Abstract
The signal sequence trap method was used to isolate cDNAs corresponding to proteins containing secretory leader peptides and whose genes are expressed specifically in the salivary glands of the malaria vector Anopheles gambiae. Fifteen unique cDNA fragments, ranging in size from 150 to 550 bp, were isolated and sequenced in a first round of immunoscreening in COS-7 cells. All but one of the cDNAs contained putative signal sequences at their 5′ ends, suggesting that they were likely to encode secreted or transmembrane proteins. Expression analysis by reverse transcription–PCR showed that at least six cDNA fragments were expressed specifically in the salivary glands. Fragments showing a high degree of similarity to D7 and apyrase, two salivary gland-specific genes previously found in Aedes aegypti, were identified. Of interest, three different D7-related cDNAs that are likely to represent a new gene family were found in An. gambiae. Moreover, three salivary gland-specific cDNA fragments that do not show similarity to known proteins in the databases were identified, and the corresponding full length cDNAs were cloned and sequenced. RNA in situ hybridization to whole female salivary glands showed patterns of expression that overlap only in part those observed in the culicine mosquito A. aegypti.
One century after the discovery of the role of mosquitoes in the transmission of Plasmodium parasites, malaria still kills >1.5 million people every year, mostly in sub-Saharan Africa. Malaria control strategies are facing increasing complications because of the acquired resistance of Plasmodium parasites to antimalarial drugs and the growing insecticide-resistance in vector populations. In the last years, research efforts have been focusing on the development of tools for the genetic alteration of mosquito vectors, with the final goal to block the parasite life cycle within mosquitoes, making them incapable of transmitting malaria (1, 2). Recent reports of the successful transformation of the yellow fever mosquito Aedes aegypti (3, 4) make more imminent the prospect of genetically engineering mosquitoes that are refractory to the growth and development of the malaria parasite.
The development of transgenic mosquitoes refractory to malaria transmission requires not only the development of appropriate germ line transformation techniques but also the identification of potentially vulnerable steps in the life cycle of Plasmodium in mosquito and the isolation of promoters able to drive the tissue-specific expression of a chosen gene. Mosquito infection by malaria-causing parasites initiates with the ingestion of gametocytes during blood-feeding on an infected host. Fertilization takes place in the insect midgut, and the mobile zygotes, ookinetes, cross the peritrophic matrix, invade the midgut epithelium, and differentiate into oocysts. Sporozoites develop within the oocysts and then are released into the haemolymph. Haemocoel sporozoites selectively invade salivary glands, where they may be transmitted to a vertebrate host on the next blood feeding (5, 6). The strategy is to produce a transgenic mosquito in which parasite development is blocked at one, or better, two different stages: for example, at the midgut level, interfering with ookinete invasion or oocyst differentiation, and at the salivary gland level, blocking invasion of gland cells or the transmission of infective sporozoites from the glands.
We have focused our initial efforts on the salivary glands of Anopheles gambiae, the most efficient malaria vector in sub-Saharan Africa. The mechanisms by which haemocoel sporozoites locate, recognize, and penetrate salivary glands are presently unknown. However, several observations suggest that specific interactions between ligand(s) on the parasite surface and receptor(s) on the basal lamina and/or plasma membrane of gland cells may be involved in the invasion process. Specific recognition is suggested by the refractoriness to sporozoite invasion of salivary glands exhibited by some mosquito species, presumably because they lack the appropriate receptor (7). Also, the invasion-blocking activity exhibited by antisalivary gland antibodies and by some lectins suggests that sporozoites may interact with glycosylated salivary gland receptor molecules (8). The isolation and characterization of the sporozoite receptor(s) on the mosquito salivary glands may lead to a way to block these interactions: for example, mislocalizing the receptor by expressing its binding domain as a haemolymph-secreted peptide (1).
Salivary glands of mosquito vectors are also important because they express genes whose products are involved in the ability of mosquitoes to feed efficiently on blood. Several secreted factors able to counteract the host haemostatic response, such as vasodilators, inhibitors of platelet aggregation, and anticoagulants, have been identified by classical biochemical techniques in the salivary glands of mosquitoes and other haematophagous insects (9, 10). Six salivary gland-specific genes, four of which are expressed specifically in the female glands, have been isolated and characterized from the mosquito Ae. aegypti (refs. 11 and 12; B. T. Beerntsen, D. E. Champagne, J. Coleman, Y. A. Campos, and A.A.J., unpublished work).
We used a cloning procedure suited to the isolation of sequences that encode secreted and membrane-bound proteins to screen a 5′ end-enriched cDNA library from the salivary glands of the mosquito An. gambiae, and we report here on the identification of fifteen cDNA fragments. All of them, with one exception, appear to have a signal peptide fused to the reporter gene and are likely to encode secreted or membrane-linked proteins. At least six of these cDNA fragments are expressed specifically in the glands, and four of them appear to be female-specific because the corresponding cDNAs cannot be detected in males. These represent the only salivary gland-specific genes identified so far in An. gambiae.
MATERIALS AND METHODS
Mosquito Colony and Tissue Dissections.
The An. gambiae strain used for dissections and polytene chromosome preparations was the homokaryotypic GASUA reference strain (Xag, 2R, 2La, 3R, 3L) selected in our insectary in 1989 from a 2Rd-2La polymorphic colony that, in turn, originated from adult females collected in the village of Suakoka Liberia in 1986. Salivary glands from 0- to 3-day-old An. gambiae adult females were dissected in PBS and were kept on ice for a maximum of 1 hour. After a low speed centrifugation step, the medium was removed, and the glands were frozen in liquid nitrogen and were stored at −80°C. Anterior portions of the thorax containing the salivary glands were dissected from 4-day-old females by a surgical blade, were placed in an Eppendorf tube on dry ice, and were stored at −80°C.
RNA Purification.
General nucleic acids manipulations were performed according to standard procedures (13, 14) if not otherwise specified. Poly(A)+ RNA from the salivary glands of 0- to 3-day-old females and from thoracic tissues of 4-day-old females was purified by, respectively, Dynabeads oligo(dT)25 (Dynal, Oslo) and the Oligotex Direct mRNA Kit (Qiagen, Chatsworth, CA).
cDNA Library Construction.
The thoracic cDNA library was constructed by using 7 μg of poly(A)+ RNA and the ZAP Express kit (Stratagene). A PCR-based method was used for the screening of the thoracic cDNA library (15). The signal sequence-enriched cDNA library was constructed as described (16) with minor modifications. First-strand cDNA was synthesized by using 5 ng of random hexamers and ≈1 μg of salivary gland poly(A)+ RNA. Reaction conditions for the addition of 10–20 deoxycytosine residues to the 3′ end of the first strand cDNA were determined empirically by using a 5′ end-labeled 25-mer oligonucleotide. Second strand cDNA was synthesized by the ESLG primer containing an EcoRI linker and an oligo(dG) (5′-GCGGCCGCGAATTCTGACTAACTGAC(G)17-3′). After second strand synthesis, the SacI adapters were ligated, and fragments in the range of 200 to 600 bp were size-selected by gel electrophoresis. These fragments were amplified (25 cycles of 94°C, 30 sec; 49°C, 1 min; and 72°C, 3 min) by using the primers LLHES (5′-GAGGTACAAGCTTGATATCGAGCTCGCGG-3′) and ESL (5′-GCCGCGAATTCTGACTAACTGAC-3′). Amplified cDNA fragments were purified by gel electrophoresis, were digested with SacI and EcoRI, and were cloned directionally into the EcoRI- and SacI-digested pcDL-SRα-Tac(3′) vector (kindly provided by T. Honjo, Kyoto University). An aliquot of the cDNA library was used to transform XL1-Blue super-competent cells (Stratagene).
Screening and Immunostaining.
Forty-nine individual colonies were transferred from the master plate to an agar plate in a matrix format of seven rows by seven columns and were designated as a pool. Five-hundred nanograms of plasmid DNA preparation from each pool were used for transfection of 105 COS-7 cells by the LipofectAmine Reagent (GIBCO/BRL). Forty-eight to seventy-two hours after transfection, the cells were stained with the fluorescein isothiocyanate-conjugated anti-human interleukin 2 receptor (Anti-Tac) mAb (Dako), and the immunofluorescent positive pools were identified microscopically. Positive pools were divided in 14 smaller pools and were rescreened, leading to the identification of single positive clones. Pools were judged as positive or negative by comparison to transfections with the positive and negative control plasmids pcDL-SRα-hGCSF(5′)-Tac(3′) and pcDL-SRα-hRAR(5′)-Tac(3′) (16).
Signal Peptide Prediction and Sequence Comparison.
cDNA clones were sequenced by the T3, T7, SRA (5′-TTTACTTCTAGGCCTGACG-3′), TAC (5′-CCATGGCTTTGAATGTGGCG-3′), and SLG9 (5′-GACTAACTGACGGGGGGGGG-3′) primers. Signal peptide prediction analysis was performed by using the psort worldwide web server (http://psort.nibb.ac.jp/). Sequence comparison was performed by using the wisconsin package 9.1 (Genetics Computer Group, Madison, WI) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the European Bioinformatics Institute (http://www.ebi.ac.uk/ebi_home.html) worldwide web servers.
In Situ Hybridization to Polytene Chromosomes.
Polytene chromosome spreads were obtained from half-gravid female mosquitoes (17), and probe-labeling and in situ hybridization procedures were performed as described (18). Sites of hybridization were localized to specific bands within numbered and lettered subdivisions of the polytene chromosome map (M.C., A. Sabatini, M. A. Di Deco, and V. Petrarca, unpublished work) revised from that of Coluzzi and Sabatini (19).
Expression Analysis by Reverse Transcription (RT)–PCR.
About 100 ng of DNase-treated total RNA (DNAaseI RNase-free, Boehringer Mannheim) was used for the RT-PCR amplifications with gene-specific primers and the SuperScript one-step RT-PCR system (GIBCO/BRL). After the reverse transcription step (50°C for 30 min) and heat-inactivation of the reverse transcriptase (2 min at 94°C), 35 cycles were performed as follows: 94°C, 30 sec; 55°C, 30 sec; and 72°C, 45 sec. The sequence of the gene-specific oligonucleotide primers is not reported because of space limitations and will be made available on request.
In Situ Hybridization to Salivary Glands.
The cDNA clones of interest were subcloned as SacI-EcoRI fragments into the polylinker of the plasmid vector pBCKS(+) (Stratagene). Antisense and sense RNA probes were synthesized by using digoxigenin-substituted ribonucleotides. Salivary glands were dissected from 3- to 7-day-old adult female mosquitoes and were fixed in 4% paraformaldehyde for 30 min in PBS supplemented with 67 mM EGTA. The glands were dehydrated in four washes of methanol followed by five washes in 100% ethanol and were stored at −20°C in ethanol for several days before use. Glands were postfixed for 30 min in 5% formaldehyde in PBS with 0.1% Tween 20, were digested for 10 min in 33 μg/ml proteinase K, were rinsed twice in PBS with 0.1% Tween 20 to stop proteinase digestion, and, finally, again were postfixed for 30 min in 5% formaldehyde. Hybridizations were carried out in the presence of formamide, and an alkaline wash with 0.1 M Tris (pH 9.5) was used to remove background. Preparations were visualized and photographed with compound microscopy.
RESULTS
Signal Sequence Trap (SST).
To identify cDNAs encoding secreted and membrane-bound proteins specifically expressed in the salivary glands of the malaria vector An. gambiae, we used the SST method. The SST technique originally was developed by Tashiro et al. (16) to take advantage of the observation that secreted and transmembrane proteins are targeted to the secretory pathway by amino-terminal signal peptides that are encoded by signal sequences, usually located within 400 base pairs of the 5′ termini of the mRNAs. In the SST system, a 5′ end-enriched cDNA expression library is cloned directionally between a promoter that confers high level of expression in COS-7 cells and a “reporter” gene, the α chain of the human interleukin 2 receptor (Tac). cDNAs that are cloned in-frame with Tac and contain signal sequence produce fusion proteins that can be expressed on the surface of transfected COS-7 cells and are easily detectable by immunostaining with an anti-Tac monoclonal antibody.
Salivary glands dissected from An. gambiae females were used to construct a 5′ end-enriched cDNA expression library. Fragments from 200 to 600 bp in length were cloned directionally in the pcDL-SRα-Tac(3′) vector (16). Twenty-two pools (each one consisting of 49 clones) were screened initially by transfection in COS-7 cells, and 21 of them were positive after immunostaining with the anti-Tac mAb. Six pools were chosen randomly, were divided into subpools, and were rescreened in COS-7 cells, leading to the isolation of 18 immunofluorescent positive clones. Sequence analysis showed that 15 cDNA fragments were unique and that their size ranged from 164 to 523 bp in length (Table 1). Three additional cDNA fragments represented shorter versions of the clones aB1, cE5, and iC5 and were not characterized further. The putative peptide sequences were deduced from the ORFs fused to Tac, assuming that the first methionine was the start codon. Scores obtained by the von Heijne (20) and McGeoch (21) methods are reported in Table 1. At least 12 of the 15 cDNA fragments have large positive scores with both methods, suggesting that they likely encode secreted or membrane-bound proteins.
Table 1.
Properties of the cDNAs isolated by SST
| Clone | Accession no.* | Size† | Similarity‡ | Signal scores§ | Division¶ |
|---|---|---|---|---|---|
| aA5 | Y17688 | 523 | 3.97/7.44 | 1D | |
| aB1 | Y17689 | 366 | 0.8/20.7 | ||
| bB2 | Y17699 | 299 | 7.96/11.99 | 25A | |
| bD3 | AJ00037 | 316 | 6.63/7.66 | ||
| cB1 | Y17700 | 406 | 4.24/3.85 | 1D | |
| cE5 | Y17717 | 382 | 5.58/14.25 | ||
| cF1 | Y17690 | 164 | 3.94/11.46 | 25A | |
| cF3 | Y17701 | 262 | Apyrase 5′ nucleotidase | 5.28/6.96 | 44D-45A |
| cF6 | Y17702 | 332 | AG5-AG3 | 8.12/9.79 | |
| dB1 | Y17703 | 400 | D7 | 3.48/8.72 | 30B |
| dF2 | Y17704 | 396 | 8.57/6.83 | 1D | |
| dG5 | Y17705 | 375 | Opsin | −0.61/8.93 | 7A |
| iB6 | AJ000036 | 479 | D7 | 4.1/7.89 | 30A |
| iC5 | AJ000035 | 502 | D7 | 8.28/7.22 | 30B |
| iC6 | AJ000034 | 320 | Apyrase 5′ nucleotidase | −0.96/2.86 | 41A |
European Molecular Biology Laboratory accession numbers.
Length of the cDNA fragments expressed in base pairs.
Amino acid similarities to known sequences deposited in GenBank or European Molecular Biology Laboratory databases.
Scores obtained by the signal peptide prediction methods of von Heijne and McGeoch.
Locations on polytene chromosomes as obtained by in situ hybridization.
Only half of the clones gave significant similarity to previously identified proteins when the sequences of the cloned cDNA fragments were used for database searches (Table 1). Five different cDNAs showed similarity to D7 and apyrase, two genes that are specifically expressed in the female salivary glands of the yellow fever mosquito Ae. aegypti (22–24). The cDNA fragment dG5 encoded an opsin; the cF6 clone shared similarity with venom allergens from ants and wasps (25), and we will refer to it as gVAG.
Anopheles gambiae Apyrase and 5′-Nucleotidase.
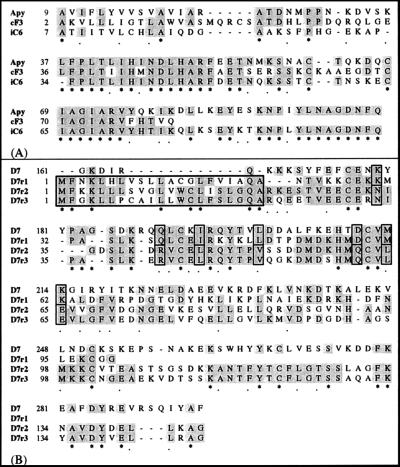
The clones cF3 and iC6 both showed similarity to the Ae. aegypti apyrase and to several members of the 5′-nucleotidase family. An amino acid alignment of the An. gambiae cF3 and iC6 cDNA fragments with the Ae. aegypti apyrase is shown in Fig. 1A. The two clones show, respectively, a 45 and 56% identity and a 70 and 67% similarity to the Ae. aegypti gene products. The clones also share high similarity, 50 and 65%, respectively, with members of the 5′-nucleotidase family (data not shown).
Figure 1.
Alignment of the Ae. aegypti apyrase (A) and D7 (B) proteins to the translated An. gambiae cDNA fragments identified by the SST. Asterisks mark identities in all of the sequences. Dots identify conservative substitutions. Identities in at least two of the aligned sequences are shaded. In B, the putative signal peptides of the D7-related cDNA fragments are boxed with a thick line.
D7-Related.
The cDNA fragments dB1, iB6, and iC5 show similarity to D7, a secreted protein that is expressed abundantly and exclusively in the salivary glands of Ae. aegypti adult females (22). The D7 function is unknown, but the stage-, sex-, and tissue-specific patterns of expression suggest that it may have a role in blood feeding. Because of the sequence similarity to the Ae. aegypti gene, we propose to refer to these cDNAs as D7-related and to identify them individually as D7r1 (dB1), D7r2 (iB6), and D7r3 (iC5). Fig. 1B shows an alignment of the deduced peptide sequence of these three partial cDNAs with the Ae. aegypti D7. D7r1, with a 35% identity and a 52% similarity, is the most similar to D7 whereas D7r2 and D7r3 show, respectively, a 23 and 26% identity (44 and 45% similarity) to the Ae. aegypti protein. D7r2 and D7r3 share 62% identical amino acid residues between themselves and are by far the most closely related to each other. These three An. gambiae cDNA clones hybridize relatively closely to chromosomal positions on the right arm of the third chromosome in the divisions 30A (D7r2) and 30B (D7r1 and D7r3) (Table 1).
cDNA Clones Expressed Specifically in the Salivary Glands.
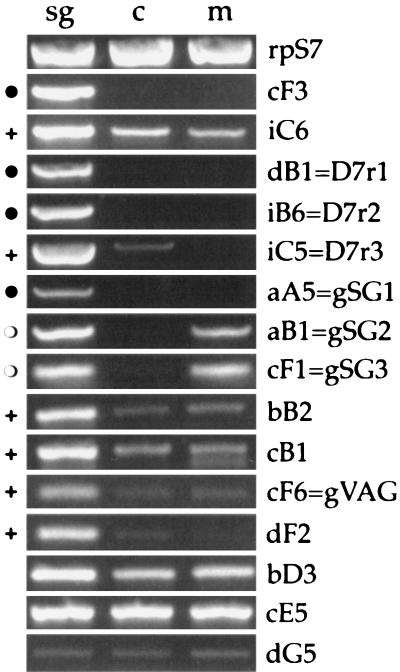
To evaluate the tissue-specific expression of the 15 cDNAs identified by SST, we amplified the corresponding messenger RNAs by RT-PCR using, as template, total RNA extracted from female salivary glands and carcasses (adult females with salivary glands removed) and from adult males. As summarized in Fig. 2, at least six cDNAs were specifically expressed in the salivary glands, and five other clones may be expressed at higher level in female glands than in other tissues or in males.
Figure 2.
RT-PCR analysis of the 15 An. gambiae cDNA fragments. sg, salivary glands of adult females; c, carcasses (adult females with salivary glands removed); m, adult males. The different cDNA clones are indicated on the right side. rpS7, ribosomal protein S7. The specificity of expression is annotated as follows: ●, specifically in female salivary glands; ○, in female salivary glands and males; +, enriched in female salivary glands.
RT-PCR analysis clearly showed that cF3 is specifically expressed in female glands whereas iC6 is detectable also in carcasses and in adult males (Fig. 2), suggesting that cF3 is most likely the An. gambiae apyrase whereas iC6 may represent a 5′-nucleotidase. The three D7-related cDNA fragments all showed high levels of expression in female salivary glands. However, although D7r1 and D7r2 are specifically expressed in female glands, D7r3 is detectable also at a very low level when RNA from carcasses is used as template (Fig. 2).
Finally, three cDNAs, aA5, aB1, and cF1, were salivary gland-specific (Fig. 2). The aA5 cDNA was detectable only when female salivary gland RNA was used as template whereas aB1 and cF1 were detectable also in males. The simplest interpretation is that aA5 is expressed only in female glands and aB1 and cF1 are expressed also in male glands. As previously mentioned, these three cDNA fragments did not show significant similarity to any other protein in the databases. We screened a thoracic cDNA library and cloned the full length cDNAs corresponding to aA5, aB1, and cF1. They are, respectively, 1,372, 436, and 721 bp in length and encode putative proteins of 401, 114, and 189 amino acids. We propose to refer to these cDNAs as gSG1 (aA5), gSG2 (aB1), and gSG3 (cF1), for gambiae salivary genes 1, 2, and 3 (Fig. 3). In all three cases, the additional sequence information did not allow further insight into their possible function. gSG2 is a small protein, rich in glycine and phenylalanine, with a highly hydrophobic profile (data not shown). The gSG3 putative protein contains a serine-rich domain toward the amino terminus and, at the carboxy terminus, a threonine-rich region composed of repeats of the motif EEATTT or closely related hexapeptides (Fig. 3). Other, less obvious, repeats are found between the serine- and threonine-rich regions. The functional significance of these motifs is still to be elucidated.
Figure 3.
Peptide sequences deduced from the gSG1, gSG2, and gSG3 cDNA clones are shown from the top to the bottom. The putative signal peptides at the amino terminus are underlined. In the gSG3 sequence, the serine-rich region is boxed and shaded, the motif RP(P/W)(F/W) is underlined by a thick line, and the repeated motifs at the carboxy terminus are boxed.
Hybridization in Situ to Salivary Glands.
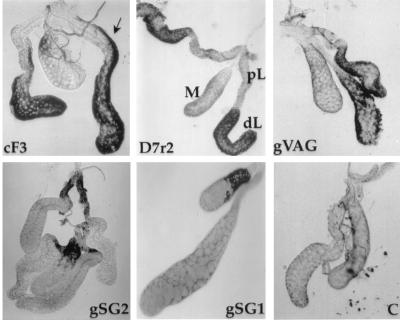
A selected set of cDNAs, which includes the six salivary gland-specific clones, were expressed as antisense and sense RNA probes and were hybridized in situ to female salivary glands to determine the localization of expression of their corresponding genes (Table 2). In all cases, signals were obtained only with antisense RNA probes and not with control sense RNA probes. The apyrase-like cDNA cF3 hybridizes to mRNA localized in the distal–lateral lobes with the signal extending along the length of the proximal–lateral lobes (Fig. 4). In addition, hybridization was seen with this probe to mRNA in some of the proximal regions of the medial lobe. The three D7-related cDNAs hybridize to mRNA in the distal-lateral lobes, with occasional hybridization to the medial lobe. The gVAG probe also hybridizes to the distal-lateral lobes of the female glands. The cDNA gSG1 is peculiar because it is specifically expressed in female glands and hybridizes to mRNA in subsets of cells in the medial lobes of female glands. Finally, two cDNAs, gSG2 and gSG3, with sequences that are expressed in both males and females, are expressed specifically in the proximal–lateral lobes of female glands. The gSG2 cDNA also hybridizes occasionally to the proximal medial lobe.
Table 2.
Expression patterns of genes in adult female salivary glands
| Clone | n* | Expression product† | Expression pattern |
|---|---|---|---|
| cF3 | 3 | apyrase-like | distal–lateral lobes; strips in proximal–lateral lobes; weak in proximal–medial lobe; |
| D7rl | 7 | D7-related | weak, distal–lateral, and medial lobes; |
| D7r2 | 3 | D7-related | distal–lateral lobes; |
| D7r3 | 7 | D7-related | distal–lateral lobes; |
| gVAG | 10 | venom allergen | distal–lateral lobes; |
| gSG1 | 12 | novel‡ | variable in the proximal–medial lobe; |
| gSG2 | 7 | novel‡ | proximal–lateral and proximal–medial lobes; |
| gSG3 | 6 | novel‡ | proximal–lateral lobes; |
Number of salivary glands with a positive signal.
The nature of the expression product is based on the primary aminoacid sequence.
Novel implies no known similarity to other protein sequences deposited in the databases.
Figure 4.
Hybridizations in situ to adult female salivary glands with antisense probes as indicated. C, control glands hybridized with a sense probe; pL, proximal-lateral lobes; dL, distal-lateral lobes; M, medial lobe. The arrow points to the region of the proximal–lateral lobes that hybridizes to the apyrase-like cF3 probe. The gSG2 probe hybridizes to the proximal–lateral lobes (upper gland) and to the proximal–medial lobe (lower gland). Note limited signal over the medial lobe in the hybridization with the gSG1 probe.
DISCUSSION
We constructed a 5′ end-enriched cDNA library from the salivary glands of the malaria vector An. gambiae and used the SST technique to screen for cDNAs coding for secreted and type I membrane proteins. The SST method worked efficiently with the salivary glands, and 6% of the clones analyzed were positive. Taking into consideration that only one cDNA fragment in every three is expected to be cloned in frame with the reporter gene, the average number of cDNAs containing a signal peptide is estimated to be 18%. This number is relatively high but perhaps not surprising for a tissue involved in intense secretory activity. Moreover, the 5′ end-enriched salivary gland cDNA library was shown to be complex, with 15 of the 18 clones isolated being unique. In addition, insect signal peptides were efficiently recognized in COS-7 cells, as shown both by the signal peptide prediction methods and by the sequence comparison analysis. At least 6 of the 15 cDNA clones identified were expressed specifically in the salivary glands.
Two cDNA fragments showed similarity to the Ae. aegypti apyrase and to several members of the 5′-nucleotidase family (24). Apyrase activity is generally rare in animal tissues, and salivary glands of haematophagous insects are the richest source known. The apyrase is a secreted protein that hydrolizes ATP and ADP to AMP and Pi. Its function is to help mosquitoes blood-feed efficiently by inhibiting the ADP-induced platelet recruitment and aggregation. 5′-nucleotidases are membrane-linked ubiquitous proteins that hydrolyze extra cellular nucleotides to nucleosides making them permeable to cell membranes. It has been suggested that the apyrase, a secreted protein, evolved by gene duplication and subsequent divergent evolution from the membrane-bound 5′-nucleotidase by loss of a carboxy terminal domain involved in membrane anchoring (24). We were able to show by RT-PCR that one of these cDNAs, cF3, is expressed only in the female salivary glands of An. gambiae whereas the other one, iC6, is also detectable, at a lower level, when RNA from carcasses or from adult males was used as template. These patterns of expression strongly suggest that cF3 may be the An. gambiae apyrase and that iC6 may be a 5′-nucleotidase. The identification of cDNA fragments corresponding to these genes in An. gambiae offers the opportunity to investigate the evolutionary relationship between these two genes within the same organism.
We also have identified three members of a family of genes related to the Ae. aegypti D7. Two cDNAs, D7r1 and D7r2, were expressed specifically in female salivary glands whereas the third, D7r3, also was detectable at a very low level when RNA from carcasses was used as template. It is possible that D7r3 is expressed in female glands and, at a much lower level, in some other female tissue. However, because of the apparently higher abundance of the corresponding mRNA as compared with D7r1 and D7r2 (Fig. 2), we cannot rule out the possibility that the weak band detectable from carcass RNA preparations is attributable to a small contamination with gland tissue. In any case, the redundancy in copy number of these D7-like genes suggests that they must play some essential role in An. gambiae, most probably in connection with blood feeding.
The Ae. aegypti D7 gene is encoded in five exons separated by small introns with the signal peptide that is encoded by exon 1 (22). As shown in Fig. 1B, the An. gambiae D7-related cDNA fragments can be aligned for their entire length to the carboxy terminal half of the D7 protein with the similarity starting immediately after the signal peptides and involving a region corresponding to exons 4 and 5. If their function is homologous to the one that D7 plays in Ae. aegypti, then the domain(s) encoded by exons 4 and 5 of D7 must be especially important. The cloning of the full length cDNAs and their molecular characterization may help elucidate the function and the structure of this intriguing protein family.
The efficacy of the SST technique as applied to the salivary glands of An. gambiae is confirmed further by the identification of the three salivary gland-specific cDNAs gSG1, gSG2, and gSG3. Two of them are expressed both in male and female glands, but their sequence is unrelated to Maltase-like I (26) and Amylase I (27), the only genes previously identified as expressed in both male and female Ae. aegypti salivary glands.
The cDNA clones bB2, cB1, gVAG, and dF2 showed a pattern of expression that may be compatible with higher expression in female glands than in other tissues or in males. The gVAG cDNA fragment showed similarity to allergens from ants and wasps (25). These allergens belong to a family of secreted proteins whose functions are still poorly understood and is sometimes named the CAP family (28) because it counts members from mammals (cysteine-rich secretory proteins), insects (AG5/AG3), and plants (pathogenesis-related proteins). Finally, the dG5 cDNA fragment, which shows very high similarity to opsins, most likely represents a cloning artifact caused by tissue contamination that occurred during dissection because opsin genes are characteristically expressed in the eye.
Hybridization of cDNA probes to whole female salivary glands produced a number of interesting results. In general, the pattern of expression was consistent with what is seen in the culicine mosquito, Ae. aegypti (11). The cDNAs corresponding to genes expressed both in males and females, gSG2 and gSG3, are expressed in the proximal–lateral lobes. This expression pattern reflects that of two genes, Maltase-like I (26) and Amylase I (27), that are expressed in both male and female Ae. aegypti and whose products most likely are involved in sugar feeding. The proximal–lateral lobes of female salivary glands are postulated to overlap functionally the complete male salivary glands (11). Expression of the gene corresponding to gSG2 in the proximal–medial lobe was not anticipated based on its sex-specific expression and may reflect some fundamental differences in the organization of the salivary glands between anopheline and culicine mosquitoes.
Genes expressed specifically in the female salivary glands of Ae. aegypti are expressed only in the distal–lateral and medial lobes (11). Consistent with this observation, the three cDNAs D7r1, D7r2, and D7r3, related by sequence structure to the D7 gene of Ae. aegypti (22), are expressed in the distal–lateral lobes of the female salivary glands. However, we could detect clear signal corresponding to expression in the medial lobe with the D7r1 cDNA only. Similarly, the cDNA cF3, with a conceptual translation product similar to the apyrase gene of Ae. aegypti (23, 24), also shows expression in the distal-lateral lobes and not in the medial lobe. In addition, it appears to be expressed in some regions of the proximal–lateral lobes. This pattern is unexpected from the results of Ae. aegypti, but, considering the long evolutionary separation of the families to which these species belong, perhaps it is not so unlikely that the biology and gene expression in the glands is different.
The expression pattern of the gSG1 cDNA is remarkable in that it is expressed only in the medial lobe of female salivary glands. The sialokinin gene of Ae. aegypti was shown by hybridization in situ to be expressed only in the medial lobe (B. T. Beerntsen, D. E. Champagne, J. Coleman, Y. A. Campos, and A.A.J., unpublished work). However, there is no similarity between the conceptual translation product of gSG1 and the sialokinin gene. Furthermore, we were never able to observe hybridization of the gSG1 probe to the whole medial lobe. Although it is possible that gene expression in the medial lobe cannot be fully evaluated because it is refractory to our hybridization technology, it is possible that only variable but distinct regions of the medial lobe actually express the corresponding gene.
In conclusion, we applied the SST method to the salivary glands of An. gambiae, and we were able to identify at least six genes specifically expressed in this organ. These represent the first salivary gland-specific cDNAs identified from the most efficient malaria vector in sub-Saharan Africa. The isolation of the corresponding genes should provide tissue-specific promoters that may be helpful for the development of future vector control strategies. The efficiency of the trapping method and the low number of clones that were screened suggest that a more exhaustive screening of the library should allow for the identification of several other tissue-specific genes. This will certainly result in a better knowledge of the salivary gland functions and will allow a deeper understanding of the molecular interactions occurring both between Plasmodium and mosquito and between mosquito and the vertebrate host.
Acknowledgments
We thank Prof. Christos Louis (Institute of Molecular Biology and Biotechnology, Crete, Greece) for the suggestions, the discussions, and the encouragement since the early stages of this work; Prof. F. C. Kafatos (European Molecular Biology Laboratory, Heidelberg) for helpful comments on an earlier version of the manuscript; Prof. Tasuku Honjo (Kyoto University) for kindly providing the pcDL-SRα-hRAR(5′)-Tac(3′) and pcDL-SRα-hGCSF(5′)-Tac(3′) plasmid vectors and the detailed protocols for the SST technique; Dr. Manlio Di Cristina for the help with cell culture; Gianni Pietrangeli and Maria Calzetta for the expert technical assistance; and L. Olson for helping in typing the manuscript. B.A. and A.d.T. were supported by postdoctoral fellowships of the Fondazione “Istituto Pasteur-Cenci Bolognetti”. G.D. was supported by a Training and Mobility of Researchers postdoctoral fellowship. This work was supported by the European Union Grant ERBFMRXCT960017 to M.C. and grants from the Burroughs-Wellcome Fund and John D. and Catherine T. MacArthur Foundation to A.A.J.
ABBREVIATIONS
- RT
reverse transcription
- SST
signal sequence trap
Footnotes
References
- 1.Collins F H, Paskewitz S M. Annu Rev Entomol. 1995;40:195–219. doi: 10.1146/annurev.en.40.010195.001211. [DOI] [PubMed] [Google Scholar]
- 2.O’Brochta D A, Atkinson P W. Parasitol Today. 1997;13:99–104. doi: 10.1016/s0169-4758(97)01006-5. [DOI] [PubMed] [Google Scholar]
- 3.Jasinskiene N, Craig J, Coates C J, Benedict M Q, Cornel A J, Rafferty C S, James A A, Collins F H. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Touray M G, Warburg A, Laughinghouse A, Krettli A U, Miller L H. J Exp Med. 1992;175:1607–1612. doi: 10.1084/jem.175.6.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warburg A, Miller L H. Parasitol Today. 1991;7:179–181. doi: 10.1016/0169-4758(91)90127-a. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg R. Am J Trop Med Hyg. 1985;34:687–691. doi: 10.4269/ajtmh.1985.34.687. [DOI] [PubMed] [Google Scholar]
- 8.Barreau C, Touray M, Pimenta P F, Miller L H, Vernick K D. Exp Parasitol. 1995;81:332–343. doi: 10.1006/expr.1995.1124. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro J M C. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 10.Stark K R, James A A. In: The Biology of Disease Vector. Beaty B J, Marquard W C, editors. Niwot, Colorado: University Press; 1996. pp. 333–348. [Google Scholar]
- 11.James A A. Bull Inst Pasteur (Paris) 1994;92:133–150. [Google Scholar]
- 12.Stark K R, James A A. J Biol Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsh E F, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Ausubel F M, Brent R, Kingston R F, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 15.Israel D I. Nucleic Acids Res. 1993;21:2627–2631. doi: 10.1093/nar/21.11.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tashiro K, Hideaki T, Heilker R, Shirozu M, Nakano T, Honjo T. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Collins F H. Insect Mol Biol. 1994;3:41–47. doi: 10.1111/j.1365-2583.1994.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 18.della Torre A, Favia G, Mariotti G, Coluzzi M, Mathiopoulos K D. Genetics. 1996;143:1307–1311. doi: 10.1093/genetics/143.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coluzzi M, Sabatini A. Parassitologia. 1967;9:73–88. [Google Scholar]
- 20.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch D J. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 22.James A A, Blackmer K, Marinotti O, Ghosn C R, Racioppi J V. Mol Biochem Parasitol. 1991;44:245–254. doi: 10.1016/0166-6851(91)90010-4. [DOI] [PubMed] [Google Scholar]
- 23.Smartt C T, Kim A P, Grossman G L, James A A. Exp Parasitol. 1995;81:239–248. doi: 10.1006/expr.1995.1114. [DOI] [PubMed] [Google Scholar]
- 24.Champagne D E, Smartt C T, Ribeiro J M C, James A A. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman D R. J Allergy Clin Immunol. 1993;91:71–78. doi: 10.1016/0091-6749(93)90298-t. [DOI] [PubMed] [Google Scholar]
- 26.James A A, Blackmer K, Racioppi J V. Gene. 1989;75:73–83. doi: 10.1016/0378-1119(89)90384-3. [DOI] [PubMed] [Google Scholar]
- 27.Grossman G L, James A A. Insect Mol Biol. 1993;1:223–232. doi: 10.1111/j.1365-2583.1993.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber M C, Karlo J C, Kovalick G E. Gene. 1997;191:135–141. doi: 10.1016/s0378-1119(97)00010-3. [DOI] [PubMed] [Google Scholar]