Abstract
We developed a system for complete replication of encephalomyocarditis virus (EMCV) in a test tube by using an in vitro translation extract from Krebs-2 cells. Efficient virus synthesis occurred in a narrow range of Mg2+ and EMCV RNA concentrations. Excess input RNA impaired RNA replication and virus production but not translation. This suggests the existence of a negative-feedback mechanism for regulation of RNA replication by the viral plus-strand RNA or proteins.
Encephalomyocarditis virus (EMCV) is the prototype Cardiovirus within the family Picornaviridae (31). The genome of EMCV is a ∼7,900-nucleotide-long positive-strand RNA. Within cells, EMCV RNA sequentially functions as a template for virus-specific translation and negative-strand RNA synthesis; it is also packaged into the virion at the late stage of infection. EMCV RNA recruits ribosomes with the aid of an internal ribosome entry site (10, 19), which acts as a binding site for translational initiation factor eIF4F (26, 28). Translation proceeds from a single initiation site to yield a large polyprotein. The polyprotein is cleaved in a series of steps by the viral protease 3Cpro or 3ABCpro into mature virus proteins (14, 18, 25). RNA replication involves negative and positive RNA synthesis and requires functions of at least two viral proteins, the RNA polymerase 3Dpol and VPg. The detailed mechanisms of these processes are controversial with respect to the involvement of particular cis-acting replication elements (cre) and trans-acting cellular factors (1).
Synthesis of an infectious picornavirus in a test tube has been achieved but only for poliovirus (PV) (4, 21, 22). Such a system is paramount for the understanding of mechanisms of virus replication and for screening antiviral drugs. EMCV RNA is translated accurately, with the formation of authentic viral proteins and capsid intermediates, in a nuclease-treated Krebs-2 cell extract (S10) (2, 32, 33). Here, we show that this system also supports other virus-specific processes required for complete virus replication.
Krebs-2 ascites cells (7) were washed twice with Earle's solution (80 × g for 8 min at 4°C), suspended in methionine-free Dulbecco's minimal essential medium (5 × 106 to 10 × 106 cells/ml), and incubated at 37°C for 2 h on a rotary shaker (20). Incubation of cells in nutritionally rich medium (i.e., Dulbecco's minimal essential medium) increased translation efficiency, in agreement with earlier data (17, 20). After incubation, cells were harvested and washed three times with cold isotonic buffer (35 mM HEPES-KOH [pH 7.3], 146 mM NaCl, 11 mM d-glucose) (4), with a final centrifugation at 750 × g. Packed cells were suspended in 2 volumes of hypotonic buffer (25 mM HEPES-KOH [pH 7.3], 50 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol), allowed to swell on ice for 20 min, and lysed with ∼30 strokes of a tightly fitting Dounce homogenizer (Kontes). Concentrated buffer solution (1/9 volume; 25 mM HEPES-KOH [pH 7.3], 1 M KCH3COO, 30 mM MgCl2, 30 mM dithiothreitol) was then added, and the homogenate was centrifuged at 18,000 × g for 20 min at 4°C. The supernatant (35 to 45 A260 U per ml) was stored at −70°C. Before translation, the extract was treated with micrococcal nuclease (150 U/ml; Roche) essentially as described previously (32). EMCV (K2) RNA was extracted from the purified virus (34). Translation reaction mixtures (50 μl) contained 25 μl (50% by volume) of S10, 5 μl of a master mix (10 mM ATP, 2 mM GTP, 2 mM CTP, 2 mM UTP, 100 mM creatine phosphate [di-potassium salt], 1 mg of rabbit muscle creatine phosphokinase [Calbiochem]/ml, 19 unlabeled l-amino acids [without l-methionine; 0.2 mM each], 125 mM HEPES-KOH [pH 7.3]), 5 μl of a salt solution (0.75 M KCH3COO, 10 mM MgCl2, 2.5 mM spermidine), 2.5 μl of [35S]methionine (10 mCi/ml [>1,000 Ci/mmol]), and EMCV RNA (2 to 30 μg/ml). Translation was at 32 to 33°C for the times indicated in the figure legends. Proteins were analyzed by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) and autoradiography. RNA replication and virion production were assayed in similarly programmed reactions, except that unlabeled 0.4 mM l-methionine was substituted for [35S]methionine in the reaction cocktail.
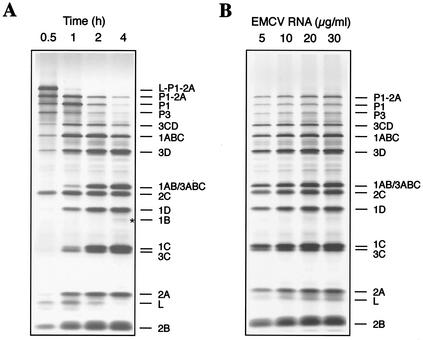
In agreement with previous data (32), translation of EMCV RNA in Krebs-2 S10 yielded all the known virus proteins (Fig. 1A). A time course of the appearance of l-P1-2A (the N-terminal portion of the viral polyprotein), 2C (the central portion), and P3 (the C-terminal portion) was consistent with the complete translation of the viral genome over 0.5 h of incubation time. During the subsequent incubation, the l-P1-2A and P3 polypeptides were cleaved and the mature viral structural and nonstructural proteins appeared. The magnesium concentration range for optimum translation was relatively broad, and variations within the 2 to 4 mM concentration range did not significantly affect labeling or the pattern of the products. Polypeptide synthesis was EMCV RNA dose dependent (Fig. 1B). Thus, Krebs-2 S10 prepared by the method described translates EMCV RNA efficiently and accurately.
FIG. 1.
(A) Time course of synthesis and processing of EMCV-specific proteins in EMCV RNA-programmed Krebs-2 S10. EMCV RNA (20 μg/ml) was translated in the presence of [35S]methionine under standard reaction conditions. At the times indicated, 5-μl aliquots of the reaction mixture were withdrawn, supplemented with SDS sample buffer, and subjected to SDS-15% PAGE. An autoradiogram of the dried gel is shown. The assignment of polypeptides is based on a comparison with proteins synthesized in EMCV-infected BHK-21 cells (34). An asterisk indicates the position of 1B. (B) EMCV RNA dose response of protein synthesis. Reaction mixtures (12.5 μl) were programmed by the indicated concentrations of EMCV RNA at 32°C for 2 h. Conditions for translation and PAGE were as described for panel A.
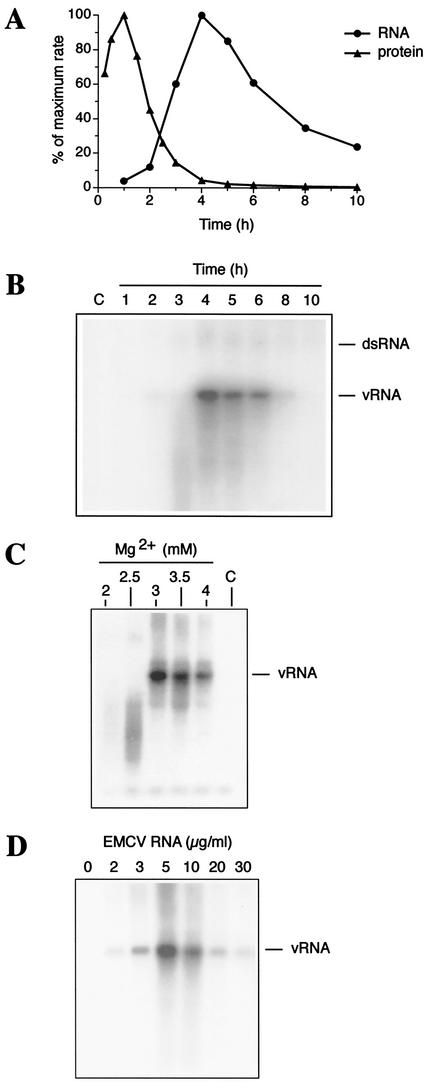
In vitro translation of EMCV RNA yields, among other viral proteins, high amounts of the RNA-dependent RNA polymerase 3Dpol (Fig. 1). To demonstrate that 3Dpol (in combination with other factors) supports RNA replication, we pulse-labeled the reaction mixtures with [α-32P]CTP at different times after the beginning of incubation. The newly synthesized RNA was extracted and quantified after trichloroacetic acid precipitation. RNA synthesis was first detectable at ∼2 h and reached a maximum at 4 to 5 h of incubation (Fig. 2A). On the other hand, pulse-labeling of reaction mixtures with [35S]methionine during the same time course revealed a maximum rate of viral translation at ∼1 h of incubation. By the time of maximal RNA synthesis (4 h), the rate of virus translation had declined dramatically (to less than 5% of the maximal rate level). Thus, our in vitro system recapitulates the switch from viral translation to RNA replication characteristic of many picornavirus systems in vivo (12). The major RNA product comigrated with the full-length EMCV RNA in an agarose gel (Fig. 2B). To examine the possibility that this product results from labeling of the input RNA by 3Dpol-associated terminal nucleotidyltransferase activity (29), we performed its digestion with RNase T1. This digestion, similarly to that of virion RNA (8, 35), liberated a large (∼150-nucleotide) poly(C) tract-containing fragment (data not shown). This strongly suggests that the vast majority of the in vitro product was uniformly labeled and was represented by the newly synthesized plus-strand RNA. It is noteworthy that small amounts of slowly migrating RNA species were generated in EMCV RNA-programmed but not in mock-programmed reactions (Fig. 2B). This product comigrates with the double-stranded replicative-form RNA (3). The propensity of the system for the synthesis of plus-strand RNA accords well with the known asymmetry of picornaviral replication in vivo, where the ratio of newly synthesized positive- to negative-strand RNAs is 30 to 70:1 (13, 24). Efficient RNA synthesis required Mg2+ in a narrow (3 to 3.5 mM) range of concentrations (Fig. 2C), in sharp contrast to the broad range observed for translation. Lowering the magnesium salt concentration to 2 mM resulted in almost no RNA synthesis (Fig. 2C) while causing only marginal inhibition of translation (data not shown). An EMCV RNA dose-response experiment showed that saturation of RNA synthesis is reached at a surprisingly low level of input RNA (5 μg/ml; Fig. 2D). Higher RNA concentrations (10 to 30 μg/ml) markedly inhibited RNA synthesis. Also surprising was that this inhibition occurred at RNA concentrations which stimulated rather than inhibited viral translation (Fig. 1B). A plausible explanation is that a viral protein(s), when accumulated to a certain level, can inhibit RNA replication. For example, RNA interaction with the structural proteins would engage it in a packaging complex and would preclude replication. Alternatively, EMCV RNA excess may sequester a host factor(s), which would facilitate RNA replication (3, 9). It would be interesting to determine whether the negative-feedback mechanism for regulation of RNA replication by virus-specific products governs the switch from replication to encapsidation of the viral RNA in vivo.
FIG. 2.
EMCV RNA replication. (A) Kinetics of EMCV RNA and protein synthesis. EMCV RNA (10 μg/ml)-programmed reaction mixtures (50 μl) were pulse-labeled with 1.5 μl of [α-32P]CTP (10 mCi/ml [3,000 Ci/mmol]) for 1 h (the label was added 30 min before the times indicated in the figure) (4). Samples were treated with proteinase K, and RNA was extracted with a mixture of phenol and chloroform. Viral proteins were pulse-labeled with [35S]methionine for 30 min (15 min before the times indicated in the figure) and dissolved in an SDS sample buffer. Portions of each sample were assayed for trichloroacetic acid-insoluble radioactivity. Data (average of two independent determinations) are given as percentages of maximum incorporation. (B) Products of RNA synthesis (from the reaction described for panel A) analyzed by native Tris-borate-EDTA-1% agarose gel electrophoresis and autoradiography (16). The positions of single-stranded (vRNA) and double-stranded (dsRNA) EMCV RNA are indicated. (C) Mg2+ concentration dependence of RNA synthesis. Reaction mixtures contained 10 μg of EMCV RNA/ml and the indicated concentrations of MgCl2. (D) EMCV RNA dose response of RNA synthesis. Reaction mixtures contained 3 mM MgCl2 and the indicated concentrations of EMCV RNA. For panels C and D, RNA was labeled with [α-32P]CTP between 4 and 5 h of incubation and analyzed as described above.
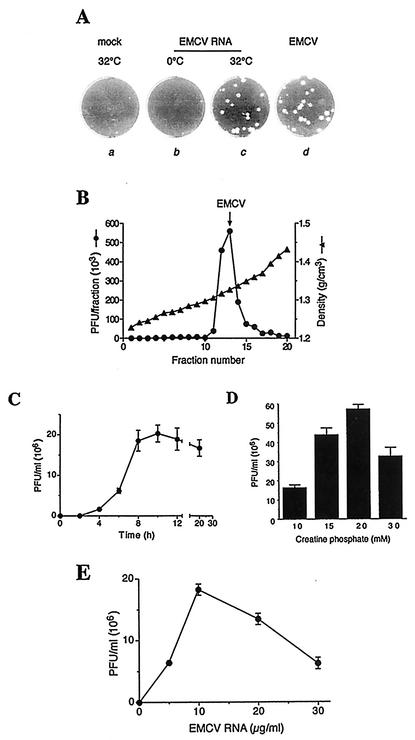
After a 4-h incubation period, a small amount of 1B was evident in a protein-synthesizing reaction (Fig. 1A), indicative of the final cleavage of 1AB, which accompanies virion morphogenesis (15). To determine whether the EMCV RNA-programmed reactions yield infectious virus, aliquots of translation samples withdrawn after 20 h of incubation were assayed for virus infectivity on BHK-21 cell monolayers. The in vitro reactions generated virus, and plaque morphologies were indistinguishable between in vitro- and in vivo-generated virus (Fig. 3A). No plaques were detected in samples that were incubated at 0°C. Thus, infectivity of the agent generated in in vitro reactions is dependent on a temperature-sensitive process and cannot be accounted for by the presence of the infectious input RNA. CsCl gradient analysis demonstrated that the in vitro-generated infectious agent was associated with the characteristic buoyant density of mature cardiovirus particles (1.33 g/cm3) (Fig. 3B) (30). To conclusively demonstrate the generation of intact virus particles, it is imperative to perform electron-microscopic examination. To this end, the peak fraction from the CsCl gradient (Fig. 3B) was subjected to dialysis and concentration. These procedures, however, resulted in significant loss of virus, probably because of its adsorption to plastic surfaces (30). Thus, electron-microscopic studies are not technically feasible now. Similar to RNA synthesis, EMCV production was more Mg2+ concentration dependent than translation and was maximal at 3 to 3.5 mM MgCl2 (data not shown). The infectivity of the in vitro-synthesized virus was already detectable between 2 and 4 h and reached maximum at 8 to 10 h of incubation (Fig. 3C). Subsequent incubation somewhat decreased infectivity. We also found that raising the creatine phosphate level from 10 to 20 mM stimulated virus production more than threefold (up to 5 × 107 to 6 × 107 PFU/ml virus titer; Fig. 3D), apparently by preventing the depletion of the nucleoside triphosphate pool (23). Virus yield reached a maximum at 10 μg of RNA/ml (Fig. 3E). Higher RNA concentrations resulted in reduced virus synthesis, most likely because of the inhibition of RNA replication (Fig. 2D).
FIG. 3.
Synthesis of EMCV in vitro. (A) Krebs-2 S10 extracts were incubated for 20 h either at 32°C (incubations a and c) or 0°C (incubation b) in the absence (incubation a) or presence (incubations b and c) of EMCV RNA (20 μg/ml). The samples were treated with a mixture of RNase A and T1 (21). Using serial dilutions of samples, a plaque assay was carried out on confluent BHK-21 cells (30). Plaques characteristic for EMCV, which was propagated in Krebs-2 cells in vivo, are shown for comparison (incubation d). (B) Isopycnic banding of in vitro-synthesized EMCV in a cesium chloride density gradient. A 250-μl translation reaction mixture was programmed with EMCV RNA for 20 h at 32°C and treated with the RNase A/T1 cocktail as described above. Triton X-100 was added to 0.5%, and the mixture was diluted to 5 ml with CsCl/phosphate-buffered saline (1.34 g/cm3 solution density) (30). The mixture was subjected to centrifugation at 45,000 rpm for 18 h at 6°C in a SW55 rotor. Fractions (0.25 ml) were collected and assayed for infectivity following serial dilutions. The arrow indicates the location of the virus formed in EMCV-infected Krebs-2 cells. (C) Time course of the infectivity titer of in vitro-synthesized EMCV. (D) Effect of creatine phosphate concentrations on EMCV yields. Reaction mixtures containing the indicated concentrations of creatine phosphate and 3.4 mM MgCl2 were programmed with EMCV RNA for 20 h and assayed for infectivity as described above. In panels B, C, and D, programming was with 10 μg of EMCV RNA/ml. (E) EMCV RNA dose response of virus yield. Reaction mixtures were programmed with the indicated concentrations of EMCV RNA for 20 h at 32°C, treated with RNase A/T1, and assayed for infectivity following serial dilutions. In panels C, D, and E, the data are averages (with standard deviations from the means) of three independent titer determinations.
PV replication both in vivo and in vitro occurs in association with membranes (6, 11, 22, 37). Although not investigated in this study, it is likely that EMCV replication in vitro is also a membrane-dependent process. Indeed, we failed to show EMCV RNA replication and virus synthesis in a rabbit reticulocyte lysate (unpublished observations), which synthesizes EMCV proteins fairly well (25, 27) but is deficient in membranous structures compared to Krebs-2 S10 (5, 36, 38).
In summary, we have described an efficient system for picornavirus synthesis. This system might be applicable to the studies of replication of pathogens other than EMCV. Indeed, PV synthesis was readily demonstrated in a Krebs-2 extract under conditions optimized for EMCV replication (unpublished observations). The system eliminates an important problem associated with cell membrane permeability and provides a means to study the effects of a wide spectrum of macromolecules and drugs on virus-specific processes.
Acknowledgments
We thank Vadim Agol for fruitful discussions and Graham Belsham for critical reading of the manuscript. We also thank Sandra Perreault and Colin Lister for excellent technical assistance.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) to N.S., who is the recipient of a CIHR Distinguished Scientist Award and a Howard Hughes Medical Institute International Scholarship.
REFERENCES
- 1.Agol, V. A. 2002. Picornavirus genome: an overview, p. 127-148. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 2.Agol, V. I., K. M. Chumakov, T. M. Dmitrieva, and Y. V. Svitkin. 1980. Functional and structural studies on the genomes of picornaviruses, p. 319-370. In V. P. Skulachev (ed.), Soviet scientific reviews (biology), vol. 1. Harwood Academic Publishers GmbH, Chur, Switzerland.
- 3.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3DPol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 5.Bielinska, M., G. Rogers, T. Rucinsky, and I. Boime. 1979. Processing in vitro of placental peptide hormones by smooth microsomes. Proc. Natl. Acad. Sci. USA 76:6152-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burness, A. T. 1969. Purification of encephalomyocarditis virus. J. Gen. Virol. 5:291-303. [DOI] [PubMed] [Google Scholar]
- 8.Chumakov, K. M., and V. I. Agol. 1976. Poly(C) sequence is located near the 5′-end of encephalomyocarditis virus RNA. Biochem. Biophys. Res. Commun. 71:551-557. [DOI] [PubMed] [Google Scholar]
- 9.Dmitrieva, T. M., M. V. Shcheglova, and V. I. Agol. 1979. Inhibition of activity of encephalomyocarditis virus-induced RNA polymerase by antibodies against cellular components. Virology 92:271-277. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenfeld, E., and N. L. Teterina. 2002. Initiation of translation of picornavirus RNAs: structure and function of the internal ribosome entry site, p. 159-169. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 11.Etchison, D., and E. Ehrenfeld. 1981. Comparison of replication complexes synthesizing poliovirus RNA. Virology 111:33-46. [DOI] [PubMed] [Google Scholar]
- 12.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya, A. E., Y. V. Svitkin, Y. A. Kazachkov, and V. I. Agol. 1979. Encephalomyocarditis virus-specific polypeptide p22 is involved in the processing of the viral precursor polypeptides. FEBS Lett. 108:1-5. [DOI] [PubMed] [Google Scholar]
- 15.Harber, J. J., J. Bradley, C. W. Anderson, and E. Wimmer. 1991. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J. Virol. 65:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogan, B. L., and A. Korner. 1968. Ribosomal subunits of Landschutz ascites cells during changes in polysome distribution. Biochim. Biophys. Acta 169:129-138. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, R. J. 1986. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology 149:114-127. [DOI] [PubMed] [Google Scholar]
- 19.Jang, S. K., H. G. Kräusslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr, I. M., N. Cohen, and T. S. Work. 1966. Factors controlling amino acid incorporation by ribosomes from Krebs II mouse ascites-tumour cells. Biochem. J. 98:826-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 22.Molla, A., A. V. Paul, and E. Wimmer. 1993. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 67:5932-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morley, S. J., and R. J. Jackson. 1985. Preparation and properties of an improved cell-free protein synthesis system from mammalian liver. Biochim. Biophys. Acta 825:45-56. [DOI] [PubMed] [Google Scholar]
- 24.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmenberg, A. C., M. A. Pallansch, and R. R. Rueckert. 1979. Protease required for processing picornaviral coat protein resides in the viral replicase gene. J. Virol. 32:770-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pause, A., N. Méthot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelham, H. R. B. 1978. Translation of encephalomyocarditis virus RNA in vitro yields an active proteolytic processing enzyme. Eur. J. Biochem. 85:457-462. [DOI] [PubMed] [Google Scholar]
- 28.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plotch, S. J., O. Palant, and Y. Gluzman. 1989. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J. Virol. 63:216-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 31.Stanway, G., T. Hovi, N. J. Knowles, and T. Hyypiä. 2002. Molecular and biological basis of picornavirus taxonomy, p. 17-24. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 32.Svitkin, Y. V., and V. I. Agol. 1978. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett. 87:7-11. [DOI] [PubMed] [Google Scholar]
- 33.Svitkin, Y. V., A. E. Gorbalenya, Y. A. Kazachkov, and V. I. Agol. 1979. Encephalomyocarditis virus-specific polypeptide p22 possessing a proteolytic activity: preliminary mapping on the viral genome. FEBS Lett. 108:6-9. [DOI] [PubMed] [Google Scholar]
- 34.Svitkin, Y. V., H. Hahn, A. C. Gingras, A. C. Palmenberg, and N. Sonenberg. 1998. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 72:5811-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkin, Y. V., G. Krupp, and H. J. Gross. 1983. Heterogeneity of the poly(C) tract of encephalomyocarditis virus RNA. Bioorg. Khim. 9:1638-1643. [PubMed] [Google Scholar]
- 36.Svitkin, Y. V., V. N. Lyapustin, V. A. Lashkevich, and V. I. Agol. 1984. Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology 135:536-541. [DOI] [PubMed] [Google Scholar]
- 37.Takeda, N., R. J. Kuhn, C. F. Yang, T. Takegami, and E. Wimmer. 1986. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J. Virol. 60:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, P., and G. Blobel. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96:84-93. [DOI] [PubMed] [Google Scholar]