Abstract
Vaccinia virus, the basis of the smallpox vaccine, is one of the largest viruses to replicate in humans. We have used in situ atomic force microscopy (AFM) to directly visualize fully hydrated, intact intracellular mature vaccinia virus (IMV) virions and chemical and enzymatic treatment products thereof. The latter included virion cores, core-enveloping coats, and core substructures. The isolated coats appeared to be composed of a highly cross-linked protein array. AFM imaging of core substructures indicated association of the linear viral DNA genome with a segmented protein sheath forming an extended ∼16-nm-diameter filament with helical surface topography; enclosure of this filament within a 30- to 40-nm-diameter tubule which also shows helical topography; and enclosure of the folded, condensed 30- to 40-nm-diameter tubule within the core by a wall covered with peg-like projections. Proteins observed attached to the 30- to 40-nm-diameter tubules may mediate folding and/or compaction of the tubules and/or represent vestiges of the core wall and/or pegs. An accessory “satellite domain” was observed protruding from the intact core. This corresponded in size to isolated 70- to 100-nm-diameter particles that were imaged independently and might represent detached accessory domains. AFM imaging of intact virions indicated that IMV underwent a reversible shrinkage upon dehydration (as much as 2.2- to 2.5-fold in the height dimension), accompanied by topological and topographical changes, including protrusion of the satellite domain. As shown here, the chemical and enzymatic dissection of large, asymmetrical virus particles in combination with in situ AFM provides an informative complement to other structure determination techniques.
Due to its historical role in smallpox eradication and its relatively benign tropism (5, 10, 28), vaccinia virus has been the most intensively studied member of the Poxviridae. Vaccinia virus is the focus of renewed interest arising from the possibility of deliberate smallpox virus release. The viral DNA genome is linear, double stranded, and completely sequenced and in the Copenhagen strain is ∼191 kbp in length and contains at least 266 open reading frames (11, 18). Vaccinia virus mRNA synthesis is divided into three temporal phases (19): early (occurring within the infecting virus particle immediately upon infection but prior to genome replication) and intermediate and late (occurring sequentially after disassembly of the infecting virion and the onset of genome replication). Unlike other large DNA viruses, vaccinia virus replicates in the cytoplasmic compartment of the infected cell (19, 27). Genome replication and virion assembly occur within discrete cytoplasmic viral factories that are thought to be devoid of cellular organelles and membranes (20, 21, 27). Infectious virions can be isolated at distinct stages of the maturation pathway. These infectious forms include intracellular mature vaccinia virus (IMV), cell-associated enveloped virus, and extracellular enveloped virus.
Among the human-tropic viruses, vaccinia virus has one of the most structurally complex, markedly asymmetric virions (10, 12, 13). The structure of the vaccinia virus virion has been the subject of electron microscopy (EM) studies (comprehensively reviewed in reference 10) extending over half a century, encompassing shadowing, thin sectioning, freeze-etching, and negative staining techniques (9, 15, 26, 29-32, 40-42). The virion was initially described (10) as brick shaped with dimensions of approximately 300 by 230 nm, with inner and outer membranes sandwiching a pair of lateral bodies and enclosing a central core containing the genomic DNA.
In recent years, cryo-EM has made significant contributions to the study of vaccinia virus virion assembly and maturation (6, 12, 13, 36). Virion features prominent in conventional EM images, such as membranous surface tubules (30, 31), ridges (4), threads (29), helices (41), as well as lateral bodies and a dumbbell shape for the core, were not visible in cryo-EM images of the intact virion (6), indicating that they may have been artifacts resulting from virion dehydration and nonisotropic collapse during sample preparation. Nonetheless, structures reminiscent of the lateral bodies have been observed in some recent cryo-EM studies (12, 13), and the dumbbell shape has been argued to be bona fide, with its visualization dependent upon virion orientation during sectioning or vitrification. Due to these uncertainties, even cryo-EM does not provide an unequivocal model of the virion.
Because of its large size, lack of symmetry, and heterogeneity, vaccinia virus is not amenable to high-resolution EM image reconstruction analyses (1). Moreover, vaccinia virus virions are not well suited to cryo-sectioning because of cutting damage resulting in a loss of structural fine detail (6). Thus, despite recent advances in cryo-EM imaging of IMV, the detailed three-dimensional architecture of the virion remains in question.
We recently demonstrated that atomic force microscopy (AFM) can be used for the in situ imaging of intact virions ranging in diameter from 16 to 100 nm (22, 25) and have also shown (ex situ in the case of herpes simplex virus) that virion internal structures can be revealed by AFM imaging after chemical and enzymatic dissection (34). We have now applied in situ AFM to the direct visualization of hydrated and dried intact IMV virions, as well as virion cores, core substructures, and core-enveloping structures arising from chemical and enzymatic treatment thereof. To our knowledge, these studies have produced the first high-resolution in situ AFM images of a human virus in its hydrated state.
MATERIALS AND METHODS
Virus preparation
HeLa cells were maintained and infected with vaccinia virus (Western Reserve strain). The virus was isolated and purified, and its titer was determined as described previously (7, 8). It was stored at −80°C. The final virus titer was ∼4 × 1010 PFU/ml of suspension. By counting of particles by making AFM images of dried virion samples of known initial volume and dilution, the average particle density was estimated to be ∼8 × 1010 ml−1, leading to an estimated particle/PFU ratio of ∼2.
AFM
Droplets of IMV suspension (1 to 1.5 μl) prepared as described above, or chemically or enzymatically treated samples thereof, were deposited directly onto freshly cleaved mica substrates and allowed several minutes to settle. For imaging of fully hydrated samples in physiological buffer (in situ imaging), the mica substrates were transferred into the AFM fluid cell, which was subsequently filled with 50 mM HEPES-NaOH (pH 7.5). For imaging in air (ex situ imaging), sample droplets were allowed to dry, and mica substrates were then rinsed gently with double-distilled water and quickly dried with a stream of nitrogen gas. Images were collected with a Nanoscope IIIa AFM (Digital Instruments, Santa Barbara, Calif.) operated in tapping mode. Commercially available silicon tips and oxide-sharpened silicon nitride tips were employed for ex situ and in situ imaging, respectively. Other AFM experimental procedures and parameters were as described previously (22, 25, 34).
RESULTS AND DISCUSSION
Intact virus
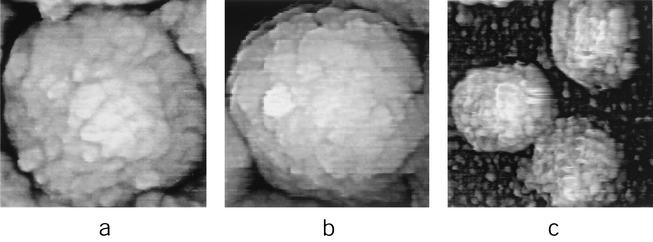
Intact, fully hydrated IMV single virions and aggregates thereof were adsorbed from suspension in Tris-HCl (pH 7.5) to mica and then imaged in situ by AFM (Fig. 1). While some virions were spherical, with a diameter of approximately 350 nm, the majority appeared more ellipsoidal, with major and minor axial dimensions in the ranges of 320 to 380 and 260 to 340 nm, respectively. Apart from being more rounded at the corners and edges, the majority of virions were not dramatically dissimilar, in lateral dimensions and shape, from the classical brick-shaped objects observed by EM (10) and cryo-scanning EM (cryo-SEM) (13). However, virion height as measured by AFM varied in the range of 240 to 290 nm, approximately double the virion heights estimated by cryo-SEM and EM (110 and 150 nm, respectively [13, 36]). Moreover, the height profile showed a distinct curvature that would be more characteristic of a three-dimensional ellipsoid than a flat brick-like surface. The accuracy of our observation of a curved virion profile is reinforced by the high precision with which height information is obtained by AFM. Thus, while estimates of vertical dimensions by cryo-SEM and EM depend on the tilt angle of the stage and are therefore error prone, height information estimated by AFM has a resolution approaching 0.1 nm (14). The heterogeneity of hydrated IMV in all three dimensions, as observed by AFM, may arise from virion-to-virion variability in the status of enveloping membranes or the presence of distinct forms in the virion population representing, perhaps, distinct stages of intracellular maturation.
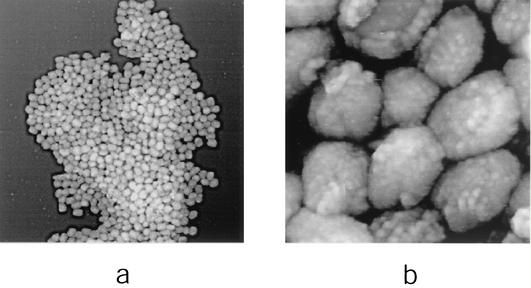
FIG. 1.
IMV virions adsorbed to mica. IMV was imaged in situ (in 50 mM Tris-HCl [pH 7.5]). Although isolated virions were observed in all experiments, the vast majority were found, irrespective of the concentration of virions in suspension, to be aggregated into two-dimensional arrays. AFM images are 10 by 10 μm (a) and 1.2 by 1.2 μm (b).
In contrast to the rather smooth appearance of the virion outer surface by cryo-EM (6, 12, 13), in situ AFM images showed the virion to have an irregular surface even at a relatively low resolution (Fig. 1b). Higher-resolution images (Fig. 2) indicated this appearance to arise from a high-density array of punctuate protrusions. Although irregular in their arrangement, these protrusions were fairly uniform in size, with a lateral diameter of 25 to 30 nm. How can the cryo-EM (smooth) and AFM (not smooth) views of the vaccinia virus virion surface be reconciled? Certainly, tip pressure on an elastic, soft biological surface such as the membrane of a living cell can lead to clearly defined images of underlying substructures such as intracellular cytoskeletal fibers, even though the latter are physically separated from the probe by an intervening ∼5-nm-thick cell membrane (23). This might be analogous to detecting the arrangement of ribs beneath the skin by passing a finger over them. At the other extreme, AFM images of relatively hard surfaces, such as the capsids of T = 3 icosahedral plant viruses (in which lateral resolution approached ∼20 Å), have proven to be remarkably consistent with the structures of the equivalent virions determined by X-ray diffraction analyses (22, 25). We think it reasonable to assume that the surface of an enveloped virus has elastic properties lying somewhere between these two extremes. Although it is possible that the apparent virion surface protrusions reflect the tracing of underlying structures, an equally if not more plausible explanation might arise from the intrinsic differences between the AFM and cryo-EM techniques. While in situ AFM is a nonpenetrating, intrinsically topographical technique, cryo-EM images provide a projection of the entire thickness of the sample (∼240 to 290 nm [see above]). Given the virion's electron-translucent properties, it would not be surprising if virion surface protrusions, which could be less than ∼20 nm in height after vitrification, were undetectable in high-resolution cryo-EM images (6, 12, 13) or even thin-section EM images (15), leading to a smooth, featureless topographical appearance for the virion.
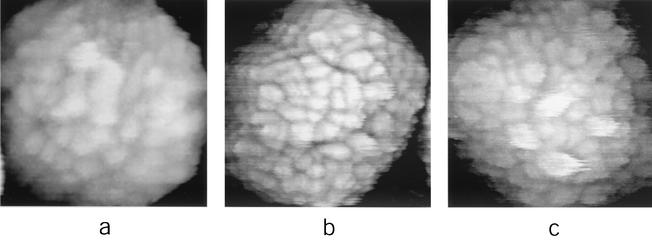
FIG. 2.
High-resolution, in situ AFM images of IMV virions (in 50 mM Tris-HCl [pH 7.5]). AFM images are 385 by 385 nm (a), 425 by 425 nm (b), and 365 by 365 nm (c).
In some of the most detailed images (Fig. 2a), fine structure comprising ∼6-nm-diameter subunits could be discerned within individual virion surface protrusions. Six nanometers corresponds to the diameter of a monomeric globular protein with a molecular mass of ∼120 to 150 kDa. Since the vaccinia virus genome encodes only three polypeptides in this size range, each of which are enzymes (the virus DNA polymerase and the two largest RNA polymerase subunits), and given that the 16 major vaccinia virus membrane proteins are all of considerably lower molecular mass (17), the ∼6-nm-diameter subunits presumably represent protein oligomers. Additional fine structure within the protrusions could not be resolved, perhaps because of slight deformations arising from the finite pressure of the AFM tip on the elastic surface of the enveloped virion, as discussed above.
Vaccinia virus is routinely dehydrated and rehydrated for storage and shipping without loss of infectivity, a property that was integral to the development of vaccinia virus as a vaccine for worldwide smallpox eradication (2). AFM imaging allows, for the first time, a side-by-side comparison of hydrated and dried vaccinia viruses. By AFM, ex situ (air-dried) IMV showed relatively uniform dimensions of 300 to 360 by 240 to 280 nm (lateral) by 120 to 130 nm (height) (Fig. 3). Thus, dehydration was associated with an overall shrinkage of the virion, with the most remarkable shrinkage (2.2- to 2.5-fold) occurring in the height dimension. Virion shrinkage was reversible upon rehydration (data not shown), indicating a remarkable deformability. Dehydration led to a more rectilinear virion shape (Fig. 3), suggesting that the classical brick shape described in EM studies (9, 10) may be a consequence of dehydration of the native ellipsoid (see above). Alternative surface features emerged upon dehydration, including a pronounced central, raised area extending approximately 30 nm above the virion surface. This feature, acting as a marker of virion orientation, indicated that the dried virions (both aggregated and nonaggregated) shared a common orientation on the imaging surface.
FIG. 3.
Air-dried IMV virions adsorbed to mica. In panel b the central raised area on the upper surface of one of the virions is indicated with an arrow. AFM images are 2.9 by 2.9 μm (a), 1.0 by 1.0 μm (b), and 350 by 350 nm (c).
Subviral structures
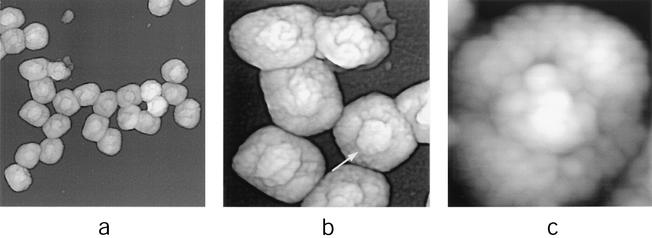
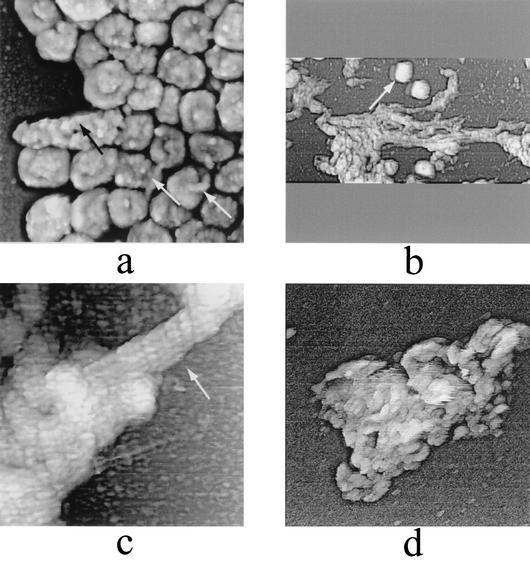
Although AFM provides information that is essentially topographical in nature, internal structural features may be probed by controlled dissection with combinations of nonionic detergent, disulfide reducing agents, protease, and ionic detergent. In an initial series of such experiments, we implemented an established procedure for the uncoating of intact, purified IMV in vitro (9), namely, treatment with nonionic detergent (Igepal, 1%) in combination with a protein disulfide reducing agent (2-mercaptoethanol, 2%). The effects of these two reagents may mimic the earliest events during infection (27), namely, the loss of the virion outer membrane(s) during cell membrane transit and arrival of the resulting (demembranated) virion in the reducing environment of the cytoplasm. Consistent with previous observations (6, 9), this treatment led not only to the presumed loss of outer membranes but also to the appearance of mica-immobilized viral coats (Fig. 4a and b) and cores (Fig. 4c and 5 a and b). Presumably, the separation of coats and cores was the result of disulfide reduction. Although comparable coats and cores have been observed previously by negative-stain EM imaging after equivalent treatment (9), AFM permitted visualization of their detailed topography in three dimensions and in clear detail. Thus, the coats seemed to retain a high degree of overall integrity, despite the possibility that structural changes may have been required for them to become separated from the cores. Their integrity was perhaps due to the resistance of significant numbers of protein cross-links to disulfide reduction by 2-mercaptoethanol. The coats were 25 to 35 nm thick and were comprised of arrays of “studs” of presumably cross-linked protein (Fig. 4a and b). The dimensions of the studs were equivalent to those of the protrusions imaged on the surfaces of intact, hydrated virions (Fig. 2), and they may represent identical structures imaged as a result of movement of the AFM tip over studs lying beneath the elastic, soft virion outer membrane(s) (as discussed above). In their location surrounding the virion core, the coats corresponded to what has been referred to in some EM studies as the virion outer membrane, despite being located within the virion (10). The structure has also been referred to as the core envelope (6).
FIG. 4.
Dissection of IMV virions with nonionic detergent in combination with a reducing agent (1% Igepal and 2% 2-mercaptoethanol in 50 mM Tris-HCl [pH 7.5]; treated for ∼30 to 45 min at 37°C). (a and b) Intact coats. (c) IMV virion cores. The satellite domain structures associated with intact and partially unfolded cores are indicated with black and white arrows, respectively. AFM images are 2.5 by 2.5 μm (a), 1 by 1 μm (b), and 1.35 by 1.35 μm (c).
FIG. 5.
High-resolution AFM images of cores after dissection of IMV virions with nonionic detergent in combination with reducing agent (1% Igepal and 2% 2-mercaptoethanol in 50 mM Tris-HCl [pH 7.5]; treated for 30 to 45 min at 37°C). (a and b) Core surface. (c) Seventy- to 100-nm-diameter particles. AFM images are 380 by 380 nm (a), 365 by 365 nm (b), and 400 by 400 nm (c).
The corresponding virion cores were imaged in both individual and aggregated forms on the mica surface. Cores had dimensions of 320 by 250 nm (lateral) by 170 to 220 nm (height) (Fig. 4c). The surface area of the core deduced from these dimensions (∼700 by ∼900 nm2) was comparable to the surface area of the coats. The average difference in dimensions between intact hydrated IMV and core was ∼70 nm, consistent with a coat of thickness of 25 to 35 nm (see above) plus an outer membrane thickness of ∼5 nm. Thus, the core and virion dimensions were mutually consistent.
The cores appeared to have an integral outer wall with very irregular outer surface pegs (Fig. 4c and 5a and b). This is consistent with EM data (9) showing that viral cores possess a ∼9-nm-thick electron-translucent outer shell (also described as a wall) decorated on the outside with what have been variously described as spicules, pegs (9), or a palisade layer (6) protruding from the core wall by ∼10 to 20 nm (9, 26, 41). The core wall is probably composed, in large part, of vaccinia virus proteins p4a and p4b (32), and the pegs may contain vaccinia virus protein p39/A4L (3, 33) as indicated by EM immunolocalization studies. A tight association of p39 with p4a/p4b has been reported (35).
Many of the viral cores possessed what we will term a satellite domain (Fig. 4c), corresponding to the central raised area on the surface of the dehydrated intact virion (Fig. 3). The former, like the latter, consistently occupied the upper surface with respect to the mica substrate. The raised area may correspond to the profile of the satellite domain beneath the coat or may be imaged via a hole in the coat. Whether or not the core's lower surface possesses an equivalent satellite domain is unclear. However, 70- to 100-nm-diameter particles (dimensions equivalent to those of the satellite domain) were observed attached to the mica surface after disassembly of intact IMV into coats and cores by treatment with nonionic detergent plus reducing agent (Fig. 5c). These particles may therefore be anchored satellite domains from which the core has become detached. Cryo-SEM images (12) of IMV undergoing cellular entry have also indicated the formation of independent, roughly spherical particles ∼90 to 100 nm in diameter along with cavities of equivalent size in the IMV, consistent with a loss of satellite domains. The satellite domains observed by AFM (this study) may correspond to the lateral bodies observed in conventional EM images. Consistent with this possibility, a nonionic detergent equivalent to the detergent (Igepal) that gave rise to the isolated ∼70- to 100-nm-diameter particles observed in Fig. 5c has been reported to be sufficient for the removal of lateral bodies from cores (26).
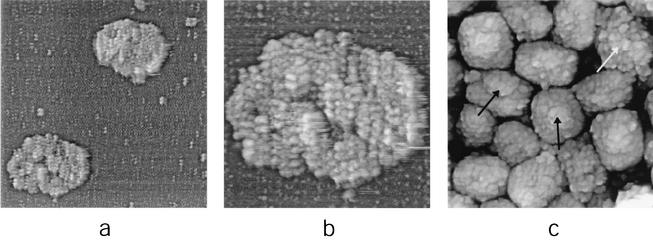
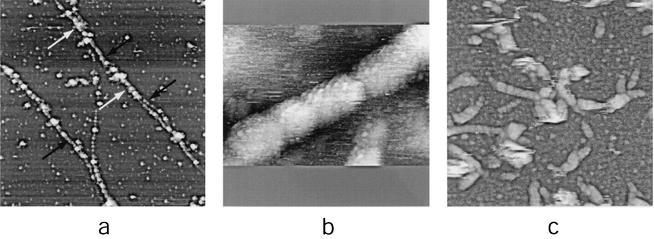
More prolonged treatment of IMV with nonionic detergent plus 2-mercaptoethanol at 37°C led to a partial unfolding of the virion core and the emergence of 30- to 40-nm-diameter tubules (Fig. 6a). After several hours of incubation at 37°C, cores unfolded more completely, revealing tangles of 30- to 40-nm-diameter tubules (Fig. 6b and c). Comparable 30- to 40-nm-diameter tubules have been observed by EM (9, 10, 12, 13) in samples of untreated IMV and upon treatment of IMV with detergent plus a reducing agent. This observation provided the basis for a prior model of IMV described as an “interconnected labyrinth of tubules and membrane cisternae” (12, 13). The combination of EM and AFM observations strengthens the view that the 30- to 40-nm-diameter tubules are integral components of the native virion as opposed to a structurally rearranged by-product of vaccinia virus proteins resulting from reducing conditions. The present study revealed the surface topography of the 30- to 40-nm-diameter tubules (such as the section of extended tubule indicated with an arrow in Fig. 6c) as a helically arranged array of protein subunits with left-handed helicity and a pitch of approximately 16 nm. Exposure of IMV to a protein disulfide reducing agent in the absence of detergent also affects its integrity (12, 24), consistent with a central role for disulfide bonding in the maintenance of core integrity (16). In our experiments, such treatment of IMV (with a slightly more active reducing agent than the 2-mercaptoethanol used above, namely, dithiothreitol [DTT]) (Fig. 6d) led to IMV core unfolding in a manner comparable to that observed with nonionic detergent plus reducing agent (Fig. 6a to c).
FIG. 6.
Unfolding of IMV virions. (a to c) Extended treatment with nonionic detergent plus disulfide reducing agent (1% Igepal and 2% 2-mercaptoethanol in 50 mM Tris-HCl [pH 7.5]; treated for 120 to 180 min at 37°C). (a) Several IMV virions (white arrows) have become partially unfolded, resulting in the appearance of 30- to 40-nm-diameter tubules. Other virions (black arrow) have unfolded completely. (b and c) Complete unfolding of IMV results in the appearance of tangled structures formed from 30- to 40-nm-diameter tubules. In panel b intact IMV virions (arrow) are also seen. (d) Thirty- to 40-nm-diameter tubular networks are seen upon treatment of IMV with the reducing agent DTT. AFM images are 1.6 by 1.6 μm (a), 6 by 3 μm (b), 570 by 570 nm (c), and 1.35 by 1.35 μm (d).
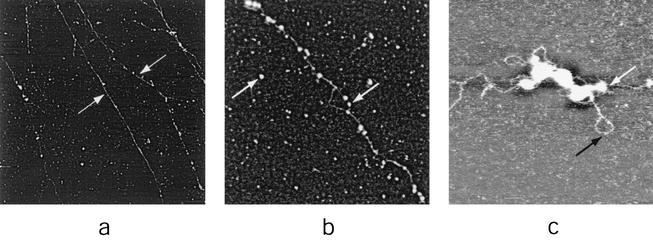
The 30- to 40-nm-diameter tubules produced under the relatively mild conditions of nonionic detergent plus 2-mercaptoethanol (Fig. 6) could be reproduced under more vigorous conditions of IMV degradation, namely, treatment with proteinase K in the presence of low concentrations (0.1%) of sodium dodecyl sulfate. Under the latter conditions, however, IMV disintegration continued beyond the 30- to 40-nm-diameter tubule stage, leading to the exposure and release of components within the tubules. The components initially revealed were comprised of filaments 16 nm in diameter (Fig. 7a). At higher resolution, the surfaces of the 16-nm-diameter filaments, like those of the 30- to 40-nm-diameter tubules, exhibited a helical geometry but were additionally linearly segmented (Fig. 7b). Large numbers of isolated 16-nm-diameter filament segments were frequently observed on the mica surface (Fig. 7c), presumably arising from filament breakage at segmental boundaries.
FIG. 7.
Dissection of IMV virions with proteinase K in combination with ionic detergent. (0.2 to 1 mg of proteinase K per ml in 0.1% sodium dodecyl sulfate-2 mM CaCl2-50 mM HEPES-NaOH [pH 7.5]; treated for ∼45 to 60 min at 37°C). (a) Partially digested portions of 30- to 40-nm-diameter tubules (white arrows) reveal 16-nm-diameter filaments (black arrows). (b and c) Sixteen-nanometer-diameter filaments. AFM images are 6 by 6 μm (a), 240 by 120 nm (b), and 1.5 by 1.5 μm (c).
Treatment of intact IMV with proteinase K at even higher concentrations, over longer periods, led to further virion disintegration, revealing extended strands of DNA on the mica surface that were associated with residual portions of the 16-nm-diameter filaments (Fig. 8). In some cases, loops were observed emerging from residual portions of partially degraded 16-nm-diameter filaments (Fig. 8c). Although the diameter of the DNA strands shown in Fig. 8c (2.2 to 2.8 nm) was appropriate for naked double-stranded DNA (helical diameter = 2.5 nm), the diameters of some portions (e.g., in Fig. 8b) were in the 5- to 8-nm range, suggesting the presence of residual filament-derived proteins. From Fig. 8 we conclude that, within the intact virion, the DNA is integral to or enclosed within the 16-nm-diameter filaments. In Fig. 8c, the DNA loop and the DNA strand to the left of the image appeared to be under superhelical stress, indicating that the packaged genome or domains thereof may be topologically restrained, perhaps by coiling. Superhelicity is controlled by the topoisomerase family of enzymes, and a vaccinia virus-encoded topoisomerase is present within the virion core (37, 39). In earlier studies, EM images of DNA complexes with purified vaccinia virus topoisomerase showed the formation of intramolecular loop structures in which noncontiguous DNA segments appeared to be interwound and synapsed within filamentous topoisomerase protein stems with right-handed helical geometry, perhaps as a plectonemic DNA supercoil (38). Our results may be consistent with the suggestion of the study that topoisomerase plays a role in packaging the vaccinia virus genome.
FIG. 8.
DNA strands formed upon extended treatment (90 to 180 min at 37°C) of IMV virions with proteinase K, as in Fig. 7. In panels b and c isolated 16-nm-diameter residual tubular segments are seen dispersed on the mica along with 16-nm-diameter tubular segments associated with DNA strands. Both are indicated with white arrows. In panel c a naked DNA strand is indicated with black arrow. AFM images are 15 by 15 μm (a), 1.5 by 1.5 μm (b), and 580 by 580 nm (c).
The physical length of the vaccinia virus genome is 65 μm. A 65-μm linear tube with a diameter of 30 to 40 nm (that of the tubules), would have a volume 4 to 6 times that of the entire virion core, which is clearly an impossibility. Our measurements are therefore consistent with a compaction of the genome within the apparently coaxial 16-nm-diameter filaments and 30- to 40-nm-diameter tubules. The upper limit on the degree of compaction would depend on the proportion of the entire core occupied by the tubule. In this regard, it is unclear whether there is an additional “viroplasm” within the core, or indeed within 16-nm-diameter filament segments themselves (whose outer diameter is ∼6.5 times that of the associated double-stranded DNA segment). An additional question would be why the 16-nm-diameter filament is segmented whereas the 30- to 40-nm-diameter enclosing tubules appear not to be. It is possible that 16-nm-diameter filament segmentation is required for folding of the tubules into the virion core, although for this to be so, the enclosing 30- to 40-nm-diameter tubules would presumably need to be flexible in the absence of corresponding segmentation. Alternatively, segmentation of the 16-nm-diameter filaments may play a role in gene expression, with segments corresponding to, say, transcriptional domains, each containing a “packet” of enzymes.
Summary
To summarize, the observations presented here are consistent with a structural model of the vaccinia virus virion comprising a hierarchy of substructures in which the linear DNA genome is enclosed within a 16-nm-diameter nucleoprotein filament that is formed from a helical assembly of protein subunits and divided into contiguous linear segments. The 16-nm-diameter filaments, in turn, appear to be enclosed within 30- to 40-nm-diameter tubules, also formed from helically arranged protein subunits but without visible segmentation. The 30- to 40-nm-diameter tubules interact with additional proteins, which fold, condense, or wind them into a compact virion core and/or comprise remnants of the enclosing core wall (9). DTT-induced disassembly indicates that disulfide bonding plays a role in the condensation or packing of the tubules. The intact core is apparently associated with at least one and possibly two 70- to 100-nm-diameter satellite domains which may correspond to the classical lateral bodies. The resulting structure is surrounded by a highly cross-linked protein coat, a number of whose internal cross-links may be resistant to nonionic detergent and mild reducing conditions. The outer surface of the IMV virions would be formed from lipid bilayer membranes, studded with transmembrane proteins (27).
As demonstrated here, in situ AFM can be an effective complement to other technologies for studying vaccinia virus virion structure and function. AFM allows direct visualization of viruses in their hydrated state and can probe surface topography in unrivaled detail. Moreover, scanning probe microscopy can be used to elucidate structural dynamics in vitro. It can readily produce single-particle, high-resolution, nonaveraged images of polymorphic and nonuniform virus populations. Although AFM does not yield images of internal structures within an intact virion as do penetrating techniques such as EM, nonetheless, by visualizing the surfaces of internal structures upon treatment with chemical and enzymatic agents, as we demonstrated here with vaccinia virus, modeling is possible.
Acknowledgments
We thank M. Plomp and A. Greenwood for technical assistance.
The National Aeronautics and Space Administration (grant NAG8-1559 to A.J.M.) supported this research. P.D.G. is supported by National Institutes of Health grant R01 GM501953 and National Science Foundation grant MCB0091260.
REFERENCES
- 1.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryoelectron micrographs. Microbol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collier, L. 1955. The development of a stable smallpox vaccine. J. Hyg. 53:76-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and J. K. Locker. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dales, S. 1963. The uptake and development of vaccinia virus in L strain cells followed with labeled viral deoxynucleic acid. J. Cell Biol. 18:51-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dales, S., and B. G. T. Pogo. 1981. Biology of poxviruses. Virology monographs. Springer-Verlag, Vienna, Austria. [DOI] [PubMed]
- 6.Dubochet, J., M. Adrian, K. Richter, J. Garces, and R. Wittek. 1994. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J. Virol. 68:1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earl, P. E., and B. Moss. 1991. Generation of recombinant vaccinia viruses, p. 16.17.6-16.17.8. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 8.Earl, P. L., and B. Moss. 1991. Preparation of cell cultures and vaccinia virus stocks, p. 16.16.1-16.16.7. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 9.Easterbrook, K. B. 1966. Controlled degradation of vaccinia virions in vitro: an electron microscopic study. J. Ultrastruct. Res. 14:484-496. [DOI] [PubMed] [Google Scholar]
- 10.Fenner, F., R. Wittek, and K. R. Dumbell. 1989. The orthopoxviruses. Academic Press, Inc., New York, N.Y.
- 11.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths, G., N. Roos, S. Schleich, and J. K. Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analysis. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, G., R. Wepf, R. Wendt, J. Krijnse-Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansma, H. G., and L. Pietrasanta. 1998. Atomic force microscopy and other scanning probe microscopies. Curr. Opin. Chem. Biol. 2:579-584. [DOI] [PubMed] [Google Scholar]
- 15.Hollinshed, M., A. Vanderplassen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichihashi, Y., M. Oie, and T. Tsuruhara. 1984. Location of protein-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J. Virol. 50:929-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus sequence. Virology 196:381-401. [DOI] [PubMed] [Google Scholar]
- 19.Joklik, W., and Y. Becker. 1964. The replication and coating of vaccinia DNA. J. Mol. Biol. 10:452-474. [DOI] [PubMed] [Google Scholar]
- 20.Kates, J., and J. Beeson. 1970. Ribonucleic acid synthesis in vaccinia virus. I. The mechanism of synthesis and release of RNA in vaccinia cores. J. Mol. Biol. 50:1-18. [DOI] [PubMed] [Google Scholar]
- 21.Krajioka, R., L. Siminovich, and S. Dales. 1964. The cycle of multiplication of vaccinia virus in Earle's strain L cells. Virology 24:295-308. [DOI] [PubMed] [Google Scholar]
- 22.Kuznetsov, Yu. G., A. J. Malkin, R. W. Lucas, M. Plomp, and A. McPherson. 2001. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 82:2025-2034. [DOI] [PubMed] [Google Scholar]
- 23.Kuznetsov, Yu. G., A. J. Malkin, and A. McPherson. 1997. Atomic force microscopy studies of living cells: visualization of motility, division, aggregation, transformation and apoptosis. J. Struct. Biol. 120:180-191. [DOI] [PubMed] [Google Scholar]
- 24.Locker, J. K., and G. Griffiths. 1999. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J. Cell Biol. 144:267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malkin, A. J., Yu. G. Kuznetsov, R. W. Lucas, and A. McPherson. 1999. Surface processes in the crystallization of turnip yellow mosaic virus visualized by atomic force microscopy. J. Struct. Biol. 127:35-43. [DOI] [PubMed] [Google Scholar]
- 26.Medzon, E. L., and H. Bauer. 1970. Structural features of vaccinia virus revealed by negative staining, sectioning, and freeze-etching. Virology 40:860-867. [DOI] [PubMed] [Google Scholar]
- 27.Moss, B. 2001. Poxviridae and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 28.Moss, B. 1991. Vaccinia virus: a tool for research and vaccine development. Science 252:1662-1667. [DOI] [PubMed] [Google Scholar]
- 29.Nagington, J., and R. W. Horne. 1962. Morphological studies of orf and vaccinia viruses. Virology 16:248-260. [DOI] [PubMed] [Google Scholar]
- 30.Noyes, W. F. 1962. Further studies on the structure of vaccinia virus. Virology 18:511-516. [DOI] [PubMed] [Google Scholar]
- 31.Noyes, W. F. 1962. The surface fine structure of vaccinia virus. Virology 17:282-287. [DOI] [PubMed] [Google Scholar]
- 32.Patrizi, G., and J. N. Middlecamp. 1968. Immature forms of vaccinia virus: morphological observations from thin sections of infected human skin. Virology 34:189-192. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. K. Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. Virology 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plomp, M., M. K. Rice, E. A. Wagner, A. McPherson, and A. J. Malkin. 2002. Rapid visualization at high resolution of pathogens by atomic force microscopy: structural studies of herpes simplex virus-1. Am. J. Pathol. 160:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risco, C., J. R. Rodriguez, W. E. Demkowicz, R. Heljasvaara, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1999. The vaccinia virus 39-kDa protein forms a stable complex with the p4a/4a major core protein early in morphogenesis. Virology 265:375-386. [DOI] [PubMed] [Google Scholar]
- 36.Roos, N., M. Cyrklaff, S. Cudmore, R. Blasco, J. Krijnse-Locker, and G. Griffiths. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 15:2345-2355. [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiguchi, J., N. C. Seeman, and S. Shuman. 1996. Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 93:785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shuman, S., D. G. Bear, and J. Sekiguchi. 1997., Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J. 16:6584-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuman, S., M. Golder, and B. Moss. 1989. Insertional mutagenesis of the vaccinia virus gene encoding a type I DNA topoisomerase: evidence that the gene is essential for virus growth. Virology 170:302-306. [DOI] [PubMed] [Google Scholar]
- 40.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westwook, J. C. M., W. J. Harris, H. T. Zwartouw, D. H. J. Titmuss, and G. Appleyard. 1964. Studies on the structure of vaccinia virus. J. Gen. Microbiol. 34:67-78. [DOI] [PubMed] [Google Scholar]
- 42.Wilton, S., A. R. Mohandas, and S. Dales. 1995. Organization of vaccinia envelope and relationship to the structure of intracellular mature virions. Virology 214:503-511. [DOI] [PubMed] [Google Scholar]