D) Viral protein and RNA levels were analyzed by immunoblotting (C) and Northern blotting (D), respectively. BHK-21 cells were infected at a multiplicity of infection of 1 with synthetic JEVs (lanes 1 to 4) or CNU/LP2 (lane 5) or mock-infected (lane 6) and cultured for 18 h. (C) Protein extracts were prepared from approximately 3 × 104 cells and separated on 10% SDS-polyacrylamide gels. Viral proteins were visualized by immunoblotting with JEV-specific mouse hyperimmune ascites (top panel). In parallel, actin protein was detected as a loading and transfer control (bottom panel). The positions of viral protein-related cleavage intermediates and actin are indicated with arrowheads on the left. Molecular mass markers (in kilodaltons) are indicated on the right. (D) Total RNA from approximately 105 cells was extracted and analyzed by Northern blotting with a 32P-labeled antisense riboprobe hybridizing to the sequence in the NS5 gene encompassing nt 9143 to 9351 (top panel). Ethidium bromide-stained 18S rRNA bands are shown as a loading control (bottom panel). The positions of full-length genomic viral RNA (11 kb) and 18S rRNA are indicated on the left.
The viral protein and RNA levels in BHK-21 cells infected with the four recovered viruses and the parental virus were also assessed. Anti-JEV hyperimmune ascites used in immunoblotting revealed that the synthetic and parental viruses produced similar amounts and identical patterns of virus-specific proteins (Fig. 5C, top panel). Actin protein was measured as an internal sample loading control and revealed equivalent levels of actin protein in mock-infected and infected cells (Fig. 5C, bottom panel). Viral RNA levels were also similar, as determined by Northern blotting (Fig. 5D). Quantification of these blots by image analysis revealed that the ratios of viral genomic RNA (Fig. 5D, top panel) to 18S rRNA (Fig. 5D, bottom panel) did not differ significantly, demonstrating that all viral genomic RNAs were produced at similar levels.
Thus, all the synthetic viruses were indistinguishable from the parental virus in terms of plaque morphology, cytopathogenicity, growth kinetics, protein expression, and RNA production. Furthermore, analyses of the 3′ end sequence did not reveal an extra three (CGA) or four (CTAG) nucleotides of virus-unrelated sequence at the 3′ end of the viral RNA genomes derived from any of the synthetic viruses (data not shown). These results validate the use of infectious JEV cDNA clones developed in this study for future molecular genetics.
Genetic marker introduced into JEV cDNA observed in the genome of recovered virus.
While the above results strongly suggest that the JEV cDNA clones can produce highly infectious RNA transcripts after SP6 or T7 polymerase transcription, the possibility that the transfected cultures were contaminated with the parental virus CNU/LP2 was not formally excluded. To assess this remote possibility, we used PCR-based site-directed mutagenesis to introduce a genetic marker (gm) into the pBACSP6/JVFLx/XbaI construct. This marker is a silent substitution (A8171→C) that results in the gain of a unique XhoI site in the NS5 gene (Fig. 6A). The mutagenized construct was designated pBACSP6/JVFLx/gm/XbaI. BHK-21 cells transfected with RNA transcripts from XbaI-linearized mung bean nuclease-treated pBACSP6/JVFLx/gm/XbaIMBN produced infectious virus containing the genetic marker (denoted JVFLx/gm/XbaIMBN) (Fig. 6A). The phenotypic characteristics of JVFLx/gm/XbaIMBN did not differ from those of the original virus JVFLx/XbaIMBN, indicating that the A8171→C substitution did not affect viral replication (data not shown).
To verify that the JVFLx/gm/XbaIMBN virus had been recovered from the cDNA template of pBACSP6/JVFLx/gm/XbaIMBN, we serially passaged the recovered virus in BHK-21 cells at an multiplicity of infection of 0.1. The viruses resulting from each passage were incubated with RNase A and DNase I to avoid the carryover of the input transcript RNA and template plasmid cDNA (21). Viral RNAs extracted from the JVFLx/gm/XbaIMBN and JVFLx/XbaIMBN viruses released at passages 1 and 3 were used in RT-PCR to amplify a 2,580-bp product that encompassed the A8171→C substitution (Fig. 6B, lanes 1, 3, and 5). Digestion of the amplified product from JVFLx/gm/XbaIMBN with XhoI resulted in two fragments of 1,506 and 1,074 bp (Fig. 6B, lanes 2 and 4). The JVFLx/XbaIMBN-derived RT-PCR product did not digest with XhoI (Fig. 6B, compared lane 5 with lane 6), demonstrating that the A8171→C substitution was indeed present in the JVFLx/gm/XbaIMBN virus. Thus, the recovered virus JVFLx/gm/XbaIMBN originated from the full-length infectious cDNA pBACSP6/JVFLx/gm/XbaIMBN.
Full-length infectious JEV cDNA as a BAC is maintained with a high degree of genetic stability in E. coli.
A previous study has shown that constructs containing full-length JEV cDNA frequently acquired stabilizing nonsense mutations in the regions encoding the structural proteins prM and E (38). Since studies into the molecular genetics of JEV will indispensably require a reliable infectious JEV molecular clone for manipulation, we manipulated pBACSP6/pJVFLx/XbaI in several ways and extensively investigated its genetic structure and functional integrity. We first assessed the clones of E. coli DH10B bacteria transformed with pBACSP6/pJVFLx/XbaI and grown on semisolid medium at 37°C. Small but homogeneous bacterial colonies appeared after 15 to 20 h, and extensive restriction analysis with 10 randomly picked clones showed no evidence of deletions or rearrangements. That is, the restriction endonuclease patterns of all 10 clones were identical when digested with NcoI (nine sites), BglI (nine sites), PstI (seven sites), HindIII (seven sites), SspI (six sites), BglII (six sites), and SacI (six sites) (data not shown). SP6 polymerase transcription with each of these 10 clones as the template consistently yielded synthetic RNAs with high specific infectivities ranging from 2.7 × 106 to 3.1 × 106 PFU/μg (Table 2 and data not shown).
We also assessed the construct stability of two independent pBACSP6/JVFLx/XbaI-containing E. coli clones propagated for 9 days. The cells were grown in liquid medium overnight and subsequently propagated by diluting them 106-fold daily as described previously (2). In our experimental conditions, each passage represented approximately 20 generations, which is consistent with observations made previously (2). The plasmids extracted from the two cultures at passages 0, 3, 6, and 9 were examined by restriction enzyme analysis. The restriction enzyme patterns at passages 3, 6, and 9 did not differ visibly from those at passage 0 (data not shown). Thus, within the resolution of agarose gel electrophoresis analysis, the two infectious BAC clones appeared to be structurally stable. Furthermore, the infectivity of the RNA transcripts made from the two cDNA clones did not differ between passage 0 and passage 9 (Fig. 7), confirming that the infectious JEV cDNA remained functionally stable during serial growth in E. coli.
FIG. 7.
Specific infectivity of synthetic RNAs transcribed from infectious JEV cDNA clones propagated for 180 generations. Two independent clones carrying pBACSP6/JVFLx/XbaI (solid and open circles) were cultivated at 37°C overnight in 2xYT with chloramphenicol. The primary cultures were propagated every day for 9 days by 106-fold dilution and adding fresh broth for overnight growth. Each passage was estimated to be about 20 generations (2). At the indicated passages, the DNA plasmids were purified, linearized by XbaI digestion, treated with mung bean nuclease, and used as templates for runoff transcription with SP6 RNA polymerase. The transcripts were then used to transfect BHK-21 cells to determine specific infectivity.
Infectious JEV cDNA as a vector for foreign gene expression.
As previously described (3), we found that JEV was able to replicate in a wide variety of eukaryotic cells originating from a number of species, including humans, mice, monkeys, swine, dogs, cats, and hamsters (data not shown). This suggests that JEV could be useful as a vector for the expression of heterologous genes in a variety of different cells. To test this, two commonly used reporter genes, the Aequorea victoria green fluorescent protein (GFP) and the Photinus pyralis luciferase (LUC), were inserted at the beginning of the viral 3′NTR of pBACSP6/JVFLx/XbaI as expression cassettes driven by the internal ribosome entry site of encephalomyocarditis virus (Fig. 8A). A deletion of 11 to 25 nucleotides exists at the beginning of the viral 3′NTR in CNP/LP2 and three other fully sequenced JEV strains (14, 25, 47), suggesting that this may be a good site to insert the foreign genes. In both cases, the insertion did not alter the infectivity of the synthetic RNA transcripts, although the level of viral replication was slightly decreased (data not shown).
To examine GFP expression, naive BHK-21 cells were transfected with infectious synthetic RNA transcribed from the pBACSP6/JVFLx/GFP/XbaIMBN template and examined by confocal microscopy. Cells expressing GFP displayed green fluorescence in both the nucleus and the cytoplasm (Fig. 8B, JVFLx/GFP/XbaIMBN) because GFP is small enough (≈30 kDa) to permit diffusion between the nucleus and the cytoplasm. As expected, this fluorescence was not observed in mock-transfected cells (Fig. 8B, mock) or in cells transfected with RNA transcripts from pBACSP6/JVFLx/XbaIMBN (data not shown).
To monitor the induction of luciferase over time in a quantitative manner, we produced not only replication-competent RNA transcripts from pBACSP6/JVFLx/LUC/XbaIMBN but also replication-incompetent RNA transcripts from pBACSP6/JVFLx/LUCREP−/XbaIMBN (Fig. 8A). The pBACSP6/JVFLx/LUCREP−/XbaIMBN template contains an 83-nucleotide deletion (nt 5581 to nt 5663) in the middle of the NS3 gene that prematurely terminates viral translation at nt 5596 (see asterisk in Fig. 8A, pBACSP6/JVFLx/LUCREP−/XbaIMBN). In BHK-21 cells transfected with the replication-competent JVFLx/LUC/XbaIMBN RNA (Fig. 8C, solid circles), the initial luciferase activity 6 h posttransfection was 2.4 × 103 ± 0.2 × 103 relative light units (RLU). This activity was dramatically increased to 5.3 × 104 ± 0.1 × 104 RLU 30 h posttransfection and reached 7.8 × 105 ± 0.6 × 105 RLU 54 h posttransfection, at which point >50% of the cells were dying. In contrast, in BHK-21 cells transfected with the replication-incompetent JVFLx/LUCREP−/XbaIMBN RNA, the initial luciferase activity 6 h posttransfection was 1.9 × 103 ± 0.4 × 103 RLU (Fig. 8C, open circles), like the JVFLx/LUC/XbaIMBN-transfected cells (Fig. 8C, solid circles), but this activity gradually decreased over time to 16 ± 1.2 RLU at 54 h posttransfection, which is at the level of background luminescence of naïve cells (Fig. 8C, open circles). Thus, the level of luciferase activity expressed over time varied depending on the presence or absence of viral replication.
We recovered the recombinant JVFLx/GFP/XbaIMBN and JVFLx/LUC/XbaIMBN viruses and infected a variety of commonly used animal cell lines, including nonneuronal and neuronal cells. The GFP or luciferase gene was expressed in all cell types tested (data not shown). Thus, recombinant JEV RNAs and viruses are useful as vectors for foreign gene expression in eukaryotic cells.
DISCUSSION
Here we report the development of a convenient and reliable reverse genetics system for JEV that can be used to generate synthetic viruses from genetically stable full-length infectious JEV cDNAs. Previous attempts to develop such a system (23, 38, 39, 51), including our own, were thwarted by the instability of the cloned JEV cDNA. The system described in this paper will greatly facilitate the study of the molecular mechanisms of replication, virulence, and pathogenesis employed by the virus. Furthermore, we showed that this system can be used as a JEV RNA vector that will rapidly express heterologous genes in a wide variety of eukaryotic cells.
We were able to overcome the instability of JEV cDNA by employing bacterial artificial chromosomes (BACs), which could even stably accommodate the full-length infectious JEV cDNA. These could be used as templates for runoff transcription that generated highly infectious synthetic RNAs with a specific infectivity of 105 to 106 PFU/μg. The transfected cells also released synthetic virus progeny with a virus titer of 106 to 107 PFU/ml. The recovered viruses were phenotypically indistinguishable from the parental virus. We also used a standard mutagenesis procedure to introduce a point mutation that acted as a genetic marker into the BAC construct. This allowed us to confirm that the recovered viruses were derived from the infectious cDNA. It also indicated that virus mutants can be generated by genetically manipulating the infectious cDNA in E. coli. Thus, our system will facilitate the genetic analysis of the replicative mechanisms of the JEV genome.
It is important to produce full-length infectious JEV cDNA that, in in vitro transcription, would generate RNA transcripts with authentic 5′ and 3′ ends because several studies have shown that both the 5′- and 3′-terminal regions are needed for the initiation of flavivirus RNA replication in vitro (49, 50) and in vivo (16). To achieve this objective, we adapted approaches used previously for other flaviviruses (29, 42). The cap structure in JEV genomic RNA is followed by the dinucleotide AG, an absolutely conserved feature of the flaviviruses (28). The authenticity of the 5′ end was ensured by placing either the SP6 or the T7 promoter transcription start at the beginning of the viral genome. Incorporating the m7G(5′)ppp(5′)A cap structure analog in the SP6 and T7 polymerase-driven transcription reactions (9) then allowed us to synthesize capped RNA transcripts with authentic 5′ ends that were highly infectious upon transfection into susceptible cells. In addition, incorporating the m7G(5′)ppp(5′)G cap structure analog in the SP6 and T7 polymerase-driven transcription reactions (9) places an unrelated extra G nucleotide upstream of the dinucleotide AG. As reported earlier (29), we did find that the extra nucleotide was lost from the genomic RNA of the recovered JEV progeny (data not shown). Furthermore, we did not observe that the infectivity or the replication of synthetic RNAs transcribed from infectious cDNA templates was altered if we added the extra nucleotide (data not shown).
The dinucleotide CU located at the 3′ end of JEV RNA is absolutely conserved among the flaviviruses except for the cell fusing agent (4, 28). This suggests that these nucleotides are important in viral replication and that transcripts from infectious cDNAs must have authentic 3′ ends. We thus designed our reverse genetics system for JEV so that the synthetic RNA would be terminated with the authentic 3′ end. We showed indeed that RNA transcripts with authentic 3′ ends were 10-fold more infectious than transcripts with three or four virus-unrelated nucleotides hanging on their 3′ ends. This is consistent with a previous study showing that unrelated sequences at the 3′ end of synthetic RNAs of the West Nile virus delayed the establishment of a productive infection (48).
A variety of vector systems were used previously in attempts to assemble a full-length JEV cDNA (23, 38, 39, 51), including a cosmid vector in E. coli (51). In all cases, the cloned cDNA became genetically unstable during its propagation in a host. Similarly, our attempts to clone stable full-length JEV CNU/LP2 cDNA in all high-copy-number pUC-derived, medium-copy-number pBR322-derived, and low-copy-number pACYC184-derived vectors tested were unsuccessful. One study attempted to overcome this problem by designing a system in which the template would be generated by in vitro ligation of two overlapping JEV cDNAs (38). This template was then used to synthesize infectious RNA transcripts in vitro. However, the specific infectivity of these transcripts was about 100 PFU/μg, which was too low to make this system useful for molecular and genetic analyses of virus biology (38).
In this report, we were able to overcome the genetic instability of JEV cDNA by cloning it into a BAC plasmid that is maintained at one or two copies in E. coli. The genetic structure and functional integrity of the infectious cDNA plasmid remained stable for at least 180 generations during its propagation in E. coli. It should be noted that the genetic stability of this system is not simply due to use of the BAC plasmid as a cloning vector, as an attempt to clone full-length cDNA of the avian infectious bronchitis virus into the same BAC plasmid was unsuccessful (6). Further studies elucidating the reason for the genetic stability of the full-length JEV cDNA in the BAC plasmid may be useful for the construction of other infectious flavivirus cDNAs.
Alphaviruses, which are also RNA viruses, can replicate in a variety of commonly used animal cells and thus have been successfully exploited as eukaryotic expression vectors in cell culture and in vivo (1, 10, 33). We and others (3) have found that JEV, like the alphaviruses, is also able to replicate in a wide variety of primary and continuous cell cultures from humans, mice, monkeys, cows, pigs, dogs, cats, and hamsters (data not shown). This makes JEV attractive as a eukaryotic expression vector. Supporting this are our studies showing that when two representative reporter genes, GFP and luciferase, were inserted into JEV genomic RNA and placed in BHK-21 cells, they were expressed. Thus, the infectious JEV cDNA can act as an expression vector for the rapid expression of a number of heterologous genes in a wide variety of eukaryotic cells.
In this regard, previous studies have shown that replicons of other flaviviruses are employed for efficient expression of heterologous genes, including GFP and luciferase (15, 24, 35, 44). In addition to a transient gene expression system, development of a noncytopathic JEV RNA replicon vector for generation of stable cell lines continuously expressing heterologous genes is desirable, if possible. When a particular type of cell is to be targeted, however, gene expression systems derived from flaviviruses might be disadvantageous due to their broad host range. The JEV cDNA clones described here would be useful in defining the minimal and optimal viral genetic elements needed to use JEV as an expression vector and to develop a packaging system for RNA vectors.
The reverse genetics system for JEV described here will greatly aid several basic and applied research areas. First, this system will help us to investigate the molecular mechanisms of JEV replication, transcription, and translation as well as to identify the JEV genetic elements involved in neurovirulence and pathogenesis. Second, the JEV RNA vector could be an invaluable genetic tool for heterologous gene expression in a wide variety of eukaryotic cells for many applications in biological research. Third, in connection with the ongoing JEV epidemic in Asia and the recent expansion of the virus to Australia, the ability to generate recombinant JEV by targeted manipulation of the infectious JEV cDNA opens up new approaches to the design of genetically modified anti-JEV vaccines.
Acknowledgments
We are very grateful to Cheong-Sun Chen for excellent technical assistance with the confocal microscopy experiments. We also thank Chan-Hee Lee for plasmid pRSGFP-C1.
This study was supported by grants from the Korea Ministry of Health and Welfare, Republic of Korea (02-PJ1-PG3-20203-0014 and 01-PJ1-PG3-20200-0060).
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Linderbach, B. M. Pragai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA replicons for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engeneering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:55-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa.
- 4.Cammisa-Parks, H., L. A. Cisar, A. Kane, and V. Stollar. 1992. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 189:511-524. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, M. S., and A. G. Pletnev. 2000. Infectious cDNA clones of Langat tick-borne flavivirus that differ from their parent in peripheral neurovirulence. Virology 269:225-237. [DOI] [PubMed] [Google Scholar]
- 6.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, T. J., C. S. Halm, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 8.Chung, Y.-J., J.-H. Nam, S.-J. Ban, and H.-W. Cho. 1996. Antigenic and genetic analysis of Japanese encephalitis virus isolated from Korea. Am. J. Trop. Med. Hyg. 55:91-97. [DOI] [PubMed] [Google Scholar]
- 9.Contreras, R., H. Cheroutre, W. Degrave, and W. Fiers. 1982. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 10:6353-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov, I., T. A. Hoffman, B. M. Pragai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gritsun, T. S., and E. A. Gould. 1995. Infectious transcripts of tick-borne encephalitis virus, generated in days by RT-PCR. Virology 214:611-618. [DOI] [PubMed] [Google Scholar]
- 12.Hanna, J. N., S. A. Ritchie, D. A. Phillips, J. Shield, M. C. Bailey, J. S. Mackenzie, M. Poidinger, B. J. McCall, and P. J. Mills. 1996. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med. J. Aust. 165:256-260. [DOI] [PubMed] [Google Scholar]
- 13.Hanna, J. N., S. A. Ritchie, D. A. Phillips, J. M. Lee, S. L. Hills, A. F. van den Hurk, A. T. Pyke, C. A. Johansen, and J. S. Mackenzie. 1999. Japanese encephalitis in north Queensland, Australia, 1998. Med. J. Aust. 170:533-536. [DOI] [PubMed] [Google Scholar]
- 14.Jan, L. R., K. L. Chen, C. F. Lu, Y. C. Wu, and C. B. Horng. 1996. Complete nucleotide sequence of the genome of Japanese encephalitis virus ling strain: the presence of a 25-nucleotide deletion in the 3′-nontranslated region. Am. J. Trop. Med. Hyg. 55:603-609. [DOI] [PubMed] [Google Scholar]
- 15.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 19.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: The viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa.
- 20.Mackenzie, J. S., M. D. Lindsay, R. J. Coelen, A. K. Broom, R. A. Hall, and D. W. Smith. 1994. Arboviruses causing human disease in the Australian zoogeographic region. Arch. Virol. 136:447-467. [DOI] [PubMed] [Google Scholar]
- 21.Mendez, E., N. Ruggli, M. S. Collett, and C. M. Rice. 1998. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J. Virol. 72:4737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishin, V. P., F. Cominelli, and V. F. Yamshchikov. 2001. A ‘minimal’ approach in design of flavivirus infectious DNA. Virus Res. 81:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molenkamp, R., E. A. Kooi, M. A. Lucassen, S. Greve, J. C. Thijssen, W. J. Spaan, and P. J. Bredenbeek. 2003. Yellow fever virus replicons as an expression system for hepatitis C virus structural proteins. J. Virol. 77:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam, J.-H., S.-L. Chae, S.-Y. Won, E.-J. Kim, K.-S. Yoon, B.-I. Kim, Y.-S. Jeong, and H.-W. Cho. 2001. Short report: genetic heterogeneity of Japanese encephalitis virus assessed via analysis of the full-length genome sequence of a Korean isolate. Am. J. Trop. Med. Hyg. 65:388-392. [DOI] [PubMed] [Google Scholar]
- 26.Polo, S., G. Ketner, R. Levis, and B. Falgout. 1997. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J. Virol. 71:5366-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214:916-919. [DOI] [PubMed] [Google Scholar]
- 28.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-960. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 29.Rice, C. M., A. Grakoui, R. Galler, and T. J. Chambers. 1989. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1:285-296. [PubMed] [Google Scholar]
- 30.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Satyanarayana, T., S. Gowda, V. P. Boyko, M. R. Albiach-Marti, M. Mawassi, J. Navas-Castillo, A. V. Karasev, V. Dolja, M. E. Hilf, D. J. Lewandowski, P. Moreno, M. Bar-Joseph, S. M. Garnsey, and W. O. Dawson. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. USA 96:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlesinger, S. 1993. Alphaviruses-vectors for the expression of heterologous genes. Trends Biotechnol. 11:18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger, S., and T. W. Dubensky. 1999. Alphaviruses vectors for gene expression and vaccines. Curr. Opin. Biotechnol. 10:434-439. [DOI] [PubMed] [Google Scholar]
- 35.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 36.Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak, Y. Tachiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli with an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon, T. 1997. Viral encephalitis in Southeast Asia. Neurol. Infect. Epidemiol. 2:191-199. [Google Scholar]
- 38.Sumiyoshi, H., C. H. Hoke, and D. W. Trent. 1992. Infectious Japanese encephalitis virus RNA can be synthesized from in vitro-ligated cDNA templates. J. Virol. 66:5425-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumiyoshi, H., G. H. Tignor, and R. E. Shope. 1995. Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J. Infect. Dis. 171:1144-1151. [DOI] [PubMed] [Google Scholar]
- 40.Tsai, T. F. 2000. New initiatives for the control of Japanese encephalitis by vaccination: minutes of a W. H. O./CVI meeting, Bangkok, Thailand, 13-15 October 1998. Vaccine 18:1-25. [DOI] [PubMed] [Google Scholar]
- 41.Umenai, T., O. Krzysko, T. A. Bektimirov, and F. Assaad. 1985. Japanese encephalitis: current worldwide status. Bull. W. H. O. 63:625-631. [PMC free article] [PubMed] [Google Scholar]
- 42.van der Werf, S., J. Bradley, E. Wimmer, F. W. Studier, and J. J. Dunn. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dinten, L. C., J. A. den Boon, A. L. Wassenaar, W. J. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varnavski, A. N., and A. A. Khromykh. 1999. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology 255:366-375. [DOI] [PubMed] [Google Scholar]
- 45.Venugopal, K., and E. A. Gould. 1994. Towards a new generation of flavivirus vaccines. Vaccine 12:966-975. [DOI] [PubMed] [Google Scholar]
- 46.Wang, K., C. Boysen, H. Schizuya, M. I. Simon, and L. Hood. 1997. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques 23:992-994. [DOI] [PubMed] [Google Scholar]
- 47.Williams, D. T., L. F. Wang, P. W. Daniels, and J. S. Mackenzie. 2000. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J. Gen. Virol. 81:2471-2480. [DOI] [PubMed] [Google Scholar]
- 48.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 49.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′ end of exogeneous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 50.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, F., Q. Huang, W. Ma, S. Jiang, Y. Fan, and H. Zhang. 2001. Amplification and cloning of the full-length genome of Japanese encephalitis virus by a novel long RT-PCR protocol in a cosmid vector. J. Virol. Methods 96:171-182. [DOI] [PubMed] [Google Scholar]