Abstract
Viroids, small circular RNAs that replicate independently and in most cases incite diseases in plants, are classified into the families Pospiviroidae, composed of species with a central conserved region (CCR) and without hammerhead ribozymes, and Avsunviroidae, composed of three members lacking CCR but able to self-cleave in both polarity strands through hammerhead ribozymes. Here we report the biological and molecular properties of Eggplant latent viroid (ELVd). Purified circular ELVd induces symptomless infections when inoculated into eggplant seedlings. ELVd can be transmitted horizontally and through seed. Sequencing 10 complete cDNA clones showed that ELVd is a circular RNA of 332 to 335 nucleotides with high variability. This RNA can adopt a quasi-rod-like secondary structure of minimal free energy and alternative foldings that permit formation of stable hammerhead structures in plus and minus strands. The ribozymes are active in vitro and, most likely, in vivo. Considering the ELVd properties to be intermediate between those of the two genera of family Avsunviroidae, we propose ELVd as the type species of a third genus with the name Elaviroid.
Viroids and satellite RNAs have been implicated in several plant diseases. Whereas viroids replicate autonomously (12), satellite RNAs depend functionally on helper viruses and are encapsidated by their coat proteins (17). A survey in Eastern Spain for detecting small infectious RNAs in vegetables identified a viroid-like RNA in eggplant (Solanum melongena L. cv. Sonja) (8). Denaturing polyacrylamide gel electrophoresis (PAGE) revealed two bands with the mobilities expected for the circular and linear forms, and treatment with RNase, but not with DNase, caused their disappearance. Inoculation of eggplant seedlings with preparations containing the viroid-like RNA showed the presence of the same RNA in some of the inoculated plants, which remained symptomless, and its absence in the noninoculated controls. On this basis, the RNA was named Eggplant latent viroid (ELVd), although the possibility that it could be a viroid-like satellite RNA could not be dismissed (8). Attempts to transmit ELVd to tomato, chrysanthemum, cucumber, and citron, which support the replication of different viroids, failed, suggesting a restricted host range, and riboprobes specific for Citrus exocortis viroid (CEVd), Hop stunt viroid, and Apple scar skin viroid did not hybridize with ELVd, suggesting that it was not a member of the genus Pospiviroid, Hostuviroid, or Apscaviroid (8). These genera belong to the family Pospiviroidae, whose members have a central conserved region and lack hammerhead ribozymes (12). These data, together with the low stability of ELVd circular forms in vitro (9), were indicative of a peculiar structure and prompted us to study the biological and molecular properties of ELVd.
Biological properties of ELVd.
Eggplant seedlings (cv. Redonda morada) inoculated with ELVd and kept in a greenhouse were used as the source of ELVd. To confirm that ELVd could replicate autonomously (8), eggplant leaves were extracted with buffer-saturated phenol. Nucleic acids were fractionated with nonionic cellulose (30) and analyzed by sequential PAGE (sPAGE) (11). After ethidium bromide staining, RNA with the slow mobility characteristic of viroid circular forms was eluted and inoculated via three incisions into the epicotyl of 5-cm-high eggplant seedlings that had been maintained in the greenhouse. Examination of 20 eggplants 6 months later revealed both ELVd forms in 4 of the plants (and their absence in the noninoculated controls), showing that ELVd behaves like a viroid. These results were confirmed by Northern hybridization when an ELVd cDNA clone became available.
To study whether ELVd had any effect on eggplants, 18 seedlings graft inoculated with ELVd-infected material (and harboring the viroid as revealed by sPAGE and silver staining 1 month later) and 18 noninoculated seedlings were grown for 3 months in an experimental plot. Weekly observations of the growing pattern, flower number, and weight, number, and aspect of fruits failed to reveal any differences, confirming previous observations by which the name Eggplant latent viroid was proposed (8). No symptoms were observed under different greenhouse conditions. ELVd was uniformly distributed in leaves, stems, and fruits (skin and pulp).
To evaluate the horizontal transmission of ELVd, a blade previously contaminated by slashing the stem of an ELVd-infected eggplant was used to slash a group of receptor seedlings. This operation was repeated 10 times per plant. After 3 months in the greenhouse, the top portions of the plants were removed, and analysis of the new shoots by sPAGE and silver staining showed that 6 of the 11 slashed plants were infected with ELVd. Regarding vertical transmission, analysis of two lots of 50 seedlings each obtained by germinating seeds from fruits of two ELVd-infected eggplants revealed ELVd infection ratios of 26 and 16%. In a third lot in which the seeds were washed with 1% sodium hypochlorite to avoid ELVd transmission from the seed coat to the plantlet during germination, the ELVd infection ratio was similar (18%), indicating that this viroid is seed transmitted. Both transmission routes, particularly the second, explain the persistence of ELVd in nature.
Primary and proposed secondary structure.
A size of 330 to 335 nucleotides (nt) for ELVd was inferred from the electrophoretic mobilities in denaturing gels of the circular forms of ELVd and known viroids (Fig. 1). ELVd cDNA clones were obtained by a reverse transcription (RT)-PCR approach that does not require prior sequence knowledge (26). Sequencing six recombinant plasmids with inserts bigger than 100 bp showed one, of 187 bp, containing the conserved motifs CUGANGA and GAAAC characteristic of the hammerhead structures (14, 20, 28), and a radioactive probe obtained by PCR from this insert hybridized with the ELVd circular and linear forms. Moreover, the sequences flanking these two conserved motifs could form the helix regions that are also characteristic of hammerhead ribozymes. This strongly suggested that ELVd was a new species of the family Avsunviroidae, whose members lack a central conserved region but are able to self-cleave in both polarity strands through hammerhead ribozymes (12, 13). To obtain further evidence, two adjacent primers of opposite polarity, PI (5′-AGTGTGCGCTTTCCCTGATGAGCCCA-3′) and PII (5′-TTCCGACGGTGAGTTCGTCGACACCTCT-3′) (Fig. 2), derived from the 187-bp insert, were used in an RT-PCR assay with Pfu DNA polymerase and the ELVd circular forms. The amplified cDNA, which in nondenaturing PAGE had the expected mobility, was cloned in a plasmid, and 50 clones were analyzed by a single-strand conformational polymorphism assay using PI and PII (27). The results indicated that ELVd was formed by a heterogeneous population of sequence variants, a point confirmed by sequencing six of the clones: they had 334 to 335 nt and presented in both polarity strands the motifs characteristic of hammerhead structures.
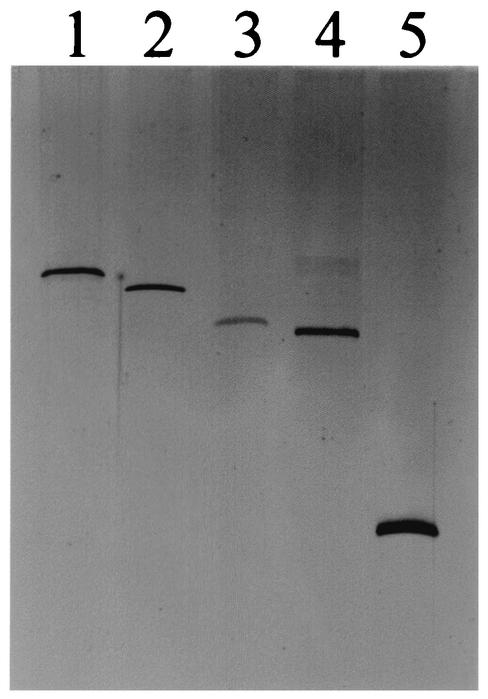
FIG. 1.
Mobilities of the purified circular forms of CEVd (371 nt), Potato spindle tuber viroid (359 nt), PLMVd (337 nt), ELVd, and ASBVd (247 nt) (lanes 1 to 5, respectively) after PAGE in 0.25× Tris-borate-EDTA-5% polyacrylamide gels containing 8 M urea and silver staining.
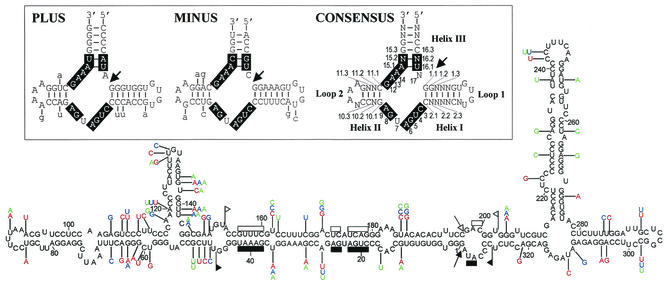
FIG. 2.
Primary and proposed secondary structure of minimum free energy of ELVd-2 variant. The heterogeneity observed in three additional representative ELVd variants (1, 3, and 4) is indicated in red, blue, and green, respectively. Sequences forming the hammerhead structures are delimited by flags, motifs conserved in natural hammerhead structures are denoted by bars, and self-cleavage sites are marked by arrows. Solid and open symbols refer to plus and minus polarities, respectively. ELVd variants 1, 3, and 4 were obtained with primers PI and PII (complementary and identical to positions 168 to 193 and 194 to 221, respectively) and the ELVd-2 variant was obtained with primers PIII and PIV (identical and complementary to positions 228 to 248 and 249 to 268, respectively). (Inset) Plus, minus, and consensus hammerhead structures of ELVd-2 RNA, with the numbering system indicated in the last. Motifs conserved in natural hammerhead structures are boxed and self-cleavage sites are marked by arrows. Substitutions found in the other eight ELVd variants are denoted with lowercase letters.
Because PI and PII contained a part of the sequences of the ELVd hammerhead structures (Fig. 2), it was impossible to generate in vitro transcripts with this self-cleaving domain being uninterrupted. Therefore, a new RT-PCR was performed with a second pair of adjacent primers of opposite polarity, PIII (5′-AGGATGTGTTCCCTAGGAGG-3′) and PIV (5′-TGAAARRGGATAGTACCTGGG-3′), where R is G or A, designed outside the regions encompassing both hammerhead structures. Sequencing of four recombinant plasmids confirmed the high variability of ELVd: 9 of the 10 sequence variants obtained with both primer pairs were different. However, three variants of 335 nt had identities higher than 98%, with ELVd-1 selected as the representative. Another three variants of 332 to 334 nt also had identities higher than 98%, although they differed (88% of identity) from those of the first group; ELVd-2 was chosen as the representative of this second group. A third group of two 335-nt variants that were 98% identical, with ELVd-3 taken as the representative, showed 88 and 91% identity with ELVd-1 and ELVd-2, respectively. Finally, one variant of 334 nt (ELVd-4) was 88, 91, and 92% identical to ELVd-1, ELVd-2, and ELVd-3, respectively. Figure 2 shows the four representative variants, which have a G+C content of 53 to 54%.
The existence in infected tissue of plus and minus ELVd circular forms was inferred from Northern hybridization (Fig. 3) and from RT-PCR amplification of the complete viroid sequence irrespective of which primer of each pair was used in the RT reaction. This indicates that ELVd probably follows the symmetric pathway of the rolling circle mechanism of replication characteristic of family Avsunviroidae (5, 19), in contrast to species of family Pospiviroidae, which replicate through the asymmetric pathway (2, 3, 10, 21). To determine which strand was accumulating at a higher level in infected tissue, to which the plus polarity is arbitrarily assigned, radioactive full-length riboprobes from the cDNA of ELVd-2 obtained by in vitro transcription with T7 and T3 RNA polymerases were used in Northern hybridization (25). The intensity of the signals (Fig. 3) allowed identification of the plus polarity strand (Fig. 2).
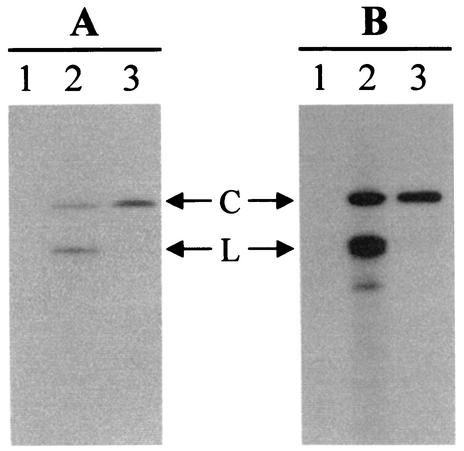
FIG. 3.
Polarity determination of ELVd strands by denaturing PAGE and Northern blot hybridization using two full-length riboprobes obtained by in vitro transcription of the linearized recombinant plasmid corresponding to variant ELVd-2 with T3 (A) or T7 (B) RNA polymerases. Lane 1, purified preparation of CEVd used as a control; lane 2, nucleic acid preparation from ELVd-infected eggplant; lane 3, purified ELVd circular forms. C and L, ELVd circular and linear forms, respectively. T3 and T7 riboprobes had the same and the complementary polarity, respectively, as the sequence presented in Fig. 2. Both riboprobes were equalized in terms of acid-precipitable counts, and the autoradiograms were exposed for the same time.
The predicted secondary structures of minimal free energy (31) for the nine ELVd variants displayed a high autocomplementarity (Fig. 2). In the four representative variants, 68 to 70% of the residues were paired (with approximately 57 to 59% G-C, 33.5 to 34.5% A-U, and 8.5 to 9.5% G-U pairs). The conformations were of the quasi-rod-like class and showed two bifurcations at both terminal domains (Fig. 2). None of the secondary structures within a 10% interval of minimal free energy were of the rod-like class characteristic of family Pospiviroidae. The predicted conformations for ELVd-1, -2, -3, and -4 accommodate most of the variability as compensatory mutations or covariations (Fig. 2), showing that these conformations, or portions thereof, most likely exist in vivo.
Hammerhead structures.
Both ELVd strands contain the 11 residues that are strictly conserved in natural hammerhead structures (14), flanked by other residues that can form helices I, II, and III characteristic of these ribozymes (Fig. 2). In plus and minus ELVd hammerhead structures, helix III is stable and helices I and II are closed by short loops 1 and 2, a morphology similar to the hammerhead structures of Peach latent mosaic viroid (PLMVd) (18) and to a lesser extent those of Chrysanthemum chlorotic mottle viroid (CChMVd), in which the minus hammerhead structure has an unusually long helix II (25), but different from the hammerhead structures of Avocado sunblotch viroid (ASBVd), which have short helices III and long loops 1 and 2 (20).
In most natural hammerhead structures positions 10.1 and 11.1 form a G-C pair and positions 15.2 and 16.2 form a C-G pair (Fig. 2). ELVd hammerhead structures conform to this rule except that of plus polarity in which positions 15.2 and 16.2 form a U-A pair, a situation found only in the plus hammerhead structure of the Satellite RNA of cereal yellow dwarf virus RPV (24) and in the minus hammerhead structure of a cherry small circular RNA (csc RNA1) (7). Interestingly, in these three hammerhead structures the residue preceding the self-cleavage site is A and not C as in most natural hammerhead structures, reinforcing the idea that these three positions are interrelated (7). Like most natural hammerhead structures, those of ELVd have a pyrimidine (U) at position 7 (Fig. 2).
The ELVd hammerhead structures are more related to each other than to other hammerhead structures, a situation reported previously for hammerhead structures of Satellite RNA of lucerne transient streak virus (15), PLMVd (18), CChMVd (25), and csc RNA1 (7). Other similarities include the hexanucleotides UGUG(U/C)(G/A) and C(A/G)AAAG, forming part of helix I, loop 1, and helix II, loop 2, in the consensus hammerhead structure of ELVd, respectively (Fig. 2), that are in equivalent positions in the consensus hammerhead structure of csc RNA1 (7), in the hammerhead structures of Satellite RNAs of arabis mosaic virus (22), Chicory yellow mottle virus (29), and Solanum nodiflorum mottle virus (15), and in the minus hammerhead structures of Satellite RNA of cereal yellow dwarf virus RPV (24) and CChMVd (25). With one exception, the substitutions in different ELVd variants preserve the stability of the hammerhead structures, either because they map at the loops or because when affecting the helices compensatory mutations or covariations restore their stability (Fig. 2), strongly indicating that these self-cleaving domains are relevant in vivo.
RNA self-cleavage.
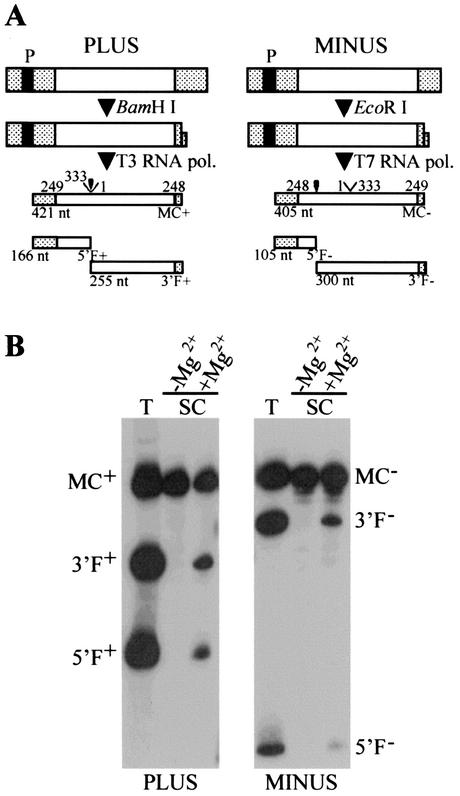
Radioactive monomeric ELVd-2 transcripts of both polarities, synthesized in vitro with T7 and T3 RNA polymerases, self-cleaved significantly during transcription (Fig. 4). The same fragments were observed when the uncleaved transcripts were eluted and incubated in a protein-free buffer containing Mg2+ (Fig. 4). The fragments 5′F+, 3′F+, 5′F−, and 3′F− showed the lengths predicted by the self-cleavage sites, which were confirmed by primer extension of the purified 3′F+ and 3′F− fragments.
FIG. 4.
In vitro synthesis and self-cleavage of both polarity RNAs of variant ELVd-2. (A) Diagram of plus and minus DNA templates and of the products generated by transcription with T3 and T7 RNA polymerases, respectively, after restriction with the indicated enzymes. Hatched boxes, vector sequences; filled boxes, RNA polymerase promoters; empty boxes, ELVd sequence. The complete primary transcripts are C+ and C− and the cleavage fragments are 5′F+, 3′F+, 5′F−, and 3′F−. Positions in the ELVd sequence are shown above the products and their expected sizes in nucleotides are shown below. Self-cleavage sites are indicated by arrowheads. (B) Analysis by denaturing PAGE (in 1× Tris-borate-EDTA-5% polyacrylamide gels containing 8 M urea) and autoradiography of the in vitro transcription (T) and self-cleavage (SC) of the purified primary transcripts incubated at 40°C for 1.5 h in 50 mM Tris-HCl (pH 8)-0.5 mM EDTA, with or without 5 mM MgCl2. The samples were previously heated in 1 mM EDTA (pH 6) at 95°C for 1 min and snap cooled in ice.
Taxonomic proposal.
Before the present report, family Avsunviroidae comprised only ASBVd (20), PLMVd (18), and CChMVd (25). ASBVd has unique properties that include a small size (246 to 247 nt), a G+C content of 38% (this ratio is higher than 50% in the other viroids) (12), and single hammerhead structures that are thermodynamically unstable (20). Because of this instability, double hammerhead structures have been proposed to mediate self-cleavage (6, 16). Considering these properties, ASBVd has been designated as the type species of genus Avsunviroid, which has no other member (12). In contrast to ASBVd, the plus and minus hammerhead structures of PLMVd and CChMVd have stable helices III and short loops capping helices I and II, and the most stable secondary structures predicted for PLMVd and CChMVd are branched and different from the rod-like or quasi-rod-like models proposed for most viroids, including ASBVd (1, 18, 20, 25). The sequence heterogeneity found in PLMVd and CChMVd supports the biological significance of their branched conformations, in which the conserved residues of both hammerhead structures are opposite each other (1, 18, 25). These common properties have served to allocate PLMVd and CChMVd to the genus Pelamoviroid (12, 25). Moreover, in vitro probing of PLMVd indicates the existence of a pseudoknot between two hairpin loops, and the possibility of a similar interaction in CChMVd has been suggested (4). The variability of natural CChMVd variants supports the in vivo existence of this element of tertiary structure (M. De la Peña and R. Flores, Abstr. XII Int. Congr. Virol., p. 248, 2002), providing further data for including PLMVd and CChMVd in one genus.
The quasi-rod-like secondary structure predicted for ELVd, supported in part by compensatory mutations and covariations, more closely resembles that of ASBVd than the branched conformations of PLMVd and CChMVd, but ELVd is more similar to PLMVd and CChMVd than to ASBVd considering its G+C-rich composition and thermodynamically stable hammerhead structures. However, although in the quasi-rod-like conformation of ELVd the conserved sequences of both hammerhead structures are opposite each other (as in PLMVd and CChMVd), these sequences hold a central position, in contrast with PLMVd and CChMVd, in which they are located in one arm. On the basis of these singular properties of ELVd, we propose to assign it to a new genus within family Avsunviroidae for which we offer the name Elaviroid (from eggplant latent viroid) conforming to previous rules (12). The sequence identity between some ELVd variants is slightly below 90%, one of the demarcation criteria separating variants of the same viroid from different viroid species (12). A similar situation has been reported for Mexican papita viroid (23), but as in this case, we consider that this sole criterion does not justify creating more than one species for ELVd.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession numbers AJ536612 to AJ536620.
Acknowledgments
This work was partially supported by grants SC97-108 and RTA01-119 from the Ministerio de Ciencia y Tecnología (to N.D.-V.) and by grants PB98-0500 from the Dirección General de Enseñanza Superior and BMC2002-03694 from the Ministerio de Ciencia y Tecnología (to R.F.). Z.F. received predoctoral fellowships from the National Palestinian Authority and the Universidad Politécnica de Valencia, C.F. received a predoctoral fellowship from the Generalitat Valenciana, and J.A.D. received a postdoctoral “Ramón y Cajal” contract of the Ministerio de Ciencia y Tecnología.
REFERENCES
- 1.Ambrós, S., C. Hernández, J. C. Desvignes, and R. Flores. 1998. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J. Virol. 72:7397-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 3.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1988. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 85:9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bussière, F., J. Ouellet, F. Côté, D. Lévesque, and J. P. Perreault. 2000. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daròs, J. A., J. F. Marcos, C. Hernández, and R. Flores. 1994. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 91:12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, C., C. C. Sheldom, and R. H. Symons. 1991. Alternative hammerhead structures in the self-cleavage of avocado sunblotch viroid RNAs. Nucleic Acids Res. 19:1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Serio, F., J. A. Daròs, A. Ragozzino, and R. Flores. 1997. A 451-nucleotide circular RNA from cherry with hammerhead ribozymes in its strands of both polarities. J. Virol. 71:6603-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagoaga, C., J. A. Pina, and N. Duran-Vila. 1994. Occurrence of small RNAs in severely diseased vegetable crops. Plant Dis. 78:749-753. [Google Scholar]
- 9.Fagoaga, C. 1995. Caracterización de viroides aislados en especies hortícolas. Ph.D. thesis. Universidad Politécnica de Valencia, Valencia, Spain.
- 10.Feldstein, P. A., Y. Hu, and R. A. Owens. 1998. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 95:6560-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores, R., N. Duran-Vila, V. Pallás, and J. S. Semancik. 1985. Detection of viroid and viroid-like RNAs from grapevine. J. Gen. Virol. 66:2095-2102. [Google Scholar]
- 12.Flores, R., J. W. Randles, M. Bar-Joseph, and T. O. Diener. 2000. Viroids, p. 1009-1024. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. 7th Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 13.Flores, R., J. A. Daròs, and C. Hernández. 2000. The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res. 55:271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores, R., C. Hernández, M. De la Peña, A. Vera, and J. A. Daròs. 2001. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 341:540-552. [DOI] [PubMed] [Google Scholar]
- 15.Forster, A. C., and R. H. Symons. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211-220. [DOI] [PubMed] [Google Scholar]
- 16.Forster, A. C., C. Davies, C. C. Sheldon, A. C. Jeffries, and R. H. Symons. 1988. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334:265-267. [DOI] [PubMed] [Google Scholar]
- 17.García-Arenal, F. 1999. Satellites, p. 2271-2273. In T. E. Creighton (ed.), Encyclopedia of molecular biology. John Wiley & Sons, New York, N.Y.
- 18.Hernández, C., and R. Flores. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 89:3711-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchins, C. J., P. Keese, J. E. Visvader, P. D. Rathjen, J. L. McInnes, and R. H. Symons. 1985. Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 4:293-304. [DOI] [PubMed] [Google Scholar]
- 20.Hutchins, C. J., P. D. Rathjen, A. C. Forster, and R. H. Symons. 1986. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14:3627-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa, M., T. Meshi, T. Ohno, Y. Okada, T. Sano, I. Ueda, and E. Shikata. 1984. A revised replication cycle for viroids: the role of longer than unit RNA in viroid replication. Mol. Gen. Genet. 196:421-428. [DOI] [PubMed] [Google Scholar]
- 22.Kaper, J. M., M. E. Tousignant, and G. Steger. 1988. Nucleotide sequence predicts circularity and self-cleavage of 300-ribonucleotide satellite of arabis mosaic virus. Biochem. Biophys. Res. Commun. 154:318-325. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Soriano, J. P., J. Galindo-Alonso, C. J. Maroon, I. Yucel, D. R. Smith, and T. O. Diener. 1996. Mexican papita viroid: putative ancestor of crop viroids. Proc. Natl. Acad. Sci. USA 93:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, W. A., T. Hercus, P. M. Waterhouse, and W. L. Gerlach. 1991. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in self-cleavage domain. Virology 183:711-720. [DOI] [PubMed] [Google Scholar]
- 25.Navarro, B., and R. Flores. 1997. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 94:11262-11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro, B., J. A. Daròs, and R. Flores. 1998. Reverse transcription polymerase chain reaction protocols for cloning small circular RNAs. J. Virol. Methods 73:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Palacio, A., and N. Duran-Vila. 1999. Single-strand conformation polymorphism (SSCP) analysis as a tool for viroid characterization. J. Virol. Methods 77:27-36. [DOI] [PubMed] [Google Scholar]
- 28.Prody, G. A., J. T. Bakos, J. M. Buzayan, I. R. Schneider, and G. Bruening. 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231:1577-1580. [DOI] [PubMed] [Google Scholar]
- 29.Rubino, L., M. E. Tousignant, G. Steger, and J. M. Kaper. 1990. Nucleotide sequence and structural analysis of two satellite RNAs associated with chicory yellow mottle virus. J. Gen. Virol. 71:1897-1903. [DOI] [PubMed] [Google Scholar]
- 30.Semancik, J. S. 1986. Separation of viroid RNAs by cellulose chromatography indicating conformational distinctions. Virology 155:39-45. [DOI] [PubMed] [Google Scholar]
- 31.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO ASI Series. Kluwer Academic Publishers, Boston, Mass.