Abstract
Adenovirus (Ad) type 5 DNA packaging is initiated in a polar fashion from the left end of the genome. The packaging process is dependent upon the cis-acting packaging domain located between nucleotides 194 and 380. Seven A/T-rich repeats have been identified within this domain that direct packaging. A1, A2, A5, and A6 are the most important repeats functionally and share a bipartite sequence motif. Several lines of evidence suggest that there is a limiting trans-acting factor(s) that plays a role in packaging. Two cellular activities that bind to minimal packaging domains in vitro have been previously identified. These binding activities are P complex, an uncharacterized protein(s), and chicken ovalbumin upstream promoter transcription factor (COUP-TF). In this work, we report that a third cellular protein, octamer-1 protein (Oct-1), binds to minimal packaging domains. In vitro binding analyses and in vivo packaging assays were used to examine the relevance of these DNA binding activities to Ad DNA packaging. The results of these experiments reveal that COUP-TF and Oct-1 binding does not play a functional role in Ad packaging, whereas P-complex binding directly correlates with packaging function. We demonstrate that P complex contains the cellular protein CCAAT displacement protein (CDP) and that full-length CDP is found in purified virus particles. In addition to cellular factors, previous evidence indicates that viral factors play a role in the initiation of viral DNA packaging. We propose that CDP, in conjunction with one or more viral proteins, binds to the packaging sequences of Ad to initiate the encapsidation process.
The assembly of adenovirus (Ad) particles has been studied using a large number of viral temperature-sensitive mutants, blocked at different stages of assembly at the restrictive temperature, and by pulse-chase kinetic analyses (7, 8, 10). The results from these studies suggest that Ad DNA is inserted into preformed empty capsids late in the viral life cycle. Whether the viral genome enters the prohead in association with viral core proteins, or as a separate entity, is unclear. The exact structure of the viral DNA before and during entry also is not known. Ad DNA packaging occurs in a polar fashion from the left end of the genome. Polarity of adenoviral DNA packaging was initially demonstrated by the finding that left end sequences were overrepresented within incomplete viral particles containing DNA of subgenomic length (6, 43). The left ends of Ad type 3 (Ad3), Ad5, and Ad16, representatives of Ad subgroups B and C, harbor conserved packaging signals (15, 19, 33, 36). These conserved cis-acting packaging domains suggest a similar mechanism of selective and polar DNA packaging for all Ad subgroups.
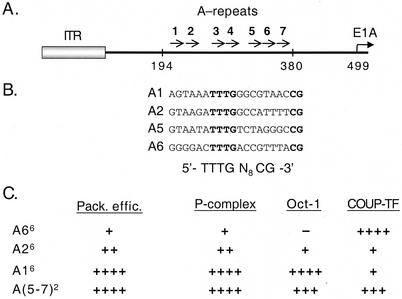
Ad5 DNA encapsidation is dependent on cis-acting sequences located on the left end of the genome between nucleotides (nt) 194 and 380 (15, 16, 22, 39). The left end of the Ad5 genome is depicted in Fig. 1A. Through deletion and linker scanning mutagenesis, seven repeats have been identified within this domain that contribute to viral DNA packaging. These repeats are called A repeats because of their A/T-rich content. Although A repeats are functionally redundant, they follow a hierarchy of importance. A1, A2, A5, and A6 are the most important repeats for packaging activity (15, 16, 39). These repeats contain a bipartite consensus motif shown in Fig. 1B. Both the first and the second half-site of the consensus motif, as well as the 8bp spacing between the half sites, are important in viral DNA packaging (39). Several lines of evidence suggest that a limiting trans-acting factor(s) plays a role in the packaging process in conjunction with the cis-acting sequences (16, 39, 40). First, an isolated packaging domain on a multicopy plasmid represses packaging of a wild type virus in vivo. Second, coinfection experiments in vivo show that viruses with a greater number of packaging repeats package more efficiently than viruses with fewer packaging repeats. These results suggest competition for a limiting packaging factor(s) in vivo. Multimerized individual packaging repeats, termed minimal packaging domains, have been used to study Ad DNA packaging (40). Minimal packaging domains were shown to support packaging in vivo at various degrees when built into viruses lacking the genuine packaging sequences (Fig. 1C). When used in binding assays in vitro as probes, two cellular activities were identified that bind these sites including P complex, a previously uncharacterized protein(s), and chicken ovalbumin upstream promoter transcription factor (COUP-TF) (40).
FIG. 1.
(A) Schematic representation of the left end of the Ad5 genome. Nucleotide coordinates, relative to the left terminus, are indicated and the ITR is represented by a gray box. A repeats 1 to 7 are represented by arrows between nt 194 and 380. (B) The packaging repeat consensus motif. Alignment of A repeats 1, 2, 5, and 6. Nucleotides comprising the bipartite sequence motif shared by these repeats are indicated in bold letters and the consensus motif is shown below in boldface type. (C) Packaging efficiencies and DNA binding affinities of the minimal packaging domains. Packaging efficiencies of the recombinant viruses carrying different minimal domains (percent packaging compared to a wild type-virus) (34) and the binding affinities of the domains when used in in vitro binding assays with P complex or COUP-TF (40) or Oct-1 (this work), are indicated. The numerical superscript refers to the number of copies of the synthetic packaging repeats present in the recombinant viruses. Symbols for in vitro binding assays: +++, competition efficiency equivalent to self-competition of the probe with identical, unlabeled competitor DNA sequences; ++++, increased binding relative to the +++ level; ++, binding reduced 3- to 5-fold relative to this level; +, binding reduced 15- to 25-fold relative to the ++ level; −, no protein binding activity detected.
A direct correlation was reported between the binding affinity of P complex for different minimal packaging domains in vitro and the ability of these domains to support DNA packaging in vivo (40) (Fig. 1C). The TTTG, but not the CG, packaging consensus half-site is critical for P-complex interaction. In addition, P complex also binds to core replication sequences in the inverted terminal repeats (ITR) (40). This dual binding activity might constitute a trans-acting link between viral replication and encapsidation. COUP-TF was determined to be functionally irrelevant for viral DNA packaging since COUP-TF binding in vitro did not correlate with the functionality of minimal packaging domains in vivo (40) (Fig. 1C). Recently, the Ad-encoded protein IVa2 was found to bind the CG-rich sequence in the A-repeat consensus motif (46), and compelling evidence for the role of IVa2 in viral packaging in vivo was described (47). Mutation of the second half-site of A-repeats significantly reduces viral DNA packaging in vivo further implicating a functional involvement of IVa2 in DNA encapsidation (39).
In this paper we report that another cellular protein, octamer-1 protein (Oct-1), binds to minimal packaging sequences. The focus of this study was to examine the functional relevance of Oct-1 and P-complex binding to the viral packaging domain. Our results demonstrate that Oct-1 binding to packaging repeats is not functionally relevant to viral DNA packaging, whereas P-complex binding to the packaging repeats directly correlates with packaging function. We show that P complex contains the cellular protein CCAAT displacement protein (CDP) and that full-length CDP is found in purified virus particles. All the data gathered to date supports the hypothesis that P complex is functionally relevant for viral encapsidation, and that CDP interacts with viral factors to direct the packaging process.
MATERIALS AND METHODS
Virus constructions.
Ad5 dl309 (24) and dl327 (42), the parents for all viruses described in this report, are phenotypically wild type in in vivo assays. Plasmid pE1A-194/814 contains the left-end Ad5 XbaI fragment (nt 1 to 1399) with a deletion between nt 194 and 814 and a unique XhoI restriction site at the junction of the deletion (40). A head-to-tail dimer of Ad3, A5-7 (5′-TCGACGGTGGAGTATTTGCCGAGGGCCGAGTAGACTTTGACCGTTTACGTGGAGGTTTC-3′), was cloned into the pE1A-194/814 background. The recombinant plasmid was subsequently rebuilt into intact virus (Ad5 dl309) by the method of Stow (41). The construction of a virus containing a head-to-tail dimer of Ad5 A5-7 was previously described (40). The sequence of the insert in Ad5 A5-7 is 5′-TCGACCGCGTAATATTTGTCTAGGGCCGCGGGGACTTTGACCGTTTACGTGGAGACTCC-3′. 293 cells were used to amplify and titer recombinant viruses. Recombinant viruses were identified by restriction analysis of viral DNA obtained from infected 293 cells, and all insertions were verified by nucleotide sequence analysis of viral DNA using PCR-based nucleotide sequencing. The CDP cDNA was inserted into pCMV-Tag2 plasmid (Stratagene) in frame with the Flag epitope tag. A hemagglutinin (HA) epitope tag was introduced at the C terminus of Flag-tagged CDP by PCR. The double-tagged CDP cDNA was substituted for the E1 region of an Ad5 transfer vector (pAd-CMV) (11) downstream of the CMV enhancer and then introduced into the dl327 virus, which lacks E3, by homologous recombination within N52.E6 cells. N52.E6 cells were used to amplify the tagged CDP-expressing virus and to determine the titer of virus stocks.
Cultured cells and infections.
Monolayer HeLa and 293 cells were maintained in Dulbecco's modified minimal essential medium (DMEM) containing 10% calf serum (HyClone) and the antibiotics penicillin and streptomycin. N52.E6 cells were maintained in α-modification of Eagle's medium containing 10% fetal bovine serum (HyClone). The N52.E6 cell line is a primary human amniocyte line that is transformed by E1 proteins of Ad5 (38). Virus infections were performed at a multiplicity of infection of 5 PFU per cell for 1 h at 37°C. Cells were then washed twice with Tris-buffered saline solution and overlaid with fresh medium. For the determination of virus yield, infected cell lysates were prepared at 4, 24, and 48 h postinfection, and the infectious virus yield was determined by plaque assays on 293 cells.
Extract preparation and gel mobility shift assays.
Nuclear extracts were prepared by the method of Dignam et al. (9) and dialyzed overnight against a solution containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 10% glycerol, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride (DB-100). The dialyzed lysate was cleared by centrifugation at 25,000 × g for 20 min. Two to five micrograms of nuclear extract was incubated with 0.5 μg of poly(dI-dC) and 20,000 to 50,000 cpm of 32P-labeled probe DNA (5 to 10 fmol of DNA) per in vitro binding reaction. The binding reaction was carried out in a total volume of 10 μl for 1 h at 4°C in 40 mM HEPES (pH 7.5), 70 mM NaCl, 0.1 mM EDTA, 0.5 mM, 10 μg of bovine serum albumin per ml, and 4% Ficoll. The complexes were resolved electrophoretically at 10 V/cm on a 4% 30:1 (acrylamide-bisacrylamide) polyacrylamide gel in 0.5× TBE (25 mM Tris[pH 8.3], 25 mM boric acid, 0.5 mM EDTA) at 4°C. For gel mobility supershift experiments, 0.3 μl of a rabbit polyclonal anti-CDP antiserum (48) or preimmune antiserum was added 0.5 h before or 0.5 h after the addition of probe. Peptide antibodies (0.5 μl) raised against different portions of CDP (α-23, α-403, α-510, α-861, and α-1300 [29; unpublished data) were added to the reaction mixes 0.5 h before or 0.5 after the addition of probe. Anti-Oct-1 antibody (catalog no. 232; Santa Cruz Biotechnology) was added after the addition of probe DNA. 0.5 to 2 μg of glutathione S-transferase (GST)-POU protein was used in binding reactions for gel mobility shift experiments.
Preparation of mobility shift probes and unlabeled competitors.
A head-to-tail dimer of Ad5 A5-7 repeat was cloned into pUC9. This fragment was liberated from the vector by digestion with EcoRI and HindIII and gel purified, and 100 ng of the fragment was 32P end labeled using Klenow DNA polymerase and [α-32P]dATP. Ad3 A(5-7)2, Ad5 A(5-7)2, Ad9 A(5-7)2 (5′-TCGACGGCGGAATATTTACCGAGGGCCGAGAGACTTTGACCGATTACGTGGGGGTTTC-3′), and Ad12 A(5-7)2 (5′-TCGACCGCGGAATATTTACCGAGGGCAGAGTGAACTCTGAGCCTCTACGTGTGGGTTTC-3′) repeats were cloned into pE1A-194/814 plasmids. These fragments were liberated from their vectors by digestion with BspE1 and SgrA1, gel-purified, and 100 ng of these fragments were 32P-labeled using Klenow DNA polymerase and [α-32P]dCTP. The ITR oligonucleotide was annealed to its respective complementary strand (flanked by Xho/Sal linkers) (5′-TCGAGTTGTCATCAATAATGGTCGAGTTGTCATCAATAATGG-3′) and was end labeled in the same way, using [α-32P]dCTP. Aliquots of radiolabeled DNA fragments were subjected to trichloroacetic acid precipitation to measure the specific activity of each probe. Quantification of bound versus unbound probe was performed using a Molecular Dynamics Storm 860 PhosphorImager and ImageQuant software. For the preparation of competitor DNAs containing packaging repeats, monomeric oligonucleotides were multimerized using T4 DNA ligase. Selection of head-to-tail multimers was achieved by subsequent digestion with SalI and XhoI, followed by phenol-chloroform extraction and ethanol precipitation. The sequences of the competitor DNAs were previously described (40).
Protein fractionation.
Uninfected HeLa cell nuclear extract (50 mg of total protein) was subjected to heparin agarose chromatography with a bed volume of 1 ml/10 mg of loaded protein. The nuclear extract was applied in buffer DB-100 and washed with 5 bed volumes of loading buffer. Bound proteins were eluted using a 0.1 to 1.0 M linear NaCl gradient in DB. Binding activities were identified by gel shift analysis using different packaging and ITR sequences as probes. Generally, 2 μl of each heparin agarose column fraction was used for gel mobility shift assays using the same binding conditions as described above.
Western blot analysis.
Western blot analysis was performed using 5 μl (10 to 15 μg protein) of uninfected, dl327-, and dl327-CMV-Flag-CDP-HA-infected N52.E6 nuclear extracts boiled in 2× sodium dodecyl sulfate (SDS) sample buffer, and electrophoresed on an SDS-8% polyacrylamide gel. Proteins were transferred to nitrocellulose and probed with anti-Flag or anti-HA antibodies (diluted 1:2,500 and 1:800, respectively). Proteins were visualized using a secondary horseradish peroxidase-conjugated antibody and chemiluminescence as recommended by the manufacturer (Amersham). dl327 and dl327-CMV-Flag-CDP-HA virus particles (1011 particles) were disrupted by boiling in 100 μl of 2× SDS sample buffer, and 20 μl of each sample was examined by Western blotting as described above.
Virion purification.
Six 100-mm-diameter plates of N52.E6 cells were infected with 5 PFU/cell dl327 virus or dl327-CMV-Flag-CDP-HA virus. Cells were harvested 48 h postinfection, pelleted, and resuspended in Tris-buffered saline. After three freeze-thaw cycles, cellular debris was removed by centrifugation at 2,500 × g for 10 min. The lysates were subjected to one CsCl step gradient and, subsequently, one or two rounds of CsCl equilibrium gradient centrifugation. Banded virions were diluted with distilled H2O and ethanol precipitated. Precipitated virions were resuspended in 2× SDS sample buffer and examined by Western blotting as described above.
Expression and purification of GST-POU protein.
pET-GST-POU plasmid (kindly provided by Winship Herr) was introduced into BL21 DE3 cells. Protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (final concentration of 0.4 mM) to a 500 ml culture at an optical density at 600 nm of 0.7. After overnight incubation, cells were pelleted by centrifugation, resuspended in phosphate-buffered saline containing protease inhibitors (2 mM phenylmethylsulfonyl fluoride, 0.5 mM pepstatin, 2 μg of leupeptin/ml, 0.5 mM benzamidine), and lysed by the addition of lysozyme (final concentration of 100 μg/ml) and NP-40 (final concentration of 0.1%). The extracts were sonicated for 30 s and clarified by centrifugation at 25,000 × g for 15 min. One milliliter of 50% glutathione agarose beads (Sigma) was added to the supernatant fluid and the slurry was incubated overnight at 4°C with gentle mixing. The beads were washed extensively with phosphate-buffered saline and bound protein was eluted using 400 μl of 50 mM Tris (pH 8.8) and 10 mM reduced glutathione. Twenty microliters of 10× buffer (100 mM KCl, 5% glycerol, 0.1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, 0.5 mM pepstain, 2 μg of leupeptin/ml, 0.5 mM benzamidine) was added to the eluate and aliquots of the sample stored at −80°C.
RESULTS
Minimal packaging domains bind to three cellular proteins.
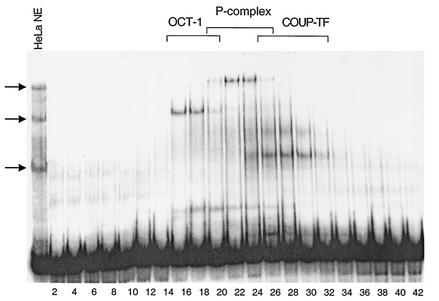
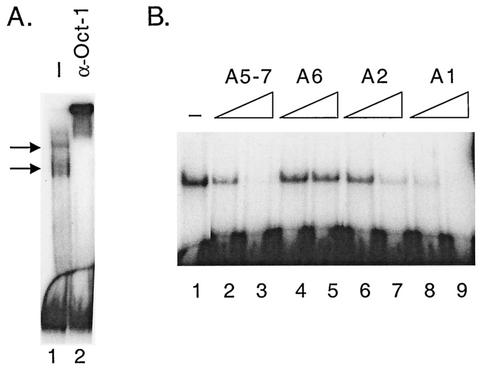
Several lines of evidence suggest that there is a limiting trans-acting factor(s) that plays a role in Ad packaging (16, 39, 40). To identify cellular proteins that may participate in Ad DNA packaging, the binding of proteins from uninfected HeLa cell nuclear extracts to minimal packaging domains with optimal activity in vivo (39) (Fig. 1, a hexamer of Ad5 A repeat 1 [A16] and a dimer of Ad5 A repeats 5 through 7 [A(5-7)2]) was examined by gel mobility shift assays (Fig. 2, lane 1). Three cellular binding activities were evident when A(5-7)2 was used as the probe. The same three activities were observed using an A16 probe and with Ad-infected HeLa nuclear extract (data not shown). HeLa cell nuclear extract was subjected to heparin agarose chromatography and every other fraction was assayed for the presence of A(5-7)2 binding activities (Fig. 2). The binding activity detected in fractions 16 and 18 corresponds to Oct-1, a ubiquitous transcription factor, as a polyclonal antibody raised against the C terminus of Oct-1 supershifted the activity detected in fraction 18 (Fig. 3A). Several complexes that contain Oct-1 binding activity were detected using the A16 probe (Fig. 3A, arrows), in comparison to the single complex detected using the A(5-7)2 probe (Fig. 2). The Oct-1 DNA binding domain contains two structurally independent domains (POUs and POUh) that cooperate to form the DNA binding domain (45). We predicted that Oct-1 would bind to Ad A-repeats since the high affinity Oct-1 consensus site (5′-ATTTGCAT-3′; (35) is identical to first half of the A-repeat consensus site (5′-ATTTG-3). Figure 3B shows the results of a gel mobility shift assay in which the Ad5 A(5-7)2 probe was incubated with the POU domain of Oct-1 fused to GST-POU. No specific competitor DNA was added to the binding reaction analyzed in lane 1, while either a 40- or 200-fold molar excess of competitor oligonucleotides representing different multimeric A-repeats was added to the binding reactions resolved in lanes 2 to 9. The A5-7 repeats (lanes 2 and 3) and A1 repeat (lanes 8 and 9) competed efficiently for GST-POU binding to the A(5-7)2 probe. A repeat 2 (lanes 6 and 7) competed weakly and A repeat 6 (lanes 4 and 5) did not compete for GST-POU binding. Thus, these results establish a direct correlation between binding of Oct-1 to the minimal packaging domains in vitro and the efficiency with which these repeats function in packaging assays in vivo (Fig. 1C).
FIG. 2.
Three cellular activities interact with the minimal packaging domains. Uninfected HeLa cell nuclear extract was fractionated by heparin agarose chromatography. Gel mobility shift experiments were performed using every other fraction (fractions 2 to 42 are represented) and the dimeric A(5-7) probe. Oct-1, P-complex and COUP-TF binding activities are indicated by arrows and the respective fractions indicated across the top.
FIG. 3.
Binding of Oct-1 to minimal packaging domains. (A) A gel mobility shift assay was performed using fraction 18 (Fig. 2) and an A16 probe. Several binding activities are evident with this probe and are indicated with arrows (lane 1). Anti-Oct-1 antibody was added to the binding reaction in lane 2. (B) Oct-1 binds to A-repeats with different apparent affinities. Bacterially expressed GST-POU domain of Oct-1 protein was used in a gel mobility shift assay with the A(5-7)2 probe (lane 1). Increasing amounts of specific competitor oligonucleotides (40- and 200-fold molar excess) were added to binding reactions in lanes 2 to 9 representing different multimeric A repeats: lanes 2 and 3, A repeats 5 to 7; lanes 4 and 5, A repeat 6; lanes 6 and 7, A repeat 2; lanes 8 and 9, A repeat 1.
The peak of the P-complex binding activity was detected in fractions 22 and 24 (Fig. 2). The molecular mass of P complex in these fractions was estimated by velocity sedimentation on a glycerol gradient at 150 to 175 kDa (data not shown). The polyclonal anti-Oct-1 antibody did not supershift the P complex, and a high-affinity Oct-1 binding site did not compete with P complex for binding the A(5-7)2 probe in gel mobility shift assays (data not shown). These two results indicated that the DNA binding activity of Oct-1 was not responsible for the formation of P complex and is likely not part of the complex. A third A(5-7)2 binding activity detected upon fractionation of HeLa cell nuclear extracts is COUP-TF (peak fractions 24 to 30), as previously reported (40). COUP-TF is a ubiquitous and highly conserved member of the steroid/thyroid nuclear hormone receptor superfamily (3). Members of this superfamily bind to their respective response elements as homo- and heterodimers. The ability of COUP-TF to bind A repeats in vitro does not correlate with packaging efficiency in vivo (40) and polyclonal antibodies directed against COUP-TF did not supershift P complex although they efficiently supershifted COUP-TF-DNA complexes (data not shown). Taken together, these results indicate that COUP-TF is unlikely to play a functional role in Ad DNA packaging. Finally, the binding activity evident in fractions 16 to 28 migrating near the bottom of the gel did not bind the A16 minimal packaging domain and was not pursued further.
Oct-1 is not functionally relevant to viral DNA packaging.
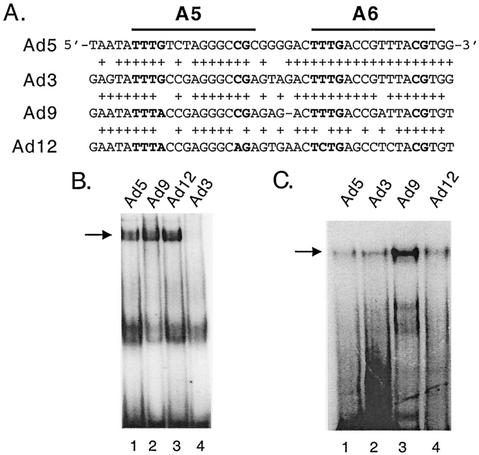
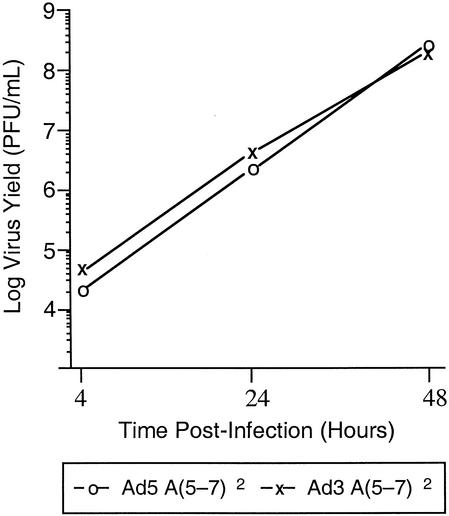
Sequences overlapping the A5 and A6 packaging repeats are highly conserved among different Ad serotypes, although several nucleotide differences within the consensus sequence are present (Fig. 4A). A(5-7)2 domains from different Ad serotypes were used as probes in gel mobility shift assays and their binding to Oct-1 and P complex was determined (Fig. 4B and C). Oct-1 bound to A-repeats 5-7 of Ad5, Ad9 and Ad12, but not to Ad3 (Fig. 4B). In contrast, P complex bound to A repeats 5 to 7 of all Ad serotypes (Fig. 4C). A hybrid virus carrying A(5-7)2 repeats from Ad3 in an Ad5 virus lacking the genuine packaging sequences was constructed and analyzed. Growth of this virus was compared to a homologous virus carrying the Ad5 A(5-7)2 packaging sequences in place of the natural packaging domain to determine if the inability of Oct-1 to bind to the packaging domain of this recombinant virus correlated with a DNA packaging defect. The Ad3 A5-7 packaging repeats supported virus growth to the same level as the Ad5 A5-7 repeats (Fig. 5). Thus, we conclude that Oct-1 does not play a functional role in viral DNA packaging since the Ad3 A5-7 A-repeats support packaging efficiently but do not bind Oct-1. In addition, P complex binding to A repeats of different Ad serotypes is consistent with packaging function.
FIG. 4.
(A) An alignment of A repeats 5 and 6 from different Ad serotypes is shown. The bold nucleotides indicate the first and the second half sites of the packaging consensus motif. The plus symbols indicate identical nucleotides to those found at that position with Ad5. (B) Gel mobility shift experiments were performed using fraction 18 (Fig. 2) with Ad5 A(5-7)2 (lane 1), Ad9 A(5-7)2 (lane 2), Ad 12 A(5-7)2 (lane 3), and Ad3 A(5-7)2 (lane 4) probes. Oct-1 binding activity is indicated by an arrow. (C) Gel mobility shift experiments were performed using fraction 22 (Fig. 2) with Ad5 A(5-7)2 (lane 1), Ad3 A(5-7)2 (lane 2), Ad9A (5-7)2 (lane 3), and Ad 12 A(5-7)2 (lane 4) probes. P-complex binding activity is indicated by an arrow.
FIG. 5.
Single step growth curve of viruses carrying Ad 5 A(5-7)2and Ad 3 A(5-7)2 packaging repeats in place of the natural packaging domain in Ad5. N52.E6 cells were infected with the two viruses and virus growth was measured by plaque assay of the virus produced at 4, 24, and 48 h postinfection.
P complex contains full-length CDP.
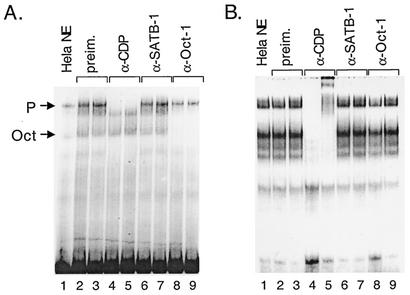
Previous results support the hypothesis that P complex plays a functional role in Ad5 viral DNA packaging (40). Based upon the size and similarities between biochemical properties of the DNA binding activity within P complex and the cellular protein CDP (29, 30), we performed gel supershift assays to determine if CDP is a component of P complex (Fig. 6). Nuclear extract from uninfected HeLa cells was used in a gel mobility shift assay with the Ad5 A(5-7)2 packaging repeat (Fig. 6A). In the absence of specific antibodies, Oct-1 and P-complex binding activities were observed (lane 1). The addition of polyclonal antibodies directed against the C terminus of CDP did not prevent the binding of Oct-1 to the probe DNA but abolished the formation of P complex (Fig. 6A, lanes 4 and 5). In contrast, the formation of P complex was not affected by the addition of preimmune serum from the same animal (lanes 2 and 3) or by polyclonal special AT-rich binding protein (SATB1) antibodies (lanes 6 and 7). These later antibodies were tested as SATB-1 protein is structurally and functionally similar to CDP (13). Polyclonal α-Oct-1 antibodies abolished Oct-1 binding to the A(5-7)2 probe (lanes 8 and 9) but did not affect the formation of P complex, as expected. The same experiment was performed using ITR sequences containing the P-complex binding site as probe (Fig. 6B). Two major binding activities were observed (lane 1). These binding activities were either abolished (lane 4) or supershifted (lane 5) upon addition of α-CDP antibodies while preimmune serum, α-SATB1 and α-Oct-1 antibodies did not have an effect.
FIG. 6.
P complex contains CDP. (A) Gel mobility shift assays were performed using uninfected HeLa nuclear extract and an Ad5 A(5-7)2 probe. Preimmune (preim.) serum (lanes 2 and 3), α-CDP antibody (lanes 4 and 5), α-SATB-1 antibody (lanes 6 and 7), and α-Oct-1 antibody (lanes 8 and 9) were added to the reactions 0.5 h before (lanes 2, 4, 6, and 8) or 0.5 h after (lanes 3, 5, 7, and 9) the addition of probe. (B) The same experiment shown in panel A was repeated using the ITR probe.
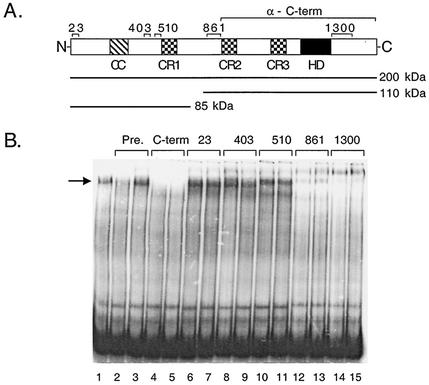
CDP is synthesized as an ∼200-kDa polypeptide that is subsequently cleaved during S phase by an unknown protease to yield ∼85-kDa and ∼110k Da proteins (29; Fig. 7A). Antipeptide antibodies raised against various regions of CDP (Fig. 7A) were used in gel mobility shift analyses to determine which form(s) of CDP was present within P complex (Fig. 7B). P complex was generated using heparin agarose fraction 22 (Fig. 2) and the Ad5 A(5-7)2 probe (Fig. 7B, lane 1). This complex was clearly supershifted upon addition of α-1300, α-861 and α-510 antibodies, before or after the addition of the probe (Fig. 7B, lanes 6 to 11) and a partial supershift was observed using the α-403 antibody (Fig. 7B, lanes 4 and 5). The proteolytic cleavage of CDP occurs downstream of CR1, between amino acids 659 and 878 (30). The α-861 and α-1300 antibodies recognize both full-length CDP and the ∼110-kDa cleavage product, whereas the α-403 and α-510 antibodies recognize full-length CDP and the ∼85-kDa cleavage product. P complex was supershifted by the α-510 antibody and there was partial supershift with α-403 antibody. Since the ∼85-kDa form of CDP does not bind to DNA (30), we conclude that P complex likely contains full-length CDP.
FIG. 7.
P complex contains full-length CDP. (A) Schematic diagram of the full-length CDP protein; a coiled coil (CC) region, the Cut repeats (CR1 to -3) and the homeodomain (HD) are indicated. Antibodies raised against the different parts of the protein are shown in brackets above the diagram. Full-length CDP and the ∼110- and ∼85-kDa proteolytic products of CDP are shown below the diagram. (B) Gel mobility shift assays were performed using fraction 22 (Fig. 2) with an Ad5 A(5-7)2 probe. Different antibodies were added to the reaction mixes, as indicated, 0.5 h before (lanes 2, 4, 6, 8, and 10) or 0.5 h after (lanes 3, 5, 7, 9, and 11) the addition of probe.
CDP is found in the virus particles.
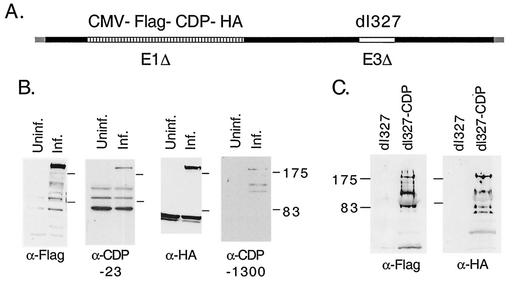
To determine if CDP is present within virions, a recombinant Ad that expresses epitope-tagged, full-length CDP was constructed (Fig. 8A). A cDNA encoding CDP tagged at its N terminus with the Flag epitope and at its C terminus with an HA epitope was constructed and substituted for E1 within an Ad containing a deletion of E3 (dl327). The deletion of E3 was necessary to accommodate the large CDP cDNA. N52.E6 cells were infected with the CDP-expressing virus, harvested 48 h postinfection, and whole-cell extract (WCE) was prepared and analyzed by Western blotting with α-Flag and α-HA monoclonal antibodies and anti-CDP 23 and 1300 polyclonal antibodies (Fig. 8B). Antibodies directed against both epitope tags detected an ∼200-kDa polypeptide in WCE prepared from infected, but not uninfected, cells and allowed us to conclude that this species represented the full-length CDP protein. Other less prominent polypeptides were also detected in the infected WCE that likely represent the cleaved portions of the protein previously described (29). Full-length CDP was detected using the 23 and 1300 polyclonal antibodies against CDP. Smaller forms of CDP that were evident using the monoclonal antibodies also were observed using these polyclonal antibodies confirming their assignment as C-terminal and N-terminal truncation forms of CDP.
FIG. 8.
CDP in found in purified virus particles. (A) Schematic diagram of the recombinant Ad expressing epitope-tagged full-length CDP. The CMV-Flag-CDP-HA expression cassette is indicated as a patterned box and is situated in place of the E1 region. The virus also contains a deletion of E3 (dl327). The schematic is not drawn to scale. (B) Western blot analysis of WCE using uninfected and dl327-CDP-infected cells. Anti-Flag and α-HA antibodies were used for detection of epitope-tagged CDP and anti-CDP antibodies 23 and 1300 were used to detect endogenous and epitope-tagged CDP forms. (C) Purified virus particles from dl327- and dl327-CDP-infected cells were isolated and examined for CDP by Western blot using α-Flag and α-HA antibodies. The mobility of molecular weight markers is indicated for each gel.
Virus particles purified from dl327- and dl327-CDP-infected cells were analyzed for the presence of epitope-tagged CDP by Western blotting using the α-Flag and α-HA antibodies (Fig. 8C). An ∼200-kDa polypeptide corresponding to full-length CDP was detected with both antibodies in the sample prepared from dl327-CDP virions but not from dl327 virions. Other smaller polypeptides were detected with the α-Flag and α-HA antibodies. These smaller polypeptides do not represent cross-reactivity of the antibodies with Ad capsid proteins since they were not detected using dl327 virus particles. They may represent cleaved forms of CDP (30). In addition, Ad virions contain an active proteinase (44) and it is possible that the smaller CDP-derived polypeptides detected in dl327-CDP virions were produced by proteolysis of full-length CDP within the virion during maturation. As a control for the specificity of packaging of CDP into virus particles, as opposed to copurification of CDP on the surface of virions, we analyzed packaging of a C-terminally truncated mutant form of CDP that does not bind DNA (30). While the CDP mutant protein was expressed at high levels in infected cells, it was not found in purified virus particles (data not shown). We conclude from these results that full-length CDP is packaged within virions.
DISCUSSION
Although limited information is available on the DNA encapsidation mechanism of eukaryotic DNA viruses, there is precedent for the binding of cellular proteins to viral packaging signals. Sp1, a ubiquitous transcription factor, participates in simian virus 40 (SV-40) assembly. It is thought that the VP1-VP2/3 (major capsid protein complex of SV-40) is recruited to ses, the cis-acting packaging signal, by Sp1 (14, 32). According to the SV-40 encapsidation model, formation of this protein-DNA structure serves as the nucleation center for capsid propagation. Ying-Yang 1 (YY1), a multifunctional cellular nuclear matrix protein, may be involved in polyomavirus DNA packaging. It is hypothesized that like Sp1, YY1 interacts with VP1 (a major capsid protein) and confers DNA binding specificity to VP1 (34). It has been previously reported that two cellular protein complexes, P complex (CDP) and COUP-TF, bind to the minimal packaging sequences of Ad5 (40). We report in this paper a third cellular activity, Oct-1, that binds to the Ad packaging sequences. Ad utilizes numerous cellular proteins during its natural life cycle for viral transcription and DNA replication and it is therefore not surprising to find that Ad may redirect a cellular factor(s) for the encapsidation of its viral DNA.
Previous results indicated that COUP-TF binding to the minimal packaging sequences does not play a functional role in viral DNA packaging (40). There is, however, a direct correlation between how well Oct-1 and P complex bind to minimal packaging domains in vitro and how efficiently they function in packaging assays in vivo (40). A(5-7)2 domains from different Ad serotypes were tested for their ability to bind P complex and Oct-1. Despite a strong homology among the Ad3, 5, 9 and 12 serotypes within this region, especially within the A-repeat consensus motif, minor differences that affect Oct-1 binding have been identified (Fig. 4A). Our goal was to establish a convincing link between DNA binding affinity and packaging efficiency. The ability of the Ad3 A(5-7)2 to substitute efficiently for the Ad5 packaging domain in a recombinant virus (Fig. 5) but not bind Oct-1 (Fig. 4B) shows that the DNA binding activity of Oct-1 is not functionally relevant to viral DNA packaging. We speculated that Oct-1 might be the DNA binding component of P complex but several lines of evidence that argue against this idea. Polyclonal α-Oct-1 antibodies, added before and after the addition of probe DNA, did not affect P-complex formation (Fig. 6A and B), and the peak of Oct-1 protein present in heparin agarose fractions did not coincide with P-complex activity (data not shown). Moreover, a high affinity Oct-1 binding site did not compete for P-complex binding in gel mobility shift assays.
Evidence gathered to this date supports the notion that P complex plays a functional role in viral DNA packaging (40). P complex binds to all the minimal packaging domains. The relative affinity of P complex for the different domains in vitro correlates well with the ability of the respective cis-acting sequences to support viral DNA packaging in vivo. We have uncovered the identity of P complex, or a component of the complex, by supershift analysis. Addition of polyclonal antibodies raised against CDP, before or after the addition of probe, abolished or supershifted P-complex binding activity depending on the probe and antibody used (Fig. 6 and 7). CDP, an ∼200-kDa protein, was first identified in Drosophila (named Cut) as a cell fate-determining factor (4). CDP contains three DNA-binding domains, termed Cut repeats (CR), in addition to a homeodomain (Fig. 7A) (29). CDP is cleaved in S phase by an unknown protease into a C-terminal ∼110k-kDa fragment and a N-terminal ∼85-kDa fragment (30). The DNA binding properties of the full-length and the 110-kDa proteins differ. The cleaved ∼110-kDa protein exhibits stable binding to DNA through its CR3 and homeodomain whereas the full-length protein binds transiently to DNA through its CR1 and CR2 domains (29, 30). The N-terminal ∼85-kDa fragment does not appear to bind to DNA (30). We have shown that P complex very likely contains full-length CDP (Fig. 7B) and that full-length CDP is found within mature virions (Fig. 8C). These results predict that CDP binds to Ad packaging A repeats via the CR1 and CR2 domains. The specificity of packaging of CDP into virus particles, as opposed to copurification of CDP on the surface of virions, is indicated by two observations. First, a truncated CDP mutant protein that lacks the C terminus was expressed at high levels in infected cells, but it was not found in purified virus particles. Since C-terminally truncated CDP mutants do not bind DNA (30), this indicates that DNA binding is required for CDP to copurify with virus particles. If CDP copurified with virus particles by sticking to the outside surface of virions, then one would expect to copurify the truncated mutant protein. Second, CDP in virions was enriched for several smaller forms compared to cellular extracts (Fig. 8). We would expect that if CDP contaminated the particles by sticking to their surface, then the CDP profile would look very similar to that seen with cellular extracts. Instead, the enrichment suggests that these forms are selectively packaged and/or full-length CDP is proteolytically cleaved within virus particles, perhaps by the Ad proteinase. We conclude that CDP copurifies with virus particles because CDP is located within virions.
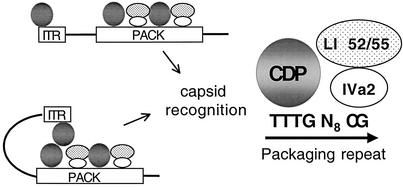
Our working model for Ad DNA packaging is depicted in Fig. 9. CDP binds to the first half site of the A repeat consensus sequence within the packaging domain along with sequences within the ITR (nt 1 to 13, Fig. 6). Recently, the Ad-encoded protein IVa2 was found to bind the CG-rich sequence in the A repeat consensus motif (46) and compelling evidence for the role of IVa2 in viral packaging in vivo was described (47). Mutation of the second half-site of A repeats significantly reduces viral DNA packaging in vivo further implicating a functional involvement of IVa2 in DNA encapsidation (39). The L1 52/55-kDa protein directly interacts with IVa2 (18). Mutant viruses that do not express L1 52/55-kDa protein accumulate empty capsids with little or no packaged viral DNA, demonstrating that this protein also plays a role in viral DNA packaging (17, 21). It is hypothesized that IVa2 binding to the packaging sequences may bring L1 52/55-kDa protein in close proximity to the packaging domain. How the L1 52/55-kDa and IVa2 proteins facilitate packaging is unknown but both proteins are found in empty capsids and at much higher levels than in mature virus particles (17, 21). Thus, the L1 52/55-kDa and IVa2 proteins may play a role as scaffolding proteins to support empty capsid structure. It also seems likely that these viral proteins are involved in specific recognition of the packaging domain based on the binding of IVa2 to A repeat sequences (46). Full-length CDP binds transiently to DNA (29, 30) and P complex has similar DNA binding properties. The requirement for a precise spacing between the first and second half-sites of A repeats for packaging function (39) is consistent with the possibility that CDP may interact with IVa2 to stabilize the protein-DNA complex.
FIG. 9.
Model for specificity of Ad DNA packaging. The left terminus of the Ad genome is schematically represented with ITR and packaging (PACK) domains denoted by boxes. CDP protein is indicated with a gray circle and binds to ITR sequences and the first half-site of the A-repeat consensus sequence. The IVa2 protein is indicated with an open oval and binds to the second half-site of the packaging A-repeats. The L1 52/55-kDa protein is indicated with a stippled oval and interacts with the IVa2 protein. The binding of CDP to the ITR and packaging sequences may direct the formation of a higher order DNA-protein structure linking these functional elements, as shown.
What is the mechanism by which CDP may facilitate viral DNA packaging? CDP represses transcription by competing for binding site occupancy of the CCAAT transcription factor (2) as well as by the recruitment of histone deacetylase HDAC-1 (26, 28). In mammals, CDP activity is generally restricted to proliferating cells and an inverse correlation exists between CDP levels and the differentiation state of the cell (31). CDP appears to function as a general repressor of genes in proliferating cells with relief of repression observed following cell differentiation. CDP binding to the Ad packaging sequences may facilitate the recruitment of proteins involved in chromatin remodeling thereby making the DNA unavailable for transcriptional activity. The packaging domain overlaps two enhancer elements which are involved in viral transcription (23). CDP may repress the activity of these enhancers thereby preparing the viral DNA for later events such as packaging. At the same time, CDP may also aid in the structural changes in viral DNA chromatin marking the genome as a substrate for packaging. CDP/Cut proteins interact with regulatory elements from a large number of genes, including matrix attachment regions (MARs) (1, 5, 27). MARs are sites where active replication, transcription, and RNA processing takes place. Previous studies have shown that the Ad terminal protein, pTP, is attached to the nuclear matrix along with other proteins necessary for viral DNA replication (12, 37). Ad DNA is recruited to these discrete sites which develop into viral replication centers. The L1 52/55-kDa protein is localized within nuclear structures that are morphologically distinct from viral replication centers (20). These structures may represent domains devoted to virus assembly. Perhaps CDP plays a role in redirecting replicated viral DNA to MARs and the virus assembly sites within the nucleus. These viral and cellular proteins may interact with the packaging sequences to mark the viral DNA as a packaging substrate leading the way to the assembly process.
Finally, these results may have implications for the development of Ad gene therapy vectors. High-capacity or “gutless” Ad gene therapy vectors have been developed that lack all viral coding sequences; these vectors rely on a helper virus for their propagation (25). The packaging of the helper virus must be regulated under conditions of gutless vector production in order to minimize contamination of the gene therapy vector with wild type helper virus. Currently, the Cre-Lox recombination system is used to excise the packaging domain from the helper virus genome in order to selectively repress helper virus packaging while allowing the packaging of the gene therapy vector (25). The selective binding of Oct-1 to Ad5 packaging repeats, but not Ad3 packaging repeats, could be used to augment the efficiency in this system. We found that A repeats 5 to 7 of either Ad5 or Ad3 efficiently supported the growth an Ad5 recombinant virus that contains these A-repeats in place of the natural packaging domain (Fig. 5). We speculate that the packaging of a helper virus that contains Ad5 A-repeats 5 to 7 could be selectively repressed by Oct-1 overexpression at late times after infection whereby the bone fide packaging factor(s) may be displaced from the packaging repeats. In contrast, the packaging of a gutted vector that contains Ad3 packaging repeats may be refractory to such repression since Oct-1 does not bind to these sites.
Acknowledgments
We thank all the members of our laboratories for many helpful discussions; Paul Freimuth, Michael Hayman, Janet Hearing, and Eckard Wimmer for expert advice; and Janet Hearing, Quan Zhu, Urmila Maitra, and Jin Seo for critical reviews of the manuscript.
This work was supported by Public Health grants AI41636 to P.H. and CA34780 to J.P.D. and by grant MT-11590 from the Canadian Institute of Health Research of Canada to A.N.
REFERENCES
- 1.Banan, M., I. C. Rojas, W. H. Lee, H. L. King, J. V. Harriss, R. Kobayashi, C. F. Webb, and P. D. Gottlieb. 1997. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem. 272:18440-18452. [DOI] [PubMed] [Google Scholar]
- 2.Barberis, A., G. Superti-Furga, and M. Busslinger. 1987. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 50:347-359. [DOI] [PubMed] [Google Scholar]
- 3.Beato, M., P. Herrlich, and G. Schutz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83:851-857. [DOI] [PubMed] [Google Scholar]
- 4.Bodmer, R., S. Barbel, S. Sheperd, J. W. Jack, L. Y. Jan, and Y. N. Jan. 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51:293-307. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay, S., C. E. Whitehurst, and J. Chen. 1998. A nuclear matrix attachment region upstream of the T cell receptor beta gene enhancer binds Cux/CDP and SATB1 and modulates enhancer-dependent reporter gene expression but not endogenous gene expression. J. Biol. Chem. 273:29838-29846. [DOI] [PubMed] [Google Scholar]
- 6.Daniell, E. 1976. Genome structure of incomplete particles of adenovirus. J. Virol. 19:685-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Halluin, J. C., M. Milleville, G. R. Martin, and P. Boulanger. 1980. Morphogenesis of human adenovirus type 2 studied with fiber- and fiber and penton base-defective temperature-sensitive mutants. J. Virol. 33:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Halluin, J. C., G. R. Martin, G. Torpier, and P. A. Boulanger. 1978. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J. Virol. 26:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edvardsson, B., S. Ustacelebi, J. Williams, and L. Philipson. 1978. Assembly intermediates among adenovirus type 5 temperature-sensitive mutants. J. Virol. 25:641-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, J. D., and P. Hearing. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed]
- 12.Fredman, J. N., and J. A. Engler. 1993. Adenovirus precursor to terminal protein interacts with the nuclear matrix in vivo and in vitro. J. Virol. 67:3384-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galande, S., L. A. Dickinson, I. S. Mian, M. Sikorska, and T. Kohwi-Shigematsu. 2001. SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell. Biol. 21:5591-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon-Shaag, A., O. Ben-Nun-Shaul, V. Roitman, Y. Yosef, and A. Oppenheim. 2002. Cellular transcription factor Sp1 recruits simian virus 40 capsid proteins to the viral packaging signal, ses. J. Virol. 76:5915-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grable, M., and P. Hearing. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grable, M., and P. Hearing. 1992. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J. Virol. 66:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustin, K. E., P. Lutz, and M. J. Imperiale. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammarskjold, M. L., and G. Winberg. 1980. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell 20:787-795. [DOI] [PubMed] [Google Scholar]
- 20.Hasson, T. B., D. A. Ornelles, and T. Shenk. 1992. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 66:6133-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasson, T. B., P. D. Soloway, D. A. Ornelles, W. Doerfler, and T. Shenk. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hearing, P., and T. Shenk. 1986. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell 45:229-236. [DOI] [PubMed] [Google Scholar]
- 24.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 25.Kochanek, S., G. Schiedner, and C. Volpers. 2001. High-capacity 'gutless' adenoviral vectors. Curr. Opin. Mol. Ther. 3:454-463. [PubMed] [Google Scholar]
- 26.Li, S., L. Moy, N. Pittman, G. Shue, B. Aufiero, E. J. Neufeld, N. S. LeLeiko, and M. J. Walsh. 1999. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J. Biol. Chem. 274:7803-7815. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mailly, F., G. Berube, R. Harada, P. L. Mao, S. Phillips, and A. Nepveu. 1996. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol. Cell. Biol. 16:5346-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon, N. S., G. Berube, and A. Nepveu. 2000. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J. Biol. Chem. 275:31325-31334. [DOI] [PubMed] [Google Scholar]
- 30.Moon, N. S., P. Premdas, M. Truscott, L. Leduy, G. Berube, and A. Nepveu. 2001. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 21:6332-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 32.Oppenheim, A., Z. Sandalon, A. Peleg, O. Shaul, S. Nicolis, and S. Ottolenghi. 1992. A cis-acting DNA signal for encapsidation of simian virus 40. J. Virol. 66:5320-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostapchuk, P., and P. Hearing. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palkova, Z., H. Spanielova, V. Gottifredi, D. Hollanderova, J. Forstova, and P. Amati. 2000. The polyomavirus major capsid protein VP1 interacts with the nuclear matrix regulatory protein YY1. FEBS Lett. 467:359-364. [DOI] [PubMed] [Google Scholar]
- 35.Pruijn, G. J., W. van Driel, R. T. van Miltenburg, and P. C. van der Vliet. 1987. Promoter and enhancer elements containing a conserved sequence motif are recognized by nuclear factor III, a protein stimulating adenovirus DNA replication. EMBO J. 6:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, C. C., and C. Tibbetts. 1984. Polar encapsidation of adenovirus DNA: evolutionary variants reveal dispensable sequences near the left ends of Ad3 genomes. Virology 137:276-286. [DOI] [PubMed] [Google Scholar]
- 37.Schaack, J., W. Y. Ho, P. Freimuth, and T. Shenk. 1990. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 4:1197-1208. [DOI] [PubMed] [Google Scholar]
- 38.Schiedner, G., S. Hertel, and S. Kochanek. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105-2116. [DOI] [PubMed] [Google Scholar]
- 39.Schmid, S. I., and P. Hearing. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid, S. I., and P. Hearing. 1998. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J. Virol. 72:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stow, N. D. 1981. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J. Virol. 37:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 43.Tibbetts, C. 1977. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell 12:243-249. [DOI] [PubMed] [Google Scholar]
- 44.Tihanyi, K., M. Bourbonniere, A. Houde, C. Rancourt, and J. M. Weber. 1993. Isolation and properties of adenovirus type 2 proteinase. J. Biol. Chem. 268:1780-1785. [PubMed] [Google Scholar]
- 45.Verrijzer, C. P., M. J. Alkema, W. W. van Weperen, H. C. Van Leeuwen, M. J. Strating, and P. C. van der Vliet. 1992. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 11:4993-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]