Abstract
Our laboratory has previously reported isolation of human rhinovirus type 16 (HRV16) mutants which depend on WIN 52035 to grow. A rapid rise of progeny virus infectivity occurred when drug was added late in growth cycles, suggesting that the drug-dependence lesion was at the step of virus assembly (W. Wang et al., J. Virol. 72:1210-1218, 1998). Here, we report that capsid subunits, 5S protomers and 14S pentamers, of a drug-dependent mutant were produced normally in the absence of drug, but mutant 80S empty capsids and 150S provirions were not formed, maturation cleavage of provirions (VP0 → VP2 + VP4) did not occur, and the unassembled mutant capsid subunits were degraded with a half-life of 15 min. Drug was not required by mutant virus for attachment, uncoating, RNA synthesis and protein synthesis, and polyprotein processing except maturation cleavage. The requirement of drug for assembly of mutant pentamers to form provirions and the rapid assembly of preformed subunits (synthesized in the absence of drug) after drug addition suggested that after native pentamers (P5) have been formed they must be converted to an assembly active state (P5∗), possibly by a conformational change induced by the binding of drug. We propose that pocket factor plays the same role in wild-type virus. In addition, we also report the construction and the properties of a full-length cDNA clone of HRV16, pR16.11, which produces in vitro transcripts with infectivity similar to that of virion RNA. This cDNA clone is available at the American Type Culture Collection.
Human rhinoviruses form the largest genus of picornaviruses, composed of 102 serotypes. They cause acute inflammatory diseases in human upper airways, known as the common cold, in billions of people worldwide each year (9). Human rhinovirus type 16 (HRV16) is one of the 91 major group serotypes using intercellular adhesion molecule-1 as a receptor; 10 minor group serotypes use the low-density lipoprotein receptor as a receptor (27, 28). HRV16 has been an important model virus for studying transmission of rhinoviruses, pathogenesis of the common cold and virus-induced asthma, and evaluation of antirhinovirus drugs in human volunteers because it reproducibly induces good symptoms in human subjects (2-4, 8, 15, 16, 24, 31). Currently, HRV16 is also widely used for in vitro studies aiming to define the molecular mechanisms by which rhinovirus infection of respiratory epithelial cells triggers the overwhelming inflammation in airways (6, 7, 13, 25, 26, 29, 30).
As a picornavirus, the HRV16 infection cycle involves attachment, uncoating, RNA synthesis, protein synthesis, polyprotein processing, and capsid assembly, but no biochemical data regarding the infection cycle of HRV16 have been documented in the literature. The complete genome sequence (15) and the virion atomic structure (10, 23) of HRV16 have been determined. Like other picornaviruses, the HRV16 virion is composed of 60 copies each of four polypeptides (VP1, VP2, VP3, and VP4) which are symmetrically organized into protomers (one copy each of VP1, VP2, VP3, and VP4), pentamers (five protomers), and icosahedral shells (12 pentamers).
Assembly of human rhinoviruses has received little attention. There is only a single report on isolation of 14S pentamers in minor group HRV1A-infected cells (18). It is assumed rhinoviruses assemble in a way similar to that of polioviruses, the most-studied picornaviruses. In poliovirus, three types of capsid precursors, 5S protomers, 14S pentamers, and 80S empty capsids, are found in infected cells. Capsid precursors encapsidate viral genomic RNA to form 150S provirions. Provirions undergo maturation cleavage (VP0 → VP2 + VP4) to form infectious mature virions (27, 28). Whether pentamers condense around a viral genomic RNA or a viral genomic RNA is inserted into an 80S empty capsid is still controversial (27, 28).
WIN compounds are synthetic organic molecules which neutralize picornaviruses at the stage of attachment or uncoating by binding to the beta-barrel (drug-binding pocket) of capsid protein VP1 (28, 33). It is believed that the bound drug locks the capsids in a conformation unfavorable for binding receptors or ejecting virion RNA (17, 33). Drug-resistant mutants have been isolated and studied for HRV14 (11, 32) and poliovirus type 3 (PV3) (20, 21). In the case of PV3, some drug-resistant mutants depend on drug to form plaques because they require drug to stabilize their capsids during cell-cell transmission (20).
Pocket factor is a natural, yet uncharacterized, molecule seen by X-ray crystallography in the drug-binding pockets of picornaviruses, including HRV16 (10, 23), HRV1A (12), HRV2 (35), PV1 and PV3 (37), coxsackievirus B3 (22), and bovine enterovirus (34). It appears as a long fatty acid-like molecule in crystallographic structure. Efforts to purify pocket factors have been futile. However, binding of WIN drugs and pocket factors to the same site makes it likely that WIN drugs are analogs of viral pocket molecules involved in assembly or uncoating of these viruses.
WIN 52035-2 blocks infection by inhibiting attachment of HRV16 (23). Drug-resistant mutants were isolated at 2 μg of drug/ml. About 10% of these mutants depended on drug to form plaques on HeLa cell monolayers (36). One of these mutants (V1210A) only formed plaques in drug at >0.5 μg/ml. Single-step growth experiments suggested that drug was required for assembly of an existing pool of capsid precursors, synthesized in the absence of drug, because addition of drug late in the infection cycle led to rapid appearance of progeny virus infectivity with a time lag as short as 5 min (36).
We report here the identification of the drug-dependent step in the life cycle of V1210A by systematically testing the drug dependency of all viral replication steps. In addition, we also report the construction and the properties of an HRV16 cDNA clone which produces highly infectious transcripts.
MATERIALS AND METHODS
Viruses, cells, and drug.
HRV16 stock has been described elsewhere (15). Mutant virus V1210A was produced and propagated as described previously (36). H1-HeLa cells, free of mycoplasma, were grown as described elsewhere (14). WIN 52035 was a gift from G. Diana (36).
Identification and quantification of assembly precursors in HRV16-infected cells.
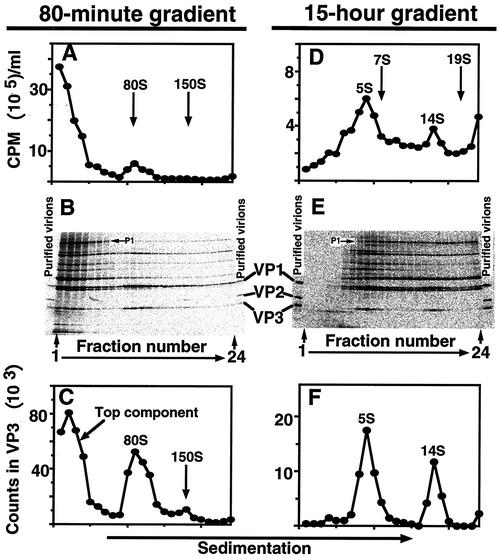
Infection and pulse-chase radiolabeling of assembly precursors were performed as follows. H1-HeLa cells, harvested at the logarithmic phase of growth, were concentrated to 4.0 × 107 per ml of PBSA (phosphate-buffered saline [PBS] with 0.1% bovine serum albumin [BSA]) as described previously (14). The cell suspension was incubated with purified V1210A (15 PFU/cell) at 23°C for 60 min to allow attachment. Infected cells were diluted 10-fold with medium BL (medium B lacking amino acids except glutamine) (14) and shaken gently at 35°C for viral growth. Drug was added to 2 μg/ml at 2 h. [35S]methionine (SJ.1015; Amersham) was added to 20 μCi/ml (0.014 μM) at 4 h (see Fig. 6 for the optimum timing of radiolabeling). Radiolabeling was stopped by addition of excess cold methionine at 4 h 15 min and chased for 15 min. After chases, infected cells were chilled in ice and pelleted in cold. Cell pellet was resuspended in a 1/10 volume of cold extraction buffer-reticulocyte standard buffer (RSB) (10 mM NaCl, 1.5 mM MgCl2, and 10 mM Tris [pH 7.4]) (18) and lysed by addition of 0.5% NP-40. Cell debris was pelleted by centrifugation in cold. Clarified cell lysate (100 μl) was sedimented in a 5-to-20% sucrose density gradient in RSB containing 0.01% BSA in an SW41 rotor at 40,000 rpm and 4°C. To identify 80S empty capsids and 150S virus particles (provirions and virions), the gradients were centrifuged for 80 min. To identify 5S protomers and 14S pentamers, the gradients were centrifuged for 15 h. The markers (80S empty capsids and 150S virions of HRV14, 7S gamma globulins, and 19S thyroglobulins) were sedimented in parallel gradients. Fractions (about 0.5 ml each) were collected from the top of the gradients into a 5-ml tube precoated with BSA. An aliquot of 20 μl from each fraction was analyzed with a 5-to-15% polyacrylamide gradient gel and a PhosphorImager. For Fig. 1A and D, below, trichloroacetic acid (TCA)-insoluble counts in each fraction were determined by the same filter disk method used for determining [3H]uridine-labeled RNA accumulation (see below).
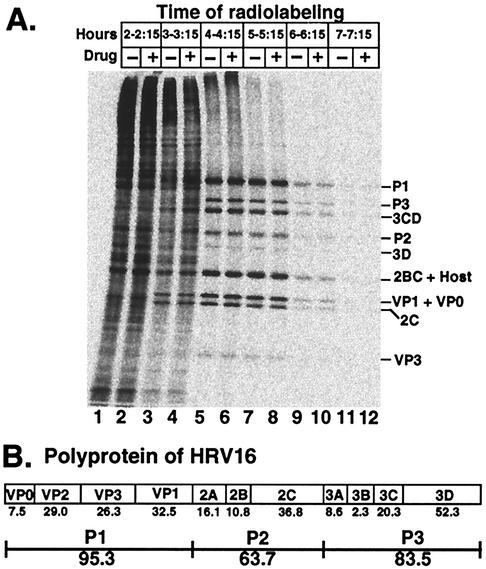
FIG. 6.
Evidence that drug had no effect on protein synthesis of V1210A as measured by 15-min incorporation of [35S]methionine at 1-h intervals through the infection cycle. (A) Electropherogram of infected HeLa cell lysates. Infection was performed as described in the legend for Fig. 1. Drug was added (+ lanes) or not (− lanes) at 2 h postattachment. Infected cells were labeled with [35S]methionine for 15 min at the indicated time. Infection was terminated by chilling cells in ice water. Proteins were released from cells by lysis with NP-40. Cell debris was pelleted. Clarified lysate was electrophoresed in a polyacrylamide gradient gel. Radioactive proteins were visualized with a PhosphorImager. The identity of each viral protein (except VP1 and VP3; see Fig. 7) is tentative, based upon electrophoretic mobility expected from the predicted molecular mass (see panel B). (B) Organization and molecular masses (numbers shown below the names of viral proteins), in kilodaltons, of viral proteins as predicted from the nucleotide sequence of the RNA genome.
FIG. 1.
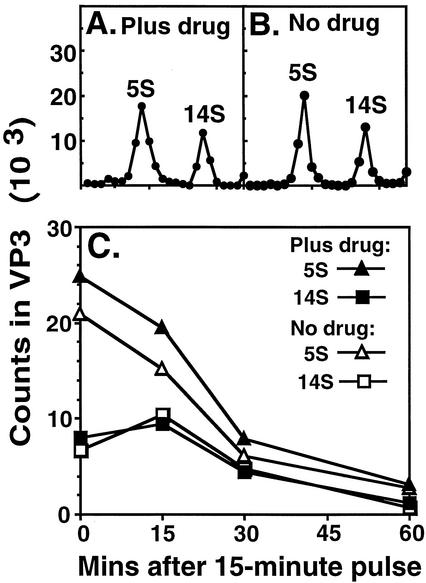
Identification and quantification of HRV16 assembly precursors. HeLa cells were infected with purified V1210A at an MOI of 15 at 23°C for 1 h in the absence of drug. Infected cells were diluted 10-fold with medium BL containing actinomycin D at 35°C. Drug was added at 2 h. [35S]methionine was added at 4 h (see Fig. 6 for the optimum timing of radiolabeling). Radiolabeling was stopped by addition of excess cold methionine at 4 h 15 min and chased for 15 min. After chase, infected cells were chilled in ice, pelleted, resuspended in cold extraction buffer, and lysed with NP-40. Cell debris was pelleted in the cold. Clarified lysate was sedimented in a sucrose density gradient for 80 min (A to C) or 15 h (D to F) at 40,000 rpm and 4°C. Fractions were collected from the top of the gradients. (A and D) TCA-insoluble counts in 10-μl aliquots from each fraction. Arrows identify positions of calibration markers used to assign sedimentation values. The markers (80S empty capsids and 150S virions of HRV14, 7S gamma globulins, and 19S thyroglobulins) were sedimented in parallel gradients. (B and E) Each lane in the electropherogram represents a 20-μl aliquot from the indicated fraction. Radiolabeled purified virions were used as markers in this gel to verify the identities of coat proteins, VP1, VP2, and VP3. (C and F) Radioactivities in VP3 in each lane of the gels in panels B and E, respectively. PhosphorImager counts (24-h exposure) in VP3 were measured by using the ImageQuant program.
Production of purified radiolabeled HRV16 wild-type and V1210A virions.
Infection was performed as above. Drug (2 μg/ml) was added to V1210A-infected cells at 2 h. Virions were harvested at 7 h and purified as described elsewhere (14). The yield of the virions was measured optically, assuming 9.4 × 1012 particles per unit of optical density at 260 nm (28). Wild-type and V1210A viruses had similar yields, typically about 50 μg of virions/108 infected cells. Virions were stored at −70°C.
Attachment and cell-mediated uncoating of HRV16.
H1-HeLa cells were prepared as described above. To attach virus, a cell pellet containing 3 × 107 cells was resuspended in 0.75 ml of PBSA containing 1.2 × 1011 35S-labeled virions at 23°C. Cells were maintained in suspensions at 23°C for 60 min in a carousel. Virus-cell complexes were washed free of unattached virus as described previously (14) and resuspended in 0.75 ml of cold PBSA (total volume, 0.9 ml).
To determine the amount of attached virus, radioactivities in aliquots of all supernatant fluids and cell suspensions were counted in a scintillation counter as described elsewhere (14). The percent attached particles was the fraction of total recovered radioactivity that appeared in the cell suspension.
To measure cell-mediated uncoating, a detergent mixture (1% NP-40, 0.5% deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) was added to the infected cell suspension at 4°C to release viral and subviral particles. Cell debris was removed by centrifugation. Cell lysate (100 μl), supplemented with sedimentation markers ([3H]leucine-labeled 80S empty capsids and 150S virions of HRV14), was layered over a 12.5-ml linear 5-to-20% (wt/vol) sucrose density gradient in PBS containing 0.01% BSA and centrifuged in an SW41 rotor at 40,000 rpm (274,000 × g) at 4°C for 80 min. Each gradient was fractionated, from the top, directly into scintillation vials. The radioactivity in each vial was used to construct the gradient profiles showing the amount of 80S empty shells and 150S virions.
HRV16 RNA synthesis.
Synthesis of viral RNA was measured by incorporation of [3H]uridine into acid-insoluble materials through the life cycle. Infection was performed as above. Infected cells were diluted 10-fold with medium B at 35°C containing actinomycin D (5 μg/ml), which inhibits cellular RNA synthesis. Drug was added to V1210A cultures (2 μg/ml) at 2 h when required. Total RNA accumulation was monitored by continuous labeling of infected cells with [3H]uridine, which was added to 1.5 μM (1 mCi/108 cells) at 1 h postattachment. To determine acid-insoluble counts, triplicates of the cell suspension (10 μl), withdrawn at the indicated times, were spotted onto filter disks and wetted with SDS and Casamino Acids. Filters were air dried and then washed with solutions containing TCA and Casamino Acids to eliminate free label. Filters were washed free of TCA with ethanol, dried, and counted for radioactivity.
HRV16 protein synthesis and polyprotein processing.
Infection and addition of drug were performed as described above. Infected cells, incubated in medium BL at 35°C, were labeled for 15 min every hour by transferring 0.8 ml of suspension into a tube containing 16 μCi of [35S]methionine at 35°C. Labeling was stopped by chilling cells in ice. Proteins were released from cells by lysis with 0.5% NP-40. Cell debris was removed by a 2-min spin in a microcentrifuge. Clarified lysate (12 μl) was electrophoresed in a 5-to-15% polyacrylamide gradient gel. Radioactive proteins were visualized with a PhosphorImager (Molecular Dynamics) after the gel was dried. PhosphorImager counts (24-h exposure) in VP3 were measured with the ImageQuant program.
To examine polyprotein processing, [35S]methionine was added to 0.014 μM (20 μCi/ml) at 4 h. Fifteen minutes later, labeling was stopped by adding a 7,000-fold excess of cold methionine (100 μM). Cell suspensions (0.4 ml), withdrawn at the indicated times, were analyzed with a 5-to-15% polyacrylamide gradient gel and a PhosphorImager as above. Radiolabeled purified virions were used as markers to verify the identities of coat proteins, VP1, VP2, and VP3.
An HRV16 full-length duplex cDNA clone produces highly infectious transcripts.
A full-length cDNA clone of wild-type HRV16 was synthesized and cloned as described previously (14). The full-length cDNA was inserted into plasmid vector pMJ3 immediately after the T7 RNA polymerase promoter, so the T7 transcripts of full-length HRV16 genome has only two extra bases (GG) at the 5′ end. These two extra Gs are the last bases of the T7 promoter and are required for promoter activity. It has been shown two extra 5′ Gs have a minimal adverse effect on the infectivity of HRV14 in vitro transcripts (14). The plasmid carrying the full-length cDNA clone of HRV16, named pR16.11, is available at the American Type Culture Collection (catalog number VRMC-8).
RESULTS
Identification and quantification of assembly precursors in HRV16-infected cells.
Earlier results (36) suggested that the drug-dependent lesion of V1210A was at the stage of assembly. To identify the assembly precursors of HRV16, V1210A growing in the presence of drug was used because its single-step growth profile and progeny virus yield were similar to those of wild-type HRV16 (36). Moreover, it was an ideal parallel positive control for V1210A growing in the absence of drug.
To produce radiolabeled HRV16 assembly precursors, V1210A-infected cells were supplied with drug at 2 h postattachment, pulsed for 15 min with [35S]methionine at 4 h (see Fig. 6 for the optimum timing of radiolabeling), and then chased for 15 min with excess cold methionine. Infected cells were concentrated 10-fold and then lysed with NP-40 to release assembly precursors.
To identify 80S empty capsids, and also 150S provirions and virions, clarified cell lysate was sedimented through a sucrose density gradient in an SW41 rotor at 40,000 rpm for 80 min. Parallel gradients containing sedimentation markers (80S HRV14 empty capsids and 150S HRV14 virions) (14) were used to calibrate the S values. To identify protomers and pentamers, cell lysate was sedimented for 15 h and 7S gamma globulin and 19S thyroglobulin were used as S value markers.
Gradient profiles were first constructed by counting TCA-precipitable radioactivity in an aliquot of each fraction. However, background counts were too high (due to weak viral translation and poor host shutoff) for accurate quantification (Fig. 1A and D). Therefore, aliquots of each fraction were electrophoresed in an SDS-polyacrylamide gel to separate coat proteins from other labeled proteins (Fig. 1B and E). Radiolabeled purified virions were used as markers in this gel to verify the identities of coat proteins, VP1, VP2, and VP3 (see Fig. 6 and 7 for assignments of the other viral proteins). The amount of radioactivity in the VP3 band was used to construct the gradient profiles shown in Fig. 1C and F. Radioactivity in VP3 was used as a measure because VP3 was well separated from the other proteins and it was present in protomers, pentamers, empty capsids, provirions, and mature virions, whereas VP1 comigrated with VP0; VP2 is only found in mature virions and, thus, it is not suitable for quantifying provirions. We will show below the existence of provirions for HRV16.
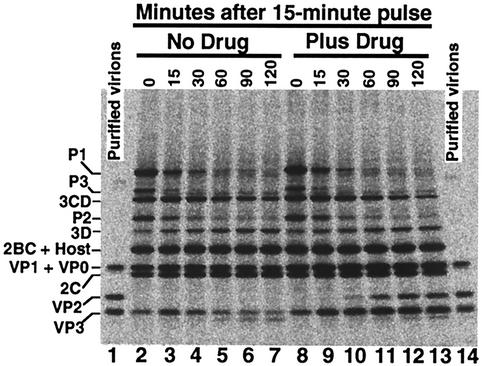
FIG. 7.
Electropherogram of infected HeLa cell lysates showing drug was not required by V1210A for polyprotein processing until maturation cleavage. Infection was performed as described in the legend for Fig. 6. Infected cells were labeled with [35S]methionine at 4 h to 4 h 15 min. Radiolabeling was stopped by addition of excess cold methionine. Cell suspensions, withdrawn at the indicated times, were processed as described in the legend for Fig. 6. Radiolabeled purified virions (lanes 1 and 14) were used as markers in this gel to verify the identities of coat proteins, VP1, VP2, and VP3.
Like PV, HRV16 produced 5S protomers, 14S pentamers and, in the presence of drug, 80S empty capsids and 150S virus particles (provirions and virions). As shown in Fig. 1B and C, the amount of VP3 (also VP1 and VP0) peaked at three locations in the 80-min gradient: the top (top component), 80S (empty capsids), and 150S (provirions and virions) positions. P1, the uncleaved precursor of VP1, VP2, VP3, and VP4 (see Fig. 6B), sedimented at the top of this gradient (Fig. 1B). When the same lysate was sedimented for 15 h, the top component was resolved into two peaks: VP3 (also VP1 and VP0) was found at 5S (protomers) and 14S (pentamers) positions (Fig. 1E and F), whereas P1 spread over the bottom two-thirds of the gradient (Fig. 1E). Therefore, the top component of an 80-min gradient was composed of protomers and pentamers.
Drug was not required for production of pentamers in mutant V1210A but was required for formation of provirions, virions, and empty capsids.
To locate the drug-dependent step in assembly of mutant V1210A, the fate of capsid precursors in the absence of drug was examined by quantifying the amount of P1 proteins (the uncleaved precursor of VP1, VP2, VP3, and VP4), capsid subunits (protomers and pentamers), 80S empty capsids, and 150S virus particles (provirions and virions) in infected cells at time intervals after a 15-min pulse with [35S]methionine. In these experiments, all steps of extraction and analysis were performed at 4°C to avoid possible artifacts due to degradation of unstable virus particles induced by a higher temperature.
To measure the flow of P1 proteins and capsid subunits into 150S virus particles and 80S empty capsids, each cell lysate was analyzed with an 80-min gradient as described in the legend for Fig. 1A to C. Then, three pools of fractions were collected for each gradient: top fractions containing P1 proteins and capsid subunits, 80S fractions containing empty capsids, and 150S fractions containing provirions and virions. The volume of each pool was equalized by addition of extraction buffer. Aliquots were electrophoresed in an SDS-polyacrylamide gel. Radioactivity in VP3 bands and radioactivity of VP3 in P1 bands were used to construct the kinetics curves in Fig. 2C. The radioactivity of VP3 in P1 was calculated according to the methionine content predicted from the nucleotide sequence (15).
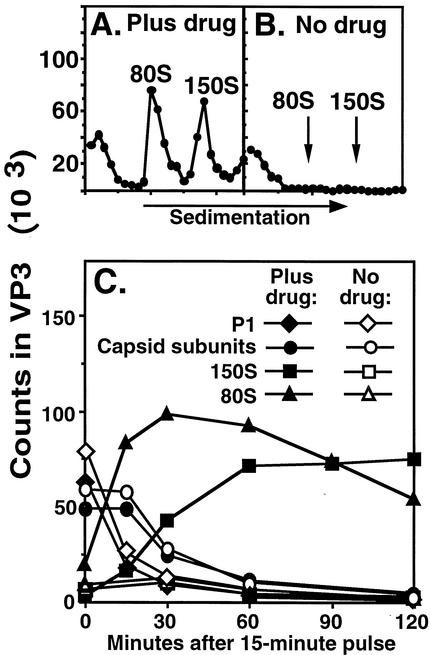
FIG. 2.
Evidence that drug was required for assembly of V1210A virus particles. Infection and pulse-chase labeling were performed as described in the legend for Fig. 1. Infected cell suspensions, withdrawn at the indicated times after 15-min pulse, were analyzed as described for Fig. 1A to C. (A and B) Eighty-minute sucrose gradient profiles. Virus particles were quantified by measuring radioactivity in VP3 in each fraction after sedimenting infected cell lysates in sucrose density gradients and electrophoresis. (C) Kinetics of formation of 150S virus particles and 80S empty capsids. The fractions containing P1 and capsid subunits (top), empty capsids (80S), or virus particles (150S) were pooled. The volume of each pool was measured and then equalized by addition of extraction buffer. Aliquots were electrophoresed in an SDS-polyacrylamide gel. The amount of radioactivity in the VP3 band (measured with a PhosphorImager, as described in the legend for Fig. 1) of each lane was used to construct the kinetics curves of capsid subunits and 80S and 150S virus particles. Capsid subunits, measured by the amount of VP3 sedimented at the top of the gradient, included both protomers and pentamers (see Fig. 3). To have a fair comparison, the amount of P1 proteins, sedimented at the top of the gradients, was expressed as the radioactivity of VP3 in P1, which was calculated according to the predicted methionine content of P1.
Figure 2A and B show the gradient profiles of cell lysates harvested at 60 min after pulse. This pair of profiles shows conclusively that 150S virus particles and 80S empty capsids were only formed in the presence of drug (Fig. 2A) and not in the absence of drug (Fig. 2B).
Figure 2C shows that P1 proteins flowed into capsid subunits and then into 150S virus particles and 80S empty capsids in the presence of drug. In the absence of drug, P1 protein flowed normally into capsid subunits which, however, were unable to assemble into 150S virus particles and 80S empty capsids. It was not known from this result if pentamers were formed in the absence of drug because they were not resolved from protomers and P1 at the top of the 80-min gradients.
That the 150S fraction shown in Fig. 2 contained provirions at the beginning of the chase and mature virions at the end was suggested by the increasing VP2/VP3 ratios in the 150S fractions during the chase (data not shown), since provirions do not have VP2 and mature virions have equal amounts of VP2 and VP3. At 0 min, a small amount of VP3 but no VP2 could be detected at the 150S fractions. At 15 min, the VP2/VP3 ratio was about 1/3. The ratio of VP2 to VP3 increased to about 1 after 60 min, indicating complete maturation cleavage, because VP3 and VP2 have the same number (eight) of methionine residues.
To determine if pentamers were formed in the absence of drug, each cell lysate was analyzed with a 15-h gradient as described for Fig. 1D to F. The radioactivity in VP3 that sedimented at the 5S or 14S position of each gradient, measured as described for Fig. 2C, was used to construct the kinetics curves in Fig. 3C.
FIG. 3.
Evidence that drug was not required for production of V1210A mutant capsid subunits. Aliquots of infected cell lysates from the experiments shown in Fig. 2C were analyzed as described in the legend for Fig. 1D to F. (A and B) Fifteen-hour sucrose gradient profiles. (C) Kinetics of formation and disappearance of 5S protomers and 14S pentamers. The amount of protomers or pentamers in each lysate was quantified as described in the legend for Fig. 2. The radioactivity in VP3 bands in an SDS-polyacrylamide gel was used to construct the kinetics curves.
Figure 3A and B show the gradient profiles of cell lysates harvested at 15 min after pulse. This pair of profiles shows that the drug was not required for the assembly of pentamers. Figure 3C shows that the drug had no apparent effect on the rate of formation and disappearance of protomers and pentamers.
In summary, pentamers were produced normally in the absence of drug; however, they were unable to assemble into provirions and empty capsids. The disappearance of capsid subunits without producing provirions, virions, and empty capsids in the absence of drug (Fig. 2C and 3C) showed that the unassembled subunits were unstable and degraded in infected cells.
Attachment and cell-mediated uncoating of V1210A do not require added drug.
Our laboratory previously reported that the multiplication profile and progeny virus yield of V1210A in a single-step growth cycle was similar to that of wild-type virus when drug was added to V1210A-infected cells after the attachment and uncoating period. Thus, drug was not required for attachment and uncoating of V1210A (36). Here we confirmed this conclusion by direct measurement of the attachment rate and uncoating rate of V1210A virions.
Since infectious virus was produced only in the presence of drug, the radiolabeled V1210A virions used for the attachment and uncoating experiments were first purified extensively (washing of infected cell pellets and sucrose cushion and sucrose density gradient purification of virions) to ensure they were free of unbound drug. It is not known if the purified virions retain drug in their drug-binding pockets. However, even if each of all 60 pockets of a V1210A virion had a drug molecule, the virus samples used for the attachment and uncoating experiments contained only 5 ng of drug/ml, 100-fold lower than the minimum level required for the growth of V1210A.
To measure attachment, purified virions were incubated with HeLa cells in suspension at room temperature (a standard condition for rhinovirus attachment), and the amount of cell-associated virions was determined after 60 min. As shown in Table 1, in the absence of added drug, attachment of V1210A was similar to that of wild-type virus: 88 and 86%, respectively, of virus radioactivity became cell associated in 60 min. Moreover, the presence of drug in the attachment buffer did not enhance but decreased the attachment of V1210A. Therefore, drug was not required for attachment of V1210A. Table 1 also shows that the V1210A mutation enabled HRV16 to overcome the complete attachment block of the drug.
TABLE 1.
Effect of WIN 52035 on attachment of HRV16
Purified 35S-labeled virions were incubated in PBSA containing the indicated amount of drug at 23°C for 1 h. Then, attachment was carried out by incubating the viruses with HeLa cells at 23°C for 60 min (a standard condition for rhinovirus attachment) in the presence of the indicated amount of drug. The virus-cell complexes were separated from free virus by centrifugation, washed free of unattached virus, and resuspended in PBSA. Radioactivities in supernatant fluids and virus-cell complexes (released with NaOH) were counted. The percent attached virions is the fraction of total recovered radioactivity that appeared in the virus-cell complexes.
Represents background nonspecific binding, since drug at 2 μg/ml completely inhibited HRV16 infection and it remained as 1% in the presence of 10 μg of drug/ml (data not shown).
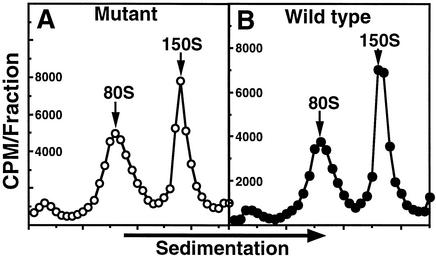
Cell-mediated uncoating (receptor-induced release of VP4 and genomic RNA from attached 150S virions, resulting in formation of 80S empty shells) was measured by the amount of 80S empty shells produced in infected cells during the 1-h attachment period, since HRV16 underwent cell-mediated uncoating at room temperature (W.-M. Lee, unpublished data). Figure 4 shows that similar amounts of 80S empty shells were formed in this period for the mutant (48% of cell-associated radioactivity [Fig. 4A]) and wild-type virus (44% [Fig. 4B]). Thus, cell-mediated uncoating of mutant virus was normal in the absence of added drug.
FIG. 4.
Evidence that uncoating of V1210A mutant virus did not require added drug. Purified 35S-labeled virions were incubated with HeLa cells at 23°C for 60 min. The virus-cell complexes were separated from free virus by centrifugation, washed free of unattached virus, resuspended in PBSA, and lysed with detergents to release virus. Clarified cell lysates were sedimented on sucrose density gradients. Radioactivity in each fraction, collected from the top of the gradients, was used to construct the sedimentation profiles. The sedimentation values of 35S-labeled peaks in each gradient were assigned according to the positions of the 3H-labeled 80S and 150S markers sedimented in the same gradient. (A) Mutant virus; (B) wild-type virus.
The conclusion that added drug was not required for attachment and uncoating of V1210A was further supported by its plating efficiency, which was similar to that of wild-type virus (150 to 200 particles/PFU) when the mutant was attached to the HeLa cell monolayer in the absence of added drug and subsequently grown in the presence of 2 μg of WIN 52035/ml.
The drug had no effect on RNA synthesis of V1210A.
Our laboratory previously reported that drug was not required for the accumulation of infectious V1210A RNA in a single-step growth experiment, leading to the conclusion that the V1210A mutation did not render viral RNA synthesis and integrity (36). However, this result depended on transfection assays, which registered only 1 PFU per 60,000 RNA molecules (see below). Therefore, we reexamined V1210A RNA synthesis in its life cycle by an alternative method, the incorporation of [3H]uridine into acid-insoluble materials. To ensure that incorporation reflected only virus replication, actinomycin D was included in the medium to inhibit cellular RNA synthesis. Actinomycin D (5 μg/ml) had no adverse effect on HRV16 replication (data not shown).
To measure total viral RNA accumulation, insoluble counts were determined at 1-h intervals after [3H]uridine was added at 1 h postattachment. As shown in Fig. 5, actinomycin D completely inhibited cellular RNA synthesis. Drug had no notable effect on the kinetics of accumulation of total 3H-labeled mutant viral RNA. Total RNA accumulation peaked at 7 h postattachment, independent of drug. This result was similar to that of infectious mutant viral RNA accumulation.
FIG. 5.
Evidence that drug had no effect on RNA synthesis of V1210A as measured by incorporation of [3H]uridine into acid-insoluble materials in a single-step growth cycle. Infection was performed as described in the legend for Fig. 1, except complete medium B was used here. Viral RNA accumulation was monitored by continuous labeling of infected cells with [3H]uridine, which was added at 1 h postattachment.
The drug had no effect on protein synthesis of V1210A.
Synthesis of viral proteins was monitored through the life cycle by 15-min incorporation of [35S]methionine at 1-h intervals. Infection was performed as described for Fig. 5. Infected cells were lysed with NP-40. Cell lysates were electrophoresed in an SDS-polyacrylamide gel to separate proteins. The assignments of most viral proteins (including intermediates of polyprotein cleavage, P1, P3, 3CD, P2, 2BC, and VP0, and final products 3D and 2C) on the electropherogram were based on their expected mobilities in a gel according to their predicted molecular weights (Fig. 6B). The identities of VP1 and VP3 were further verified with purified capsid protein makers (Fig. 1B and E and 7). The magnitude of protein synthesis was estimated by the intensity of viral protein bands.
The electropherogram in Fig. 6A shows that drug had no effect on viral protein synthesis through the infection cycle. In the absence or presence of drug, at 2 to 2.25 h (lanes 1 and 2) only cellular proteins were detected. Viral proteins became detectable at 3 to 3.25 h (lanes 3 and 4), while synthesis of cellular proteins declined. At 4 to 4.25 h (lanes 5 and 6), synthesis of viral proteins reached a maximum and host protein synthesis was essentially shut off. Thus, the proteins detected were exclusively viral in origin, in contrast to those synthesized at 2 to 2.25 h. Viral protein synthesis declined drastically at 6 to 6.25 h (lanes 9 and 10) and was almost undetectable at 7 to 7.25 h (lanes 11 and 12). Thus, drug was not required for viral protein synthesis and host protein synthesis shutoff.
In the absence of drug, polyproteins of V1210A were processed normally until maturation cleavage.
Figure 6A also shows that the viral proteins that accumulated in the 15-min labeling period were mainly intermediates of polyprotein cleavage (P1, P3, 3CD, P2, 2BC, and VP0) and a small amount of final products (3D, VP1, 2C, and VP3). Since no protein larger than P1 was detected, drug was not required for the cleavage of full-length polyprotein into P1, P2, and P3.
The cleavage of P1, P2, and P3 into final products was examined with a pulse-chase labeling experiment. Infected cells were labeled with [35S]methionine at 4 to 4.25 h. Labeling was stopped by addition of an excess of cold methionine. Aliquots of the cell suspension were removed at time intervals during the chase for protein analysis as described for Fig. 6. Purified 35S-labeled virions were included in the polyacrylamide gel to verify the identities of VP1, VP2, and VP3.
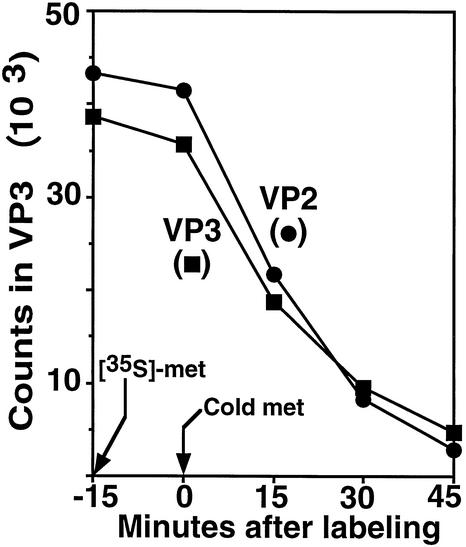
The flow of P1, P2, and P3 into final products in the absence (lanes 2 to 7) and in the presence (lanes 8 to 13) of drug is shown in the electropherogram of Fig. 7. Drug had no apparent effect on the initial amount of P1, P2, and P3 at time zero (consistent with the results shown in Fig. 6) and their rate of disappearance during chase. For the detectable final products, drug had no effect on the amount of 3D and 2C; however, drug was required for the production coat protein VP2 and accumulation of VP3.
As shown in Fig. 7, no trace amount of VP2 was evident in the absence of drug, even at 2 h after radiolabeled capsid subunits were made (lanes 2 to 7). In the presence of drug, significant amounts of VP2 were present at 30 min (lane 10) after the pulse. Figure 7 also shows that the amount of VP3 peaked at 15 min and then diminished to a low level over time in the absence of drug, but in the presence of drug VP3 accumulated to a plateau about twofold of the maximum level of VP3 that accumulated in the absence of drug. Moreover, absence of drug also decreased the accumulation of VP1 and VP0. Since they comigrated in this gel, it was not clear if one or both were degraded.
The result that drug was required for the last step of polyprotein cleavage (maturation cleavage) shows that provirions were not formed in the absence of drug. The degradation of VP3 shows that coat proteins did not assemble into more stable virus particles. In conclusion, failure of mutant virion to appear in the absence of drug was not due to virion instability; this reinforces the conclusion that V1210A was defective in assembling mutant pentamers into provirions.
The half-life of V1210A capsid subunits was about 15 min in the absence of drug.
As shown in Fig. 3, mutant capsid subunits were degraded over time in the absence of drug. To measure the half-life of capsid subunits, drug was added to infected cells at 15-min intervals after a 15-min labeling period started at 4 h. Infected cells were harvested 1 h later to allow the complete flow of radiolabeled subunits into mature virions in the presence of drug. The amount of radiolabeled virions was measured by VP3 and VP2 radioactivity sedimented at 150S position in sucrose density gradients, as described above for Fig. 2C.
Figure 8 shows that the yield of radiolabeled virions was unaffected when the drug was added before the termination of labeling. Preformed subunits, made in the absence of drug, were able to assemble into virions once the drug was added. However, the production of radiolabeled virion decreased as addition of drug was delayed during chase. Since virions are assembled from functional subunits in the infected cells, the courses in Fig. 8 are equivalent to the decay curves of subunits. From these curves, the half-life of subunits was estimated as 15 min. Thus, capsid subunits (protomers and pentamers) were short-lived, supporting the above conclusion that capsid proteins were unassembled in the absence of drug. Furthermore, Fig. 8 also suggests the maturation cleavage was completed in HRV16 virions within 1 h, since the VP2 and VP3 curve almost overlaps and VP3 and VP2 have the same number (eight) of methionine residues, as predicted by nucleotide sequence.
FIG. 8.
Determination of the half-life of capsid subunits. Infection and pulse-chase labeling were performed as described in the legend for Fig. 7. Cell suspensions, withdrawn at the indicated times, were supplemented with drug to 2 μg/ml and then incubated at 35°C for 1 h to allow formation and maturation of virions. The amount of virions in each cell suspension was measured by radioactivity in VP3 and also VP2 in each pool of gradient fractions as described in the legend for Fig. 2C. Since VP3 and VP2 each have eight methionine residues, as predicted by the nucleotide sequence, the fact that the VP2 and VP3 curves almost overlap suggests that the maturation cleavage was completed in the virions within 1 h. The decay of wild-type capsid subunits was not measurable because they assembled rapidly into stable virus particles.
An overview of the biochemical characterization of the HRV16 infection cycle.
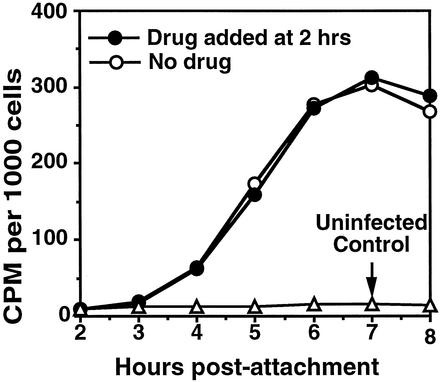
In vitro studies using HRV16 are currently being carried out in a number of laboratories to identify the viral infection step and viral protein responsible for activating the expression of proinflammatory genes in respiratory epithelial cells (6, 13, 25, 26, 29, 30). A comprehensive summary of the above biochemical results with respect to the infection cycle of HRV16 could be helpful for these studies. When HeLa cells were infected with a multiplicity of infection (MOI) of 15 at 23°C, about 90% of input virus attached to the cells (Table 1). After attachment, HRV16 started uncoating at 23°C (Fig. 4), and the eclipse of virus infectivity continued for another hour after the infected cells were warmed to 35°C for virus growth (Fig. 9). At 2 h postattachment, progeny virus infectivity started to appear and then grew exponentially until about 5 h postattachment (Fig. 9). The peak period of infectivity growth (3 to 6 h postattachment) matched that of viral RNA synthesis (Fig. 5, 3 to 6 h postattachment) and viral protein synthesis (Fig. 6, 3 to 6 h postattachment). RNA and protein synthesis and so infectivity production declined rapidly after 6 h.
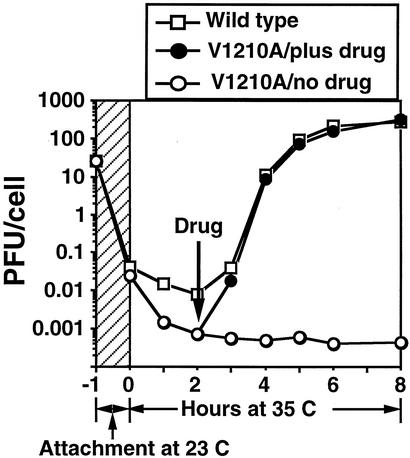
FIG. 9.
Single-step growth curves of HRV16 in HeLa cell suspension, showing that the multiplication profile and progeny virus yield of V1210A were similar to that of wild-type virus when drug was added to V1210A-infected cells after the attachment and uncoating period. The 1-h virus attachment step (hatched zone) was carried out at 23°C in PBSA in the absence of added drug (6 × 107 cells in 1.5 ml of PBSA, with an MOI of 15). Zero time marks the transfer of washed virus-cell complexes to growth medium (15 ml) at 35°C. One of the V1210A-infected cell cultures was supplied with drug to 2 μg/ml at 2 h. Infectivity (PFU per cell) was determined by plaque assay on HeLa cell monolayers after freeze-thawing and clarifying samples (0.4 ml) withdrawn at indicated times. The curves show that wild-type virus started to eclipse during attachment at room temperature and continued at 35°C until 2 h postattachment. Progeny virus infectivity started to appear after 2 h, grew rapidly until 5 h, and then leveled off at 6 h. The trend of eclipse of V1210A was similar to that of the wild type but with a deeper dip at 35°C, probably because V1210A was less stable in the absence of drug. Mutant progeny virus was produced only in the presence of drug. Its yield was similar to that of the wild type when drug was added at the end of the eclipse period (2 h postattachment).
A full-length cDNA clone of HRV16, pR16.11, produces highly in vitro transcripts.
The construction of pR16.11 is described in Materials and Methods. Sequencing analysis confirmed that the first base of the HRV16 genome was ligated to the last base of the T7 promoter and also identified a poly(A) track of 52 As at the 3′ end of the HRV16 genome. The full-length T7 transcripts of HRV16 generated from pR16.11 had a specific infectivity of about 5 × 106 PFU per μg, which was similar to that of the virion RNA (data not shown). The complete 7,124 bases of the HRV16 genome in pR16.11 were reexamined by automatic sequencing, and 18 bases were found to be different from the sequence of HRV16 in GenBank (accession number L24917) (15). The differences are 715C (in pR16.11, HRV16 genome base number 715 is a C), 2266G, 2267G, 2393G, 2419G, 2620G, 2795A, 3324A, 3640C, 3652C, 4678A, 4878G, 4914A, 5341G, 6150G, 6394T, 6457A, and 6511A. Our finding has been independently confirmed by Dan Tenney, Bristol-Myers Squibb, and Ann Funkhouser, Department of Pediatrics, the University of Chicago (personal communication). Plasmid pR16.11 was used for reconstructing mutant V1210A virus by site-directed mutagenesis, and so confirming the single V1210A mutation was sufficient to confer the drug dependency of HRV16 (36). This plasmid is now available at American Type Culture Collection (catalog number VRMC-8).
DISCUSSION
We report here identification of the drug-dependent step in the life cycle of HRV16 mutant V1210A by quantitative biochemical analysis of the effect of WIN 52035 on all viral replication steps. The assembly of mutant pentamers to form empty capsids and provirions and maturation cleavage of provirions (VP0 → VP2 + VP4) were the only steps found to be drug dependent (Fig. 2, 3, and 7). Drug was not required by the mutant virus for attachment although it retained a small inhibitory effect at this step (Table 1). Drug was not required for other steps, including uncoating (Fig. 4), RNA synthesis (Fig. 5), protein synthesis (Fig. 6), polyprotein processing until maturation cleavage (Fig. 7), and capsid subunit synthesis (Fig. 3). Mutant pentamers were produced normally in the absence of drug (Fig. 3), but they were unable to assemble into provirions until the drug was added (Fig. 2 and 8). The unassembled pentamers were degraded with a half-life of 15 min in the absence of drug (Fig. 3 and 8).
It has been shown that the drug-dependent phenotype of PV3 Sabin mutant strains is linked to the requirement of drug for stabilizing the mutant virions (20). But we show conclusively here that the drug-dependent phenotype of HRV16 is due to a defect in assembly of mutant pentamers into provirions and is not an artifact of virion instability, based on the following results. First, no trace amounts of 150S and 80S virus particles were found in the absence of drug (Fig. 2), even when extracts were kept at 4°C throughout the experiment and even when drug was included (2 μg/ml) in the extraction buffer (data not shown). Second, maturation cleavage, which occurs once provirions are made, did not take place in the absence of drug. As clearly shown in Fig. 7 (lanes 2 to 7), no VP2 was produced in the absence of drug, even at 2 h after radiolabeled capsid subunits were made (lane 7). But in the presence of drug, a significant amount of VP2 was produced at 30 min (lane 10) after capsid subunits were produced. Third, in the absence of drug, VP3 was unstable (Fig. 7, lanes 2 to 7 versus 8 to 13), showing that coat proteins did not assemble into stable virus particles. In contrast, when virus particles were formed in the presence of drug, VP3 accumulated to a very high level without any sign of degradation (Fig. 7, lanes 8 to 13). In fact, in the absence of drug, V1210A unassembled capsid proteins had a half-life of 15 min (Fig. 8).
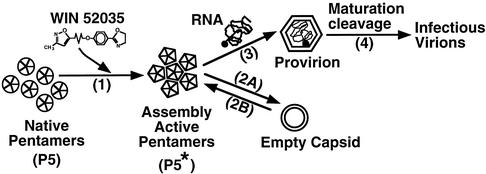
In the absence of drug, not a trace amount of mutant virus particle was formed (Fig. 2) although mutant pentamers were produced normally (Fig. 3). The unassembled pentamers were degraded with a half-life of 15 min in the absence of drug (Fig. 3 and 8). These preformed pentamers, however, were able to assemble rapidly into virus particles once the drug was added (Fig. 8 and reference 36). These results suggested that drug was required for converting the unstable native mutant pentamer (P5) to an assembly-competent state (P5∗), possibly by conformation change induced by the binding of drug. These competent pentamers (P5∗) were able to interact with each other to form icosahedral shells (Fig. 10).
FIG. 10.
A model for explaining how WIN drug was required for assembly of dependent mutant pentamers. In the absence of drug, mutant pentamers were in the unstable native structure (P5) unfavorable for pentamer-pentamer interaction required for building icosahedral shells. The binding of drug to native pentamers induced a conformation change which enabled the pentamer-pentamer interaction required for assembly (step 1). Therefore, these drug-bound pentamers (P5∗) were able to self-polymerize to form empty capsids (step 2A) or also encapsidate viral RNA to form provirions (step 3). The empty capsids may dissociate back to pentamers (step 2B) for producing provirions. The provirions readily undergo maturation cleavage (cleaving VP0 to VP2 and VP4) to become infectious mature virions (step 4). Since capsid-binding drugs are inhibitory analogs of natural pocket factor, natural pocket factor may have the same role in capsid assembly of wild-type virus.
In this report, we show for the first time that the assembly of picornaviral pentamers into provirions can be stopped completely by a single mutation in a coat protein. This block was totally reversed by a synthetic organic compound. This result raises the possibility of synthesizing new drugs for controlling the assembly pathway of mutant capsids, so we may be able to resolve the most elusive problem in picornavirus synthesis: how the RNA becomes packaged into virions (27, 28). Moreover, crystallographic study of V1210A mutant pentamers could provide useful information for understanding the structural requirement for assembling pentamers of picornaviruses.
Isolation of temperature-sensitive mutants defective in assembly has been reported for PV(5, 19) and echovirus 12 (1). These mutants were normal in protein synthesis but did not produce empty capsids and virions at the nonpermissive temperature. However, the mutations and the nature of the defects have not been defined.
Acknowledgments
This work was completed in the laboratory of Roland R. Rueckert at the Institute for Molecular Virology, University of Wisconsin—Madison. We are grateful for his guidance and financial support.
REFERENCES
- 1.Adrian, T., B. Rosenwirth, and H. J. Eggers. 1979. Isolation and characterization of temperature-sensitive mutants of echovirus 12. Virology 99:329-339. [DOI] [PubMed] [Google Scholar]
- 2.Bush, R. K., W. W. Busse, D. Flaherty, D. Warshauer, E. C. Dick, and C. E. Reed. 1978. Effects of experimental rhinovirus 16 infection on airways and leukocyte function in normal subjects. J. Allergy Clin. Immunol. 61:80-87. [DOI] [PubMed] [Google Scholar]
- 3.Calhoun, W. J., E. C. Dick, L. B. Schwartz, and W. W. Busse. 1994. A common cold virus, rhinovirus 16, potentiates airway inflammation after segmental antigen bronchoprovocation in allergic subjects. J. Clin. Investig. 94:2200-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Alessio, D. J., J. A. Peterson, C. R. Dick, and E. C. Dick. 1976. Transmission of experimental rhinovirus colds in volunteer married couples. J. Infect. Dis. 133:28-36. [DOI] [PubMed] [Google Scholar]
- 5.Drescher-Lincoln, C. K., J. R. Putnak, and B. A. Phillips. 1983. Use of temperature-sensitive mutants to study the morphogenesis of poliovirus. Virology 126:301-316. [DOI] [PubMed] [Google Scholar]
- 6.Gern, J. E. 2002. Rhinovirus respiratory infections and asthma. Am. J. Med. 112:19S-27S. [DOI] [PubMed] [Google Scholar]
- 7.Gern, J. E., and W. W. Busse. 1999. Association of rhinovirus infections with asthma. Clin. Microbiol. Rev. 12:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunberg, K., M. C. Timmers, E. P. de Klerk, E. C. Dick, and P. J. Sterk. 1999. Experimental rhinovirus 16 infection causes variable airway obstruction in subjects with atopic asthma. Am. J. Respir. Crit. Care Med. 160:1375-1380. [DOI] [PubMed] [Google Scholar]
- 9.Gwaltney, J. M. 2002. Clinical significance and pathogenesis of viral respiratory infections. Am. J. Med. 112:13S-18S. [DOI] [PubMed] [Google Scholar]
- 10.Hadfield, A. T., W. M. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 11.Heinz, B. A., R. R. Rueckert, D. A. Shepard, F. J. Dutko, M. A. McKinlay, M. Fancher, M. G. Rossmann, J. Badger, and T. J. Smith. 1989. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 63:2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, F. J. Dutko, M. A. McKinlay, and D. C. Pevear. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-227. [DOI] [PubMed] [Google Scholar]
- 13.Konno, S., K. A. Grindle, W. M. Lee, M. K. Schroth, A. G. Mosser, R. A. Brockman-Schneider, W. W. Busse, and J. E. Gern. 2002. Interferon-gamma enhances rhinovirus-induced RANTES secretion by airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 26:594-601. [DOI] [PubMed] [Google Scholar]
- 14.Lee, W. M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, W. M., W. Wang, and R. R. Rueckert. 1995. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9:177-181. [DOI] [PubMed] [Google Scholar]
- 16.Lemanske, R. F. J., E. C. Dick, C. A. Swenson, R. F. Vrtis, and W. W. Busse. 1989. Rhinovirus upper respiratory infection increases airway hyperreactivity and late asthmatic reactions. J. Clin. Investig. 83:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGregor, S., and R. R. Rueckert. 1977. Picornaviral capsid assembly: similarity of rhinovirus and enterovirus precursor subunits. J. Virol. 21:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikhejeva, A., E. Yakobson, and G. Y. Soloviev. 1970. Characterization of some poliovirus temperature-sensitive mutants and poliovirus-related particle formation under nonpermissive conditions. J. Virol. 6:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosser, A. G., and R. R. Rueckert. 1993. WIN 51711-dependent mutants of poliovirus type 3: evidence that virions decay after release from cells unless drug is present. J. Virol. 67:1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosser, A. G., J. Y. Sgro, and R. R. Rueckert. 1994. Distribution of drug resistance mutations in type 3 poliovirus identifies three regions involved in uncoating functions. J. Virol. 68:8193-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira, M. A., R. Zhao, W. M. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulos, N. G., P. J. Bates, P. G. Bardin, A. Papi, S. H. Leir, D. J. Fraenkel, J. Meyer, P. M. Lackie, G. Sanderson, S. T. Holgate, and S. L. Johnston. 2000. Rhinoviruses infect the lower airways. J. Infect. Dis. 181:1875-1884. [DOI] [PubMed] [Google Scholar]
- 25.Papi, A., and S. L. Johnston. 1999. Rhinovirus infection induces expression of its own receptor intercellular adhesion molecule 1 (ICAM-1) via increased NF-κB-mediated transcription. J. Biol. Chem. 274:9707-9720. [DOI] [PubMed] [Google Scholar]
- 26.Papi, A., L. A. Stanciu, N. G. Papadopoulos, L. M. Teran, S. T. Holgate, and S. L. Johnston. 2000. Rhinovirus infection induces major histocompatibility complex class I and costimulatory molecule upregulation on respiratory epithelial cells. J. Infect. Dis. 181:1780-1784. [DOI] [PubMed] [Google Scholar]
- 27.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, 3rd ed. Raven Press, Ltd., New York, N.Y.
- 29.Sanders, S. P., E. S. Siekierski, S. M. Richards, J. D. Porter, F. Imani, and D. Proud. 2001. Rhinovirus infection induces expression of type 2 nitric oxide synthase in human respiratory epithelial cells in vitro and in vivo. J. Allergy Clin. Immunol. 107:235-243. [DOI] [PubMed] [Google Scholar]
- 30.Schroth, M. K., E. Grimm, P. Frindt, D. M. Galagan, S. I. Konno, R. Love, and J. E. Gern. 1999. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 20:1220-1228. [DOI] [PubMed] [Google Scholar]
- 31.Seymour, M. L., N. Gilby, P. G. Bardin, D. J. Fraenkel, G. Sanderson, J. F. Penrose, S. T. Holgate, S. L. Johnston, and A. P. Sampson. 2002. Rhinovirus infection increases 5-lipoxygenase and cyclooxygenase-2 in bronchial biopsy specimens from nonatopic subjects. J. Infect. Dis. 185:540-544. [DOI] [PubMed] [Google Scholar]
- 32.Shepard, D. A., B. A. Heinz, and R. R. Rueckert. 1993. WIN 52035-]2 inhibits both attachment and eclipse of human rhinovirus 14. J. Virol. 67:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 34.Smyth, M., J. Tate, E. Hoey, C. Lyons, S. Martin, and D. Stuart. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2:224-231. [DOI] [PubMed] [Google Scholar]
- 35.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1179-1194. [DOI] [PubMed] [Google Scholar]
- 36.Wang, W., W. M. Lee, A. G. Mosser, and R. R. Rueckert. 1998. WIN 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J. Virol. 72:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeates, T. O., D. H. Jacobson, A. Martin, C. Wychowski, M. Girard, D. J. Filman, and J. M. Hogle. 1991. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J. 10:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]