Abstract
Intercellular adhesion molecule 1 (ICAM-1) functions as the cellular receptor for the major group of human rhinoviruses, being not only the target of viral attachment but also the mediator of viral uncoating. The configurations of HRV3-ICAM-1 complexes prepared both at 4°C and physiological temperature (37°C) were analyzed by cryoelectron microscopy and image reconstruction. The particle diameters of two complexes (with and without RNA) representing uncoating intermediates generated at 37°C were each 4% larger than that of those prepared at 4°C. The larger virus particle arose by an expansive movement of the capsid pentamers along the fivefold axis, which loosens interprotomer contacts, particularly at the canyon region where the ICAM-1 receptor bound. Particle expansion required receptor binding and preceded the egress of the viral RNA. These observations suggest that receptor-mediated uncoating could be a consequence of restrained capsid motion, where the bound receptors maintain the viral capsid in an expanded open state for subsequent genome release.
Human rhinoviruses (HRVs), a genus of the picornavirus family, are the major cause of the common cold in humans (25). They constitute a group of approximately 100 serotypes, all containing a single-stranded RNA genome encapsulated into an icosahedral capsid. Crystal structures of several rhinovirus serotypes demonstrated that the capsid is made of 60 protomers, each having a copy of four structural proteins (VP1 to VP4) (10, 24, 29). The three major proteins (VP1, VP2, and VP3) build the capsid shell. They have a central “jelly roll” structure, with their N-terminal regions being located close to the internal VP4 and RNA genome. Biochemical experiments showed that VP4 and internal N-terminal residues of VP1 were accessible to immobilized trypsin in HRV serotype 14 (HRV14), revealing that they became exposed spontaneously outside of the capsid (17). These findings demonstrated that the capsid shell of HRV is a dynamic, breathing entity.
The most highly conserved structural feature among HRVs is a surface depression around the fivefold axes, the so-called canyon (25). The canyon is composed of the north wall (built by VP1) around the fivefold axis and the south wall (built mainly by VP2 and VP3). While the most accessible residues along the canyon walls are hypervariable, the less exposed residues in the canyon floor are conserved and used for receptor binding, as the strategy allowing the virus to escape antibody neutralization (23). There is a hydrophobic pocket in VP1 directly underneath the canyon floor. The pocket appears empty in HRV3 and HRV14 (29) but is filled with electron density resembling a fatty acid (pocket factor) in the structures of HRV2 and HRV16 (10, 27). The pocket factor stabilizes the virion during its cell-to-cell transit (19). Hydrophobic antiviral compounds (WIN compounds) were found to bind to the VP1 pocket and to inhibit capsid breathing (17).
The major group of HRVs use the canyon region for binding to its cellular receptor, intercellular adhesion molecule 1 (ICAM-1) (16). ICAM-1 is a membrane protein with five immunoglobulin-like domains in the extracellular region. The critical virus binding epitopes are present at the tip of the N-terminal first domain (D1) of ICAM-1 (16), which penetrates into the rhinovirus depressive canyon (2, 16). The low accessibility of the receptor binding site in the major group of HRVs correlates with a low virus-receptor association rate (28). Binding of ICAM-1 to HRV3 and HRV14 (receptor-sensitive serotypes) triggers the release of the RNA genome from the capsid (uncoating), but this does not occur with other members of the major group of HRVs (13, 28). Receptor-mediated uncoating has been reported for the major group of HRV and poliovirus, showing that these viruses use their receptors for both virus attachment and uncoating (13, 15, 21). Indeed, entry of poliovirus and receptor-sensitive rhinoviruses (HRV14) can occur without endosomal acidification (20, 26).
The structural changes mediated by the receptor in the virus particles lead to the release of the RNA genome from the capsid for penetration into the cell cytoplasm (13, 15, 21). It has also been proposed that the RNA will move through a membrane pore generated by the externalized N-terminal region of VP1 and VP4 during the uncoating process (3). Externalization of those polypeptides has been supported by two structural analyses of high-temperature (>50°C)-treated poliovirus and a minor-group rhinovirus, which have provided new insights into picornavirus uncoating (3, 12). However, no structural analysis of receptor-mediated uncoating has been reported so far, in contrast to the well-documented biochemistry of this process (5, 13, 15). The precise mechanism by which a cellular receptor triggers the exit of the viral genome is currently unknown. To gain new insight into this process, we applied cryoelectron microscopy (cryo-EM) to a well-defined model, HRV3 and its ICAM-1 receptor (5, 13). Our results indicate that the receptor catalyzes uncoating by locking the viral capsid in an expanded open state for subsequent RNA release.
MATERIALS AND METHODS
Preparation of receptor-virus complexes and uncoating intermediates.
The receptor-HRV complexes were prepared by incubating purified virions with a two-domain fragment of ICAM-1 overnight at 4°C (28). Unbound receptor was removed via filtration through a 1-ml Superose-12 column in 20 mM Tris-HCl buffer (pH 8.0) with 10 mM MgCl2 and 50 mM NaCl. Fractions containing the complexes were determined by measuring the absorbance at 260 nm and were used to prepare vitrified specimens for cryo-EM. After overnight incubation, the H3RC sample was directly subjected to the gel filtration step at 4°C; the other complexes were further incubated at 37°C for 15 and 60 min for HRV3-ICAM-1 and HRV16-ICAM-1 complexes, respectively. These samples were then chilled on ice for 5 min before removal of unbound receptors at 4°C.
Structure determination and analysis.
A Philips CM120 microscope equipped with a Gatan cryotransfer system was used to photograph the HRV-receptor complex. The micrograph then was digitized at 3.1 Å/pixel on a SCAI scanner. Images of individual particles were boxed out for further analysis based on icosahedral symmetry (7). The data from the large defocus film was processed first to get a stable model map for the close-defocus data. The H3RC complex was reconstructed to a resolution of 23 Å by combining 125 individual close-defocus images. The H3RC reconstruction served as the initial model to assign the center and the orientation of the particles at 37°C (H3RH and H3Re). The single-image reconstructions were used to aid the particle selection and to help in fine-tuning the intermediate polar Fourier transform models (1). The size of individually boxed H3RH images was determined by correlating the observed data with the corresponding projection of the H3RC reference map by grid interpolation. The Gaussian distribution of the particle size of H3RH versus H3RC was shifted, with a 4% increase in the mean value. The final reconstruction of H3RH and H3Re complexes was resolved to 23 and 25 Å by using 91 and 64 particles, respectively. The resolution was determined by R-factor assessment. The completeness of the data sampling in the reconstruction was verified by obtaining eigenvalue spectra, showing that of eigenvalues were >1 or 10 (8).
In a particular experiment, the internal size standard was generated by the addition of a large portion of native HRV3 particles to the 37°C-treated HRV3-receptor mixture after it was chilled on ice. The empty HRV3-receptor complex and full particles, mainly including unheated HRV3, were boxed out from a single micrograph and were used to calculate three-dimensional maps. Comparison of these three-dimensional reconstructions gave 4% size difference. The consistent dimensions of the bound ICAM-1 molecule (∼75 Å long and ∼20 Å in diameter) in the three complexes demonstrated no detectable systematic error due to the microscope. Finally, the consistent size of the HRV16-receptor complexes at different temperatures was an additional control for the collection of low-dose imaging of microscope data.
Model fitting.
Fitting of the ICAM-1 receptor (PDB access number 1IC1) and the HRV3 crystal structures (PDB access number 1RHI) into the cryo-EM density was performed manually using the program O (14). The docking of coordinates was done based on both visual and quantitative criteria. The HRV3 coordinates and H3RC map had the same origin. Domain 2 of the ICAM-1 molecule first was fitted into the corresponding EM density, with a previously described orientation as reference (16). The position of receptor domain 1 was adjusted manually. To model the capsid movement in the H3RH structure, the alpha-carbon coordinates of ICAM-1 were fitted into the receptor density of the expanded particle and then the coordinates of the viral proteins were fitted individually into the H3RH map. VP2 and VP3 were moved out radially and tilted to best follow the density distribution. Based on the modeled ICAM-1 position with its unique density profile, contact between VP2 and ICAM-1 was maintained and was used to localize the position of VP2. Subsequently, VP1 was docked into the EM density before the position of the whole protomer was adjusted by minimizing the crash with neighboring protomers. The resulting model was used to calculate a set of structure factors from which an electron density map was generated at a resolution corresponding to the EM density. The real-space correlation between the calculated and experimental densities was computed, and the high value of the correlation coefficient in the H3RH fitting (0.79) indicated optimal positioning of the viral proteins. This value was similar to that of H3RC docking (0.80). The best correlation coefficient of H3Re fitting was 0.73 (data not shown).
RESULTS
Reconstruction of HRV3 entry intermediates.
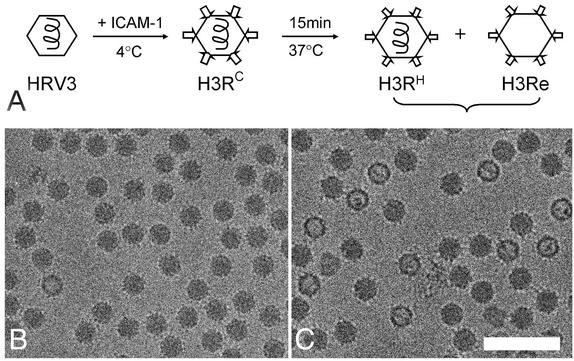
Receptor binding to HRV3 at physiological temperature generates virus-receptor intermediates, leading to the simultaneous exit of the internal RNA and VP4 and the subsequent appearance of empty viral capsids (5) (Fig. 1A). Cryo-EM micrographs showed that while HRV3 and ICAM-1 formed stable complexes at 4°C (H3RC) (Fig. 1B), a mixture of full (H3RH) and empty (H3Re) particles appeared after a 15-min incubation of the H3RC complexes at 37°C (Fig. 1C). About 40% of the particles were empty for HRV3, as judged by their lower density in the center of the capsid. In contrast, no empty particle was observed for the stable HRV16 even after 60 min of extensive incubation with ICAM-1 at 37°C (data not shown), consistent with the inability of the receptor to mediate uncoating of this serotype at neutral pH (13).
FIG. 1.
Cryo-EM of HRV3-receptor complexes and uncoating intermediates. (A) Schematic view of the pathway of HRV3 uncoating mediated by soluble ICAM-1 binding (5) and the procedure used for preparation of virus-receptor complexes for cryo-EM. Arrows and hexagons represent the ICAM-1 receptor and the HRV particles, respectively. H3RC was prepared by incubation of HRV3 with ICAM-1 at 4°C, while H3RH and H3Re were prepared at 37°C as described in Materials and Methods. H3RC and H3RH preserve all viral proteins and RNA, while H3Re lacks both VP4 and RNA (5). (B) Micrograph of H3RC. (C) Micrograph of H3RH and H3Re complexes. About 40% of the particles appear empty (H3Re), lacking the internal RNA genome represented by a spiral line in panel A. Bar, 1,000 Å.
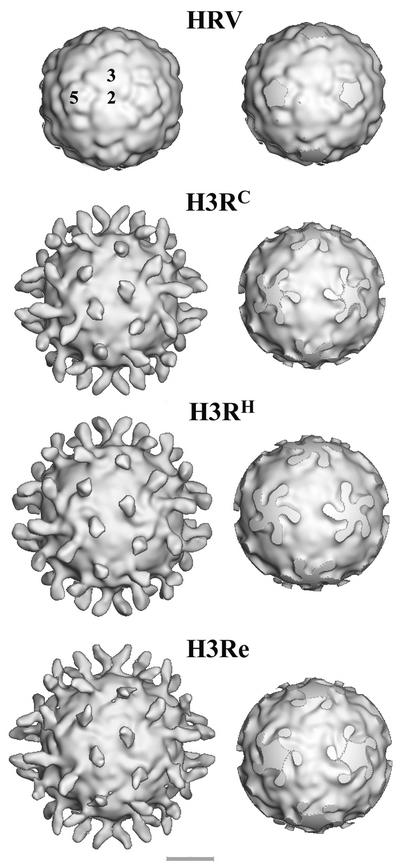
Three-dimensional reconstruction of the H3RC complexes (Fig. 2) revealed binding of the ICAM-1 receptor perpendicularly to the viral capsid into the canyon region around the 12 icosahedral fivefold axes (Fig. 2). Receptor binding occupancies were similar in the H3RC, H3RH, and HRV16-receptor (not shown) complexes and higher than in the H3Re particles. The capsid distal density of ICAM-1 corresponding to domain 2 vanished in the 3.0-σ-contoured map of H3Re. However, disappearance of the EM density of that ICAM-1 region in the H3RC and H3RH maps required contour levels of 3.5 and 3.8 σ, respectively. These data showed that the receptor molecule remained bound to the capsid during RNA release and suggest that the bound receptor slowly released from the capsid after the egress of RNA (5). The lower occupancy of ICAM-1 in H3Re could be also due to a less ordered bound receptor molecule.
FIG. 2.
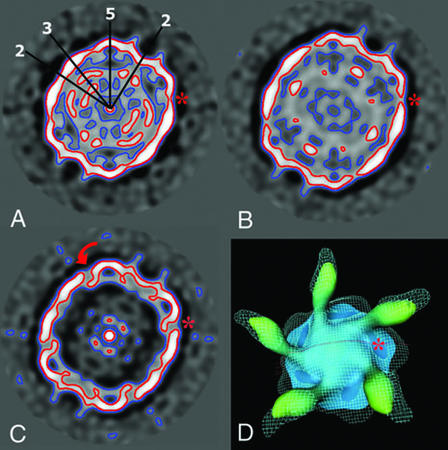
Surface rendering of native HRV3 and virus-receptor complexes. The maps are viewed along the twofold axis and contoured at 1 σ above the average. The labels of the complexes correspond to the data acquisition scheme presented in Fig. 1. In the right column, maps truncated 10 Å below the surfaces at the fivefold axis are shown. The locations of icosahedral two-, three-, and fivefold axes are indicated by the numbers superimposed on the HRV3 reconstruction, which was computed from the crystallographic coordinates (29). Bar, 100 Å.
The star-shaped fivefold plateau in H3RH is very similar in size and shape to that of H3RC and differs from the broader plateau presented by H3Re (Fig. 2, right). Unlike in H3RC, where a clear boundary between the receptor and the fivefold plateau is quite apparent (Fig. 2, right), the receptor appears connected to the side of the plateau by a protruding density in H3RH (see also Fig. 5). The bound receptor underwent a pronounced clockwise rotation of approximately 30° after RNA release in H3Re, so that the long axis of the receptor footprint was parallel to the edge of the plateau (Fig. 2, bottom right).
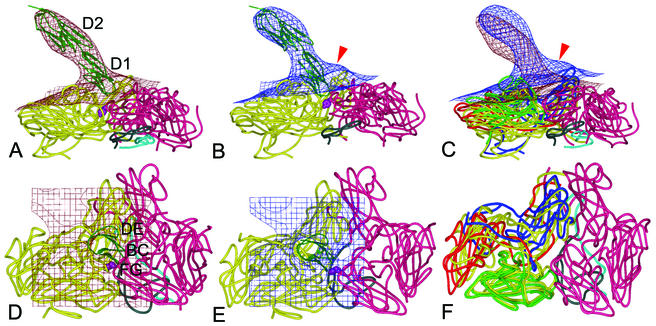
FIG. 5.
Virus-receptor binding surfaces. Side (top) and front (bottom) views of the fitted ICAM-1 and HRV3 crystal structures into the corresponding EM densities (grid) of the H3RC (A and D) and H3RH (B and E) complexes, superimposed in panels C and F. Density maps are contoured at 1.5 (top) and 2.0 (bottom) σ. A ribbon diagram of the two-domain crystal structure of ICAM-1 is shown, with the two interacting HRV3 protomers presented as worm drawings. The two connecting viral protomers are colored in yellow (left protomers in panels A, B, D, and E) and magenta (right protomers). In panels C and F, viral proteins VP1, VP2, and VP3 of H3RH are colored in blue, green, and red (the left protomer) or magenta (the right protomer), respectively; the superimposed H3RC protomer is colored in yellow. VP4 is cyan, and the N-terminal 60 residues of VP1 are gray. The red arrow points to the protruding density featuring the H3RH particle, predicted to be filled by the externalized VP4 and the VP1 N terminus. The side chain of the VP3 Tyr181 residue is indicated by purple spheres. The first (D1) and second (D2) domains of ICAM-1 are labeled, as are virus-binding BC, DE, and FG loops (6). The illustrations were prepared with the program PyMOL (http://www.pymol.org).
Capsid expansion precedes RNA release.
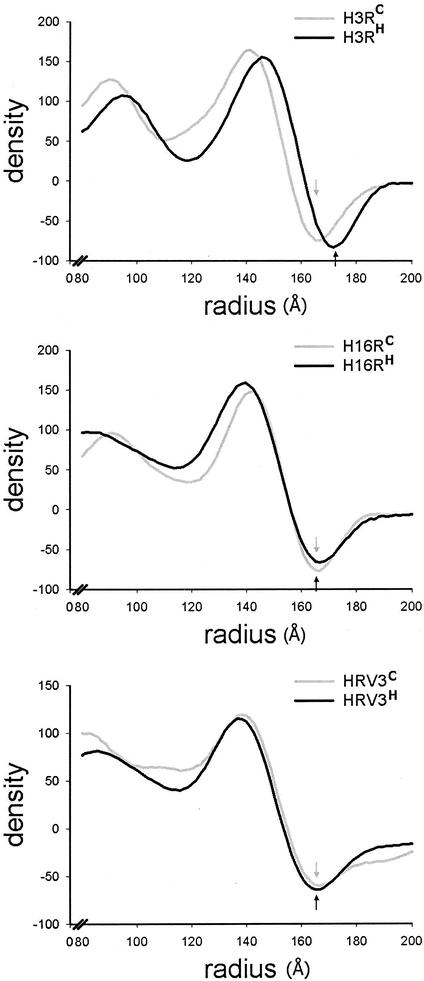
Reconstruction of HRV3-receptor complexes revealed differences in size between the viral particles exposed at 37 and 4°C (Fig. 2, right). The virus particle in H3RH expanded approximately 12 Å in diameter compared to that in H3RC (Fig. 3, top), even though this complex was reported to preserve all the viral proteins and the RNA genome (5). As a result, the C-terminal end of ICAM-1 moved radially from 216 to 222 Å in H3RH while the length of the receptor remained essentially the same. The distance between the end of the receptor density and the capsid fivefold axis for H3RC and H3RH was 49 and 48 Å, respectively. Similar expansion was observed for H3Re (data not shown). Capsid expansion was unique for the HRV3-receptor complexes since it was not observed for HRV16-receptor complexes or free HRV3 virions exposed at physiological temperature (Fig. 3). Because uncoating was also specifically observed in the RNA-containing HRV3-receptor complex, we concluded that expansion precedes the egress of the viral genome.
FIG. 3.
Size of the virus particles in the HRV-receptor complexes. Density profiles of the HRV reconstructions, presented as spherically averaged density versus particle radius, are shown. The arrows indicate the outer boundary of the viral capsid. (Top) Density distribution for the H3RC and H3RH reconstructions presented in Fig. 2. (Middle) Density distribution for HRV16-ICAM-1 complexes prepared at 4°C (H16RC) or 37°C (H16RH) as described in Materials and Methods. (Bottom) Density distribution for HRV3 native virions treated at 4°C (HRV3C) or 37°C (HRV3H) in the absence of receptor.
Capsid rearrangements in the HRV-receptor complexes.
Capsid expansion implies conformational rearrangements in the viral particles. An equatorial section of the particles showed a less compact RNA in the interior of H3RH than H3RC and an empty shell in the H3Re (Fig. 4). Lower density was observed in the two uncoating intermediates across the junction of twofold related protomers (red contour line in Fig. 4B and C). A significant decrease in the capsid density at the canyon region was observed in the H3RH and H3Re complexes compared to H3RC and appeared as a “density cleft” near the receptor binding site in the high-contour maps (asterisk in Fig. 4). The canyon region is built up by two neighboring protomers (see below), so the decrease of EM density in that region could be related to separation of the protomers in the expanded virus-receptor complexes.
FIG. 4.
Capsid rearrangements in the HRV-receptor complexes. (A to C) Equatorial sections of H3RC (A), H3RH (B), and H3Re (C) reconstructions perpendicular to the icosahedral twofold axis. The icosahedral axes (two-, three-, and fivefold) are drawn on the section of H3RC. The contour plots circle the density distribution at 1 σ (blue) and 2 σ (red). The arrow in panel C indicates the twist movement predicted for VP1. (D) Top view of the H3RC and H3Re electron density maps of a viral pentamer. The H3Re density map (cyan grid lines) contoured at 2 σ is superimposed onto the H3RC isosurface map rendered at 3 σ. The red line indicates the location of the equatorial sections presented in panels A to C. An asterisk marks the canyon position. The illustrations were prepared with the NAG Explorer program.
The exit of the viral genomic RNA led to major rearrangements at the fivefold plateau and VP1 (Fig. 4C). The canyon north wall appears more parallel to the fivefold axis in H3Re than H3RC-H, suggesting a pronounced twist movement of VP1 (red arrow in Fig. 4C). As a consequence, the plateau became broader in the empty complex (also shown in Fig. 2).
Receptor-virus binding contacts.
The crystal structure of the two N-terminal domains of ICAM-1 fits well into the corresponding density of the H3RC and the H3RH maps (Fig. 5A and B). The receptor appears more bent in H3RH than in H3RC (Fig. 5C). The side of the first domain (β-strands C and F) contacts the south wall of the canyon (the promoter is shown in yellow), and the tip (BC and FG loops of domain 1) penetrates into the canyon and sits on the junction of two closely associated promoters (Fig. 5D and E).
In H3RH, the neighboring promoters remain in contact at the fivefold axis region but are separated in the canyon (Fig. 5E); this is due mainly to the expansion of the capsid and to the slight movement of VP2 and VP3 toward the threefold axis (Fig. 5F). The docking of the viral proteins in H3RH is consistent with the preservation of the density organization at the fivefold axis as well as with the appearance of a density cleft at the canyon (Fig. 2 and 4) concomitant with capsid expansion. We have observed that residues such as Tyr181 of VP3 become exposed at the interprotomer junction of the fitted viral proteins in the expanded capsid. This residue could then interact with the critical Pro70 residue located in the FG loop of the receptor (Fig. 5B and E).
The internal N-terminal regions of VP1 and VP4 are located underneath the hole at the canyon region generated by capsid expansion. Those polypeptides then could externalize through this region and account for the protruding density connecting the receptor to the north wall of the canyon in H3RH (arrowhead in Fig. 5B and C). The externalized proteins may interact with the relatively hydrophobic DE loop of ICAM-1, which is involved in crystallographic dimeric association (6).
DISCUSSION
Complexes of the ICAM-1 molecule with HRV3 and HRV16 were analyzed to elucidate how receptor binding mediates subsequent capsid uncoating. Our results provide the first structural view of virus uncoating mediated by receptor binding under conditions similar to those found at the cell surface. Capsid expansion in receptor-bound HRV3 was observed in conjunction with uncoating. However, no such changes were observed either with HRV3 viruses treated at 37°C in the absence of receptor or with the stable HRV16-receptor complex. These observations suggest that uncoating requires receptor-mediated capsid expansion, which breaks interprotomer connections for externalization of capsid proteins and RNA (scheme in Fig. 6). The expansion observed here was similar to that reported for the 135S entry intermediates of poliovirus generated by heating (3), suggesting that viral capsid expansion may precede RNA release during virus entry.
FIG. 6.
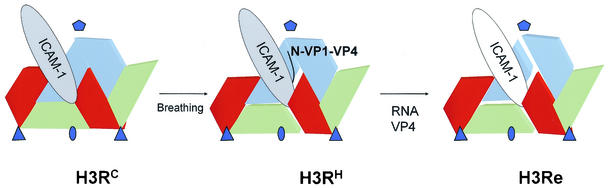
Dynamics of receptor-mediated rhinovirus uncoating. Schematic views of two neighboring protomers with a bound ICAM-1 receptor molecule, representing conformations of H3RC, H3RH, and H3Re complexes. Ovals, triangles, and pentagons indicate the positions of the twofold, threefold, and fivefold axes, respectively. VP1, VP2, and VP3 are represented as blocks colored blue, green, and red, respectively. Stable virus-receptor complexes (H3RC) are formed at low temperatures by receptor binding to a close virus conformation, represented by the crystal structure (28). The capsid dynamics (or breathing) opens the interprotomer junction, so that the internal N-terminal regions of VP1 and VP4 (labeled as N-VP1-VP4) became exposed simultaneously at physiological temperatures (17, 18, 22). This expanded conformation is maintained by receptor binding between two adjacent protomers and to the exposed region of VP1, as shown by the cryo-EM H3RH structure. Egress of the viral RNA genome and loss of VP4 result in major rearrangements in VP1 and the fivefold axis and subsequent release of the receptor from the H3Re complex.
The pocket factor molecule present in HRV16 appears to inhibit both capsid dynamics and uncoating (17). These data, along with the temperature dependence of rhinovirus uncoating (5, 13), suggest that capsid breathing might initially drive particle expansion (Fig. 6). Therefore, rather than being the trigger, the bound receptor molecules would just hold the capsid in an expanded conformation by binding within two neighboring capsid protomers in their expanded open state (Fig. 5 and 6). Our model implies receptor binding to several conformations of the canyon region, as suggested by the nonlinear kinetic binding rates (28).
A protruding and well-defined density appeared, bridging the receptor and the north canyon wall in the expanded H3RH capsid, but it was absent in H3RC and H3Re (Fig. 2 and 5). This density might represent the externalized N-terminal regions of VP1 and VP4, which have been reported to exit simultaneously and reversibly from the capsid (17, 18). These polypeptides localize beneath the interprotomer interface in the native capsid (Fig. 5D). Thus, these two peptides may exit through the open canyon region and account for the protruding density featuring the H3RH intermediate. Externalization of these polypeptides through a similar location has been proposed for related picornaviruses (3, 12). Localization of the externalized polypeptides near the hydrophobic ICAM-1 dimerization region (6) suggests that they may interact with the receptor. This interaction could lock the VP1 N terminus and VP4 in their externalized conformations so that they could bind to the membrane (9).
The release of the RNA from the capsid of picornavirus is thought to occur through one of its 12-pentameric vertices. However, there has been no evidence showing the egress of the VP3 N-terminal cylinder connection, VP1 N terminus, VP4, or viral RNA through this opening. Analyses of uncoating intermediates in HRV3 presented here and in poliovirus (135S) (3) showed little evidence for the formation of a capacious tunnel through the fivefold axis region. Changes at the fivefold axis region arise after RNA egress and might lead to capsid disassembly (Fig. 2 and 4) (12).
While the fivefold axis regions of H3RC and H3RH particles are structurally similar, significant differences were observed along the interprotomer interfaces in the canyon region and at the twofold axis junctions (Fig. 2 and 4). These observations strongly suggest an expansive movement of the pentamers along the fivefold axis. Polarized expansion of a single receptor-attached, membrane-proximal pentamer could occur during virus entry. RNA egress through the expanded pentamer could then take place close to the membrane while the virus particle remains attached to the receptor, thereby facilitating penetration into the cell cytoplasm.
The results presented here reveal for the first time that capsid expansion can be mediated by receptor binding at physiological temperature. The magnitude of capsid expansion for the H3RH (4%) uncoating intermediate is similar to that observed for related picornavirus particles (3, 12), so that it must be significant for virus entry. We propose that the ICAM-1 receptor passively catalyzes uncoating by locking the capsid in an expanded and open intermediate state for subsequent RNA release (Fig. 6). In rhinovirus and related picornaviruses, this process can be attributed to the dynamic feature of the viral capsids (17, 18, 22), as well as the fragmented nature of the receptor binding site (4, 11, 16, 28). Expansion of nonenveloped virus capsids catalyzed by receptor binding or other cellular factors could precede uncoating during virus entry into cells, as shown here for HRV and its ICAM-1 receptor.
Acknowledgments
We are grateful to Birgitta Lindqvist for assistance with tissue culture. We thank Lars Liljas, Henrik Garoff, and Maarit Suomalainen for helpful suggestions about manuscript preparation.
This work has been sponsored by grants from the Medical Research Council (MFR-12175 and MFR-12637) and the Natural Science Research Council (NFR-11691 and NFR-11994) to R.H.C. and J.M.C. L.X. is the recipient of a fellowship from the Swedish Structural Biology Network (to R.H.C.).
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Bella, J., P. R. Kolatkar, C. W. Marlor, J. M. Greve, and M. G. Rossmann. 1998. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. USA 95:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belnap, D. M., B. M. J. McDermott, D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casasnovas, J. M., and T. A. Springer. 1994. Pathway of rhinovirus disruption by soluble intercellular adhesion molecule 1 (ICAM-1): an intermediate in which ICAM-1 is bound and RNA is released. J. Virol. 68:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casasnovas, J. M., T. Stehle, J. H. Liu, J. H. Wang, and T. A. Springer. 1998. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc. Natl. Acad. Sci. USA 95:4134-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, R. H. 2000. Visualization on the grid of virus-host interactions, p. 141-153. In L. Johnsson (ed.), Simulation and visualization on the grid. Springer-Verlag, New York, N.Y.
- 8.Crowther, R. A., L. A. Amos, J. T. Finch, D. J. De Rosier, and A. Klug. 1970. Three dimensional reconstructions of spherical viruses by Fourier synthesis from electron micrographs. Nature 226:421-425. [DOI] [PubMed] [Google Scholar]
- 9.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 11.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewat, E., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317.. [DOI] [PubMed] [Google Scholar]
- 13.Hoover-Litty, H., and J. M. Greve. 1993. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol. 67:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, T. A., J. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Ser. A. 47:110-119. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan, G., M. S. Freistadt, and V. R. Racaniello. 1990. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol. 64:4697-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Q., A. G. Yafal, Y. M. Lee, J. Hogle, and M. Chow. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosser, A. G., D. A. Shepard, and R. R. Rueckert. 1994. Use of drug- resistance mutants to identify functional regions in picornavirus capsid proteins. Arch. Virol. Suppl. 9:111-119. [DOI] [PubMed] [Google Scholar]
- 20.Perez, L., and L. Carrasco. 1993. Entry of poliovirus into cells does not require a low-pH step. J. Virol. 67:4543-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Racaniello, V. R. 1996. The poliovirus receptor: a hook, or an unzipper? Structure 4:769-773. [DOI] [PubMed] [Google Scholar]
- 22.Roivainen, M., L. Piirainen, T. Rysa, A. Narvanen, and T. Hovi. 1993. An immunodominant N-terminal region of VP1 protein of poliovirion that is buried in crystal structure can be exposed in solution. Virology 195:762-765. [DOI] [PubMed] [Google Scholar]
- 23.Rossmann, M. G. 1989. The canyon hypothesis. Viral Immunol. 2:143-161. [DOI] [PubMed] [Google Scholar]
- 24.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, and A. G. Mosser. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 25.Rueckert, R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. Fields, D. Knipe, and P. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 26.Schober, D., P. Kronenberger, E. Prchla, D. Blaas, and R. Fuchs. 1998. Major and minor receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol. 72:1354-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1179-1194. [DOI] [PubMed] [Google Scholar]
- 28.Xing, L., K. Tjarnlund, B. Lindqvist, G. G. Kaplan, D. Feigelstock, R. H. Cheng, and J. M. Casasnovas. 2000. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 19:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0 A resolution. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]