Abstract
Helper-dependent (HD) adenoviral vectors devoid of all viral coding sequences provide for safe and highly efficient gene transfer with long-lasting transgene expression. High titer stocks of HD vectors can be generated by using the cre-recombinase system. However, we have encountered difficulties with this system, including rearranged HD vectors and variable efficiency of HD vector rescue. These problems represent a major hindrance, particularly with regard to large-scale production. To overcome these limitations, we have modified the system in two ways: We constructed a new helper virus with a modified packaging signal and enhanced growth characteristics. We also redesigned the vector backbones by including noncoding adenovirus sequences adjacent to the right inverted terminal repeat and by incorporated a number of different segments of noncoding DNA of human origin as “stuffer.” Comparison of these vectors showed that the nature of the stuffer sequence affects replication of the HD vector. Optimization of the system resulted in a more robust and consistent production of HD vectors with low helper contamination and high in vivo potency.
The clinical success of gene therapy depends on the development of suitable gene transfer vehicles. High efficiency gene delivery, a favorable safety profile, and feasibility of large-scale production are the main requirements.
Adenoviruses have distinct advantages over other delivery methods, including high titers, the infection of dividing and nondividing cells, and a broad natural tropism that can be altered by modifications of the capsid (1, 2). Adenoviruses have safely been used as a live vaccine and have never been shown to induce tumors in humans. However, first generation (FG) adenoviruses are not suitable for long term stable expression. Low-level expression of viral genes is, to a large extent, responsible for direct vector toxicity, inflammation in the transduced tissue, and a strong cellular immune response against the virus (3–5). Numerous efforts have been undertaken to minimize these adverse effects. Removal of E2 and/or E4 genes has been shown to reduce late gene expression in noncomplementing cells (6–8). However, it is not clear whether this results in extended gene expression in vivo (9–11). Consequently, helper-dependent (HD) vectors deleted of most or all adenovirus coding sequences have been developed (12–17) and show great promise as gene transfer vectors for gene therapy. Long term expression of therapeutic genes has been observed in mice and monkeys (18–22), and toxicity is dramatically reduced or absent.
Growth of these vectors depends on coinfection of the producer cell by a second virus, which provides all necessary adenoviral proteins in trans. The HD vector is designed to retain only the elements required in cis for replication and packaging, allowing for ≈35 kb of “genomic space” to incorporate therapeutic genes and regulation systems. Any “unused” space is occupied by noncoding stuffer sequence to build the vector to a size that is suitable for optimal packaging.
Early attempts to develop the helper-dependent system were not very practicable because of high levels of contaminating helper virus, which proved difficult to eliminate (13, 14, 23). This approach became much more practical after a strong biological selection against the helper virus was introduced. It is based on cre-mediated excision of the packaging signal from the helper genome during coinfection (12, 16). The excision does not affect replication or protein synthesis. It does, however, render the majority of helper DNA molecules nonpackagable. Although this system allows for stable rescue and propagation of some vectors, we and others have observed that rearrangements of the HD vectors and outgrowth of variant helper viruses frequently occurs.
In this study, we have addressed deficiencies in both the helper virus and the HD vector by incorporating modifications that result in a more robust system capable of producing HD virus stocks with low helper contamination without the need for physical separation of helper and HD vector. We also demonstrate long term stable expression of erythropoietin from multiple HD vectors with different stuffer sequences of human origin in immunocompetent mice.
Materials and Methods
Construction of Helper Viruses.
Helper viruses were constructed by homologous recombination in Escherichia coli BJ5183 (24). They contain either a wild-type packaging signal (element A1-A5; helper H1) or a modified packaging signal with the sequence 5′- GTACACAGGA AGTGACTTTT AACGCGCGGT TTGTTACGGA TGTTGTAGTA AATTTGTCTA GGGCCGAGTA AGATTTGACC GTTTACGCGG GGACTTTGAA TAAGAGCGAG TGAAATCTGA ATAATTTTGT TGTACTCATA GCGCGTAATC TCTAGACG-3′, both flanked by lox sites (Helper H10, H14). In addition, H14 contains 2,902 bp of human DNA from chromosome11q13 (starting 13,340 bp upstream of Genthon-STS marker 11S1337) inserted into the XbaI site at nucleotide 28,593 (Ad5wt).
Construction of HD Backbones with Alternative Stuffer.
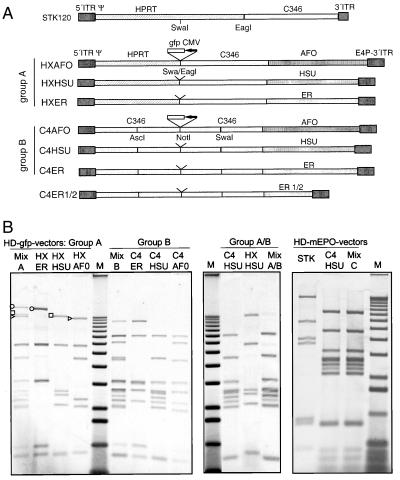
Fragments of human genomic DNA were selected by screening for the absence of genes, repeat elements, and coding sequence (program acedb, http://www.acedb.org), were amplified from genomic DNA, and were incorporated into the HD vectors after subcloning by homologous recombination in E. coli. The structure of new HD vectors is shown in Fig. 4A. In the first set of vectors, a gfp expression cassette was incorporated between the SwaI and EagI sites of pSTK120 (group A) and into the NotI site (group B). In vectors C4AFO-mEPO and C4HSU-mEPO, the gfp expression unit was replaced by a cassette containing the EF1α promoter, the mouse EPO cDNA, and the bGH poly(A) signal. This cassette was also introduced into the SwaI site of pSTK120 to create pSTK120-mEPO.
Figure 4.
Competition of HD vectors with altered stuffer sequences. (A) Structure of new HD vectors. To build HD vectors of group A, a 8.4-kb bp fragment between the unique Swa and Eag sites in STK120 was deleted. The following new sequences were introduced together with 400 bp of the right Ad5 terminus: fragment AFO (11, 3 kb, locus AF011889) and HSU (9, 3 kb, locus HSU 71148) located on Xq28 and ER [5, 4 + 5, 8 kb, estrogen receptor β region, Merck sequencing group (M.L.M., unpublished work)] located on 14q21. Vectors of group B contain 19 kb of sequence from the genomic region HUMDXS455A (cosmid C346), part of which is also present in pSTK120. This sequence was interrupted into four segments of ≈4.5 kb each. Segments were cloned in reversed orientation compared with their original position in the genome. Unique restriction sites (as shown) were created at the junction between fragments to allow for insertion of therapeutic genes. (B) P33-labeled HindIII restriction fragments of purified HD vectors. Vectors were generated individually or as mixtures by cotansfection of equimolar ratios of the respective plasmids. All amplifications are carried out with helper H14. Vectors carrying the gfp expression cassette were compared within and between groups A and B (MixA, B, A/B). One preferred vector of group B (C4HSU) was compared with the STK120-based vector (STK), both carrying the EPO expression cassette (MixC). Representative bands of each vector in group A are marked in the individual preparations and in the Mix.
HD Vector Rescue and Purification.
Vector production was carried out in 293 cre415 cells (a subclone of 293cre4) (25). HD vector genomes were released from the respective plasmids and were transfected by calcium phosphate coprecipitation in six-well plates in duplicates. Twelve hours after transfection, individual wells were infected with helper virus (75 and 150 particles/cell). Wells that reached cytopathic effect at 48 h after infection were chosen for propagation into passage 2. Cells were lysed by three freeze/thaw cycles, and 1/3 of the lysate was used to infect 8 × 105 293cre cells together with helper virus at 200 particles/cell. Infection cycles were repeated with increasing numbers of cells until passage 6 by using a helper dose of 200 particles/cell. At passage 6, HD vector from two 15-cm plates was isolated by a single CsCl step gradient and was analyzed by restriction digestion. Passage 6 was also used to produce large-scale vector stocks (10 layer Nunc cell factory). HD vectors were purified by triple CsCl banding (one step gradient followed by two continuous gradients).
Helper Quantification and Replication Assay.
The amount of helper genomes was measured relative to HD vector genomes by TaqMan real-time PCR (Perkin–Elmer). DNA was extracted from purified vectors by using the QIAamp DNA Kit (Qiagen, Hilden, Germany). Specific sets of primers and probes labeled with FAM (6-carboxylfluorescein) and TAMRA (6-carboxyltetramethylrhodamine) were designed for both helper and HD vector by using the program primer express (Perkin–Elmer). A plasmid containing both target sequences was constructed and used as a standard. Samples were analyzed with both primer/probe-sets in the absence or presence of spike. DNA copy numbers were calculated by using the 7700 software (Perkin–Elmer).The same technology was used to analyze replication of HD vectors relative to helper replication by using total DNA extracted from infected cells.
Virus Titration.
Helper viruses were titered by using a TCID50 end-point dilution method in a 96-well plates. In brief, 96-well plates were seeded with 3 × 103 293 cells and were infected after 48 h with a 3-fold serially diluted virus. Infected cells were incubated for 14 days, after which wells with cytopathic effect were scored visually.
Measurement of Infectious Titers of HDmEPO Vectors.
Infectious titers were determined by infection of HeLa cells in parallel with Ad-mEPO, an E1/E3-deleted adenovirus carrying the identical expression cassette. Serial dilutions of the vectors were used to infect 105 HeLa cells in 96-well plates. Twenty-four hours after infection, medium was harvested and mEPO concentration was measured by using the Quantikine IVD rhEPO ELISA kit (R & D Systems). Comparison of the amount of secreted mEPO allowed the estimation of the infectious titers of HD-mEPO vectors based on the plaque-forming unit titre of Ad-mEPO.
Mouse Experiments.
BALB/c mice were purchased from Charles River Italy (Lecco, Italy). The virus was diluted to the required dose with PBS, and 100 μl were injected into the tail veins of 8- to 9-week-old mice. Blood was obtained by retroorbital puncture. Hematocrits were determined by centrifugation of whole blood in heparinized tubes.
Results and Discussion
Homologies Between Helper and HD Vector Often Result in Vector Rearrangements.
The generation of HD viruses requires serial passages of the vector in cells co-infected with helper virus. Depending on the individual procedure and the amount of vector required, between 6 and 10 amplifications may be necessary (12, 15). Extended passaging increases the risk of changes in the vector genome, even for FG adenoviruses. The most likely event is the generation of wild-type viruses by homologous recombination with E1 sequences from 293 cells (26, 27).
In the helper-dependent system, the risk of rearrangements is even greater because of (i) the co-replication of two different genomes in a single infected cell resulting in extremely high concentrations of donor and target sequences within replication centers, (ii) partial sequence overlap between these genomes, and (iii) the absence of selection pressure to maintain the structure of the HD vector.
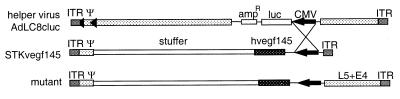
We initially constructed helper-dependent vectors based on plasmid pSTK120 (20). We introduced into this plasmid cytomegalovirus (CMV) promoter-driven expression cassettes (seap, vegf) located close to the 3′ inverted terminal repeat (ITR). The plasmid constructs were rescued into viruses (28–29 kb in length) and were amplified by coinfection with AdLC8cluc (12). The helper virus contains a CMV promoter-driven luciferase gene in place of the E3 region. When we analyzed both HD vectors after six passages, an unexpected structure was found. Restriction digestion revealed that these HD vectors incorporated the L5 and E4 regions from the helper virus (Fig. 1). The phenomenon was only observed when CMV promoters in helper and HD vector were located close to one end of the vector and the promoters from both vectors were oriented in the same direction. Sequence analysis confirmed that the exchange occurred within the CMV promoter. Five repeated amplifications of the STKvegf145 vector starting from transfection resulted in the identical product (data not shown). Consequently, recombination events between homologous sequences in co-replicating adenovirus vectors can occur at considerable frequency, and selection eventually results in a dominating new vector species.
Figure 1.
Rearrangement of HD vectors as a result of homologous recombination. Structure of the helper AdC8cluc, the HD vector STK120vegf145, and a mutant HD vector generated during amplification. ░⃞, adenovirus-derived sequences; , hvegf145-cDNA and bGH poly A; ◂, loxP-site; arrow, CMV promoter; —, CMV intron A.
The high homologous recombination rate of adenovirus is directly linked to replication (28) and can be explained by the persistence of single-stranded molecules stabilized by the DNA-binding protein, which eventually reanneal to form heteroduplexes (29). To create a stable system, homologous sequences within the HD vector and between HD vector and helper should be avoided.
Modified Helper Viruses Grow to High Yields and Avoid Recombinations with the HD Vector.
Growth characteristics of the helper virus as well as efficient suppression of its packaging have a major impact on yield and purity of helper-dependent vector preparations. Cre-mediated excision of the packaging signal was shown to efficiently suppress helper contamination (12, 16). Nevertheless, we sometimes observed outgrowth of helper virus during some HD vector amplifications generated with ADLC8cluc. The helper virus can escape selection because one of the lox sites flanking the packaging signals is lost by recombination between two identical packaging signals (230 bp) present in helper and HD vector (data not shown). The packaging signal of adenovirus serotype 5 is formed by at least seven functional units called A-repeats, with the consensus ATTTGN8GC (30). We modified the packaging signal in the helper by replacing the eight ambiguous nucleotides within each A-element by the sequence taken from a different A-element. The packaging signal in the HD vector was unaltered, thereby reducing contiguous homology between the two packaging signals to 22 bp. Table 1 shows that the helper containing the modified signal (H14) grew only slightly less efficiently in 293 cells than an identical virus with the unaltered packaging signal (H1). We found that the modification did not allow outgrowth of the helper in more than 30 independent HD- rescues.
Table 1.
Growth properties of helper viruses in 293 and 293cre cells
| dl70-3 without lox wild-type E3 | AdLC8luc | H1 wild-type Ψ E3 insert | H14 mod. Ψ E3 insert | H10 mod. Ψ wild-type E3 | |
|---|---|---|---|---|---|
| 293 | 1,267 ± 603* | 33.8 ± 5.86 | 860 ± 295 | 612 ± 141 | 930 ± 181 |
| 293cre415 | 1,450 ± 444 | 1.15 ± 0.38 | 1.48 ± 0.31 | 1.30 ± 0.23 | 1.78 ± 0.40 |
Cells (1 × 106) were infected with different HD vectors at multiplicity of infection of 2 and were lysed at 48 h, and virus was titrated as described.
*Infections units/cell.
In addition to recombination with the HD vector, the helper can also escape selection by obtaining the packaging signal and E1-sequences present in 293 cells. Parks et al. (12) have shown that large insertions placed in the E3 region prevent the formation of replication-competent viruses because viruses containing both the wild-type E1 region and an E3 insert exceed the packaging limit of Ad5 and are not viable. Unfortunately, we found (Table 1) that the insert used in helper AdLC8cluc (12) strongly affects virus growth. It has been shown that some E3 inserts can affect fiber production (31) possibly caused by the introduction of alternative splice signals. We have inserted human intronic sequence into an unaltered E3 region. The new insert had no major effect on virus yields: Growth of H14, which contains the E3 insert, is comparable to that of an identical virus lacking the insert (H10) and exceeds that of AdLC8cluc by more than one order of magnitude (Table 1). Despite different growth properties, cre-activity suppressed production of helper viruses that carry floxed packaging signals to similar levels as for AdLC8luc.
Because it is difficult to totally remove helper virus from final HD preparation, it was important to remove the reporter gene from the helper virus to avoid the production of an additional protein in target cells. The lack of an expression unit in the helper also removes areas of potential sequence overlap with the HD vector. To quantify the helper virus without a reporter gene during amplification, we developed an assay based on real time quantitative PCR.
A 400-bp Fragment from the Right End of the Adenovirus Genome Confers Growth Advantage.
As described above, recombination between identical sequences in the E3 region of the helper virus and at the right end of the HD vector repeatedly resulted in a modified HD vector with E4 and L5 regions that had a strong growth advantage and repeatedly overgrew the vector pool. The selective advantage may be attributable to either intact Ad5 transcription units (E4), size (32 kb, original vector 28 kb), or additional sequences acting in cis. However, the incorporation of potentially expressed E4 genes into the HD vector is not desirable because their expression may lead to an immune response to transduced cells.
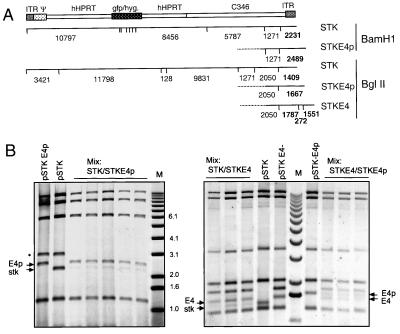
The first ORF (ORF1) of E4 starts at position 406 from the right end of the virus (32). We wanted to determine whether inclusion of the first 400 bp from the right end containing the E4 promoter but lacking any coding sequence would confer a similar selective advantage to the dependent vector. Furthermore, we compared effects of this sequence to those of the complete E4 region. Helper-dependent vectors based on pSTK120 were constructed containing (i) the 3′ ITR only (bp 1–118, STK120 gfp), (ii) the first 400 bp from the right end (STK120 gfp-E4p), or (iii) the 3′ ITR and the complete E4 region to position −3,415 (STK120 gfp-E4) (Fig. 2A). The multistep process of HD vector generation including transfection and serial passage makes it difficult to compare vectors produced individually. Competition experiments were therefore set up to compare pairs of different vectors, which were cotransfected at a 1:1 molar ratio to originate rescue of two competing HD vectors. After passage six, vectors from at least three independent rescue procedures were purified, and DNA was extracted and analyzed by restriction digestion and gel electrophoresis (Fig. 2B). Because fragments were end-labeled, the intensity of the bands directly corresponds to the molar ratio of the vectors. In five independent rescues, the 2,489-bp fragment originating from STK120 gfp-E4p was found to be 5–10 fold stronger than the fragment originating from STK120 gfp (Fig. 2B). Furthermore, the fragment from STK120 gfp-E4 was found at least 3-fold stronger than that of STK120 gfp. However, the discriminating fragments originating from STK120 gfp-E4p and STK120 gfp-E4 were of equal intensity in each rescue. These results suggest the presence of an element that provides growth advantage in both STK120 gfp-E4p and STK120 gfp-E4. The element must be entirely located in the right part of the E4 region (between position −400 from the right end and the 3′ end). E4-deleted vectors usually retain the E4 promoter sequence (7–9), but no cis-acting sequences have been reported in this area. Because HD vectors lacking this region can be amplified, the element seems not essential but is supportive to virus growth.
Figure 2.
Competition between HD vectors containing different fragments from the right end of Ad5. (A) Structure of STK120 gfp (STK) and its derivatives containing 400 bp STKgfpE4p (STKE4p) or 3,135 bp STKgfpE4 (STKE4) of the Ad5 3′ end. Fragment sizes for BamHI and BglII restriction digests are shown. Discriminating fragments are marked in bold. (B) P33-labeled restriction fragments of purified HD vectors resulting from cotransfections of two HD vectors in equimolar ratios. Five or three preparations were generated by independent transfection, amplification to passage 6, and single-step gradient banding. The respective plasmids digested with PmeI and BamHI or PmeI and BglII are shown for comparison. *, plasmid backbone.
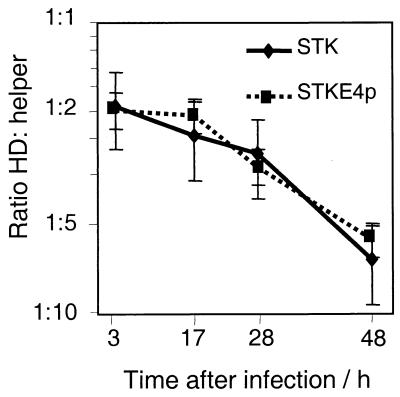
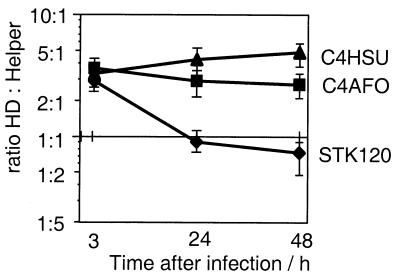
The E4 element either may allow for more effective replication of the vector DNA or it may support packaging. To test these two hypotheses, we performed genome replication studies. Cells were infected with either STK120 gfp or STK120 gfp-E4p and equal amounts of helper (H14). Plates were collected at different time points after infection, and genomic DNA was extracted. Additional plates were cultured to reach cytopathic effect. Virus was released and purified without separating helper and HD vector. Genomic DNA as well as DNA from purified virus was analyzed by real time PCR for the relative amounts of HD vector and helper (Fig. 3). For both tested vectors, the ratio between HD vector and helper changes from 1:2 at 3 h postinfection (which reflects the incoming virus) to 6:1 at 48 h. Thus, compared with the helper as an internal standard, both vectors had a similar amplification rate, and the E4 element has no effect on HD viral DNA replication.
Figure 3.
Effect of Ad5 3′ end sequences on replication or packaging. Cells were infected with helper 14 and either STKgfp or STKgfpE4p and were harvested at the indicated time points. Genomic DNA was extracted and analyzed for helper and HD vector sequences by TaqMan PCR. Changes in the ratio of HD vector and helper DNA over time indicate the replication of HD vectors relative to that of the helper. After a single-step gradient, the ratio between purified HD vector and helper was 14:1 (STKgfp) and 50:1(STKgfpE4p)
After packaging into capsids, the HD vector/helper ratio changed to 14:1 for STK120 gfp and to 50:1 for STK120 gfp-E4p as a result of selection against the helper. Helper genomes, which have escaped selection, should compete with HD genomes for packaging components. Given a similar ratio between free genomes (helper and HD vector) and identical selection against the helper in both experiments, the E4 element appears to affect the efficiency of packaging. This is rather unexpected because packaging is known to proceed from the left end, where the packaging signal is located (33).
Packaging of adenoviral DNA into capsids and even capsid formation strictly depends on presence of this signal (34). A cellular complex (P-complex) shown to bind packaging elements also interacts with the left ITR (35). The packaging signal overlaps with E1 enhancer elements. It is therefore not unlikely for proteins interacting with the right ITR and E4 promoter sequences to play a modulatory role, perhaps in completing the packaging process.
The experiment also shows that the helper virus amplifies faster than the HD vectors, a rather unexpected result because the HD genomes are shorter than the helper by >5.7 kb (18%), which should result in a faster replication cycle. However, because most of the HD sequences were chosen artificially, they may not perfectly meet the requirements of the adenovirus replication machinery.
The Choice of Stuffer Sequences Affects Vector Replication and Stability During Propagation.
HD vectors containing only a heterologous expression unit, and the required cis-acting signals would usually be much smaller than the wild-type adenovirus. Vectors of this size amplify less efficiently or multimerize (14, 36). To avoid this, stuffer sequences are added to provide a final size of 28–36 kb. The next important consideration is the source of the stuffer sequence. Vectors made with phage lambda as stuffer DNA were found to provide limited duration of transgene expression in vivo (37).
pSTK120 was thought to contain better stuffer DNA because the sequences are of human origin. But when we amplified vectors based on pSTK120 (constructed from 12 kb of the human genomic HPRT gene and 9 kb from cosmid C346), we occasionally found modified vectors that lacked part of the HPRT sequence and showed enhanced growth rates (data not shown).
The HPRT fragment is formed by introns 1 and 2 and exon 2. It contains 18 Alu repeats, two retroviral long terminal repeats, and several simple repeats and low complexity repeat regions. A matrix attachment region (MAR) is present in intron 1. The cosmid C346 sequence, which appeared to be stable during vector propagation, contains only one ALU repeat. We reasoned that repeat elements could contribute to instability and should be omitted from the stuffer. A total of 1.5 Mb of human genomic sequences was screened with regard to our criteria. Four fragments, each of ≈10 kb, were selected on the basis of being almost free of repeat units, lacking retrovirus elements and known genes. The fragments were used to construct two sets of HD vectors ranging between 29 and 31 kb in size (Fig. 4A). In one set (group A), the potentially unstable part of HPRT was replaced by new stuffer sequences. In the second set (group B), the HPRT region was completely removed, sequences from C346 sequences were extended, and new stuffer sequences were added. A vector of 25 kb that would be expected to amplify poorly because of size (36) was constructed as a control.
Whereas human DNA may be considered a suitable stuffer for human applications, it also poses concern for more frequent integration of the expression unit into the host genome by homologous recombination. To prevent integration, the contiguous stuffer sequence was interrupted. Individual fragments 4.5 kb in length were reversed when they were integrated in the HD vector backbone, and expression units were inserted only at the specific breakpoints. Using this strategy, the transgene is not flanked by regions that would support homologous recombination.
To compare stuffer sequences, vectors were rescued in competition by using the procedure described above. Instead of DNA from a single vector plasmid, an equimolar mixture of three or two vectors was used. Three “mixed rescue” experiments were carried out: one with HXAFO, HXHSU, and HXER (group A); one with C4AFO, C4HSU, and C4ER (group B); and the third with HXHSU and C4HSU (between groups A and B). Fig. 4B shows the restriction pattern of each vector amplified individually and the restriction pattern of the “Mix.” All fragments found in each of the individual vector amplifications are also present in Mix A or B. Because bands originating from different vectors have a comparable intensity in Mix A and B, no major growth differences exist between vectors within these groups. However, C4HSU (group B) clearly dominated in the competition with HXHSU (group A) (MixA/B). It appears that the retained HPRT sequences in HXHSU had an adverse effect on adenoviral vector propagation. A pronounced dominance was also found for the 31-kb vector (C4ER) in competition with a shortened version of this vector (C4ER1/2) with a size of 25 kb (data not shown). We conclude that, in addition to maintaining an appropriate size, stuffer sequence selection is also important and can have an effect on the amplification of HD vectors.
To investigate the behavior of the new HD vectors compared with pSTK120 in vivo, a murine EPO expression cassette was introduced into pSTK120, C4AFO, and C4HSU. In addition, pSTK120-mEPO and C4HSU-mEPO were cotransfected and amplified with H14. In this mixed amplification, STK120 was almost completely eliminated after six passages (Fig. 4).
In vivo performance of HD vectors is expected to depend on the amount of contaminating helper because the expression of viral genes is in part responsible for short-lived effects of FG vectors. To measure the helper content, we extracted DNA from large-scale virus preparations and determined the content of helper genomes relative to the amount of HD vector by quantitative PCR. Contamination by helper sequences in STK120-mEPO purified by step and equilibrium CsCl gradients was 1.0%. Even the crude stock of C4HSU-mEPO obtained after a single CsCl step gradient had a helper content of only 0.23%. Additional purification by equilibrium gradients reduced the helper load only slightly (to 0.18%). This helper content was equivalent to 5 plaque-forming units of helper virus in 104 infectious HD vector units. Combination of cre-recombinase-based selection with preferred growth characteristics of the optimized HD vector allows the production of HD vectors with relatively low helper content without physical separation from the helper. This is of considerable importance for larger scale production, where a chromatography-based purification process is desirable. C4HSU-mEPO and C4AFO-mEPO not only have a lower helper content but also increased yields (up to 8 × 103 part/cell).
Because only a limited number of HD vector constructs were tested, we can only speculate about factors contributing to vector amplification rates. The matrix attachment region of HPRT present in STK120 is one possible factor. Although the presence of a MAR is believed to support stability in vivo, it could negatively affect HD vector propagation. It is well established that the adenovirus genome is tightly bound to the nuclear matrix via the preterminal or terminal proteins during replication and is released for packaging (38, 39). Moreover, processing and phosphorylation of the preterminal protein seem to direct the genome to the appropriate sites within the nucleus for replication, transcription, and packaging (40). An additional MAR could interfere with either localization of the HD vector or its release from the matrix. Because the HPRT-MAR was removed from vectors of both group A and B, other differences in structure or sequence should have an impact as well. To study specific features in more detail, we carried out replication assays as described before.
Replication of STK120-mEPO, C4AFO-mEPO, and C4HSU-mEPO was measured relative to helper replication. Fig. 5 shows that, even if the amplification is started with an excess of HD vector, the ratio between vector and helper decreases 3-fold for the STK120-based vector. However, the starting ratio is maintained throughout the replication cycle for C4HSU-mEPO and only slightly decreasing for C4AFO-mEPO. This shows that the nature of the backbone has an impact on the rate of DNA replication. Considering the many years of selective evolution of the native adenovirus genome, a less efficient replication for artificially constructed sequences might be expected. Interestingly, replication is comparable for the helper and the vector C4HSU. The GC content of C4HSU is similar to that of adenovirus (52.1 and 55% respectively). C4AFO has a lower GC content (47%). STK120 has the lowest GC content (44.5%) within the series of these vectors because the HPRT fragment has a GC content of only 41%.
Figure 5.
Effect of stuffer sequences on replication. 293cre415 cells were infected with helper 14 and purified HD vectors at a ratio of 3:1 and were harvested at the indicated time points. Genomic DNA was extracted and analyzed for helper and HD vector sequences by real time PCR.
The Nature of the Stuffer Sequence Can Influence in Vivo Potency of HD Vectors.
Vector potency should be determined based on infectious titers. Because the HD vectors lack the ability to form virus plaques, we have compared expression in HeLa cells from different HD vectors to that of a FG vector containing the same expression unit. Based on this assay, ratios between infectious units and particles were calculated for STK120-mEPO, C4HSU-mEPO, and C4AFO-mEPO (1:100, 1:82, and 1:95 respectively). Because these ratios are very close to each other and similar to that of FG vectors, the nature of the stuffer did not influence potency in vitro.
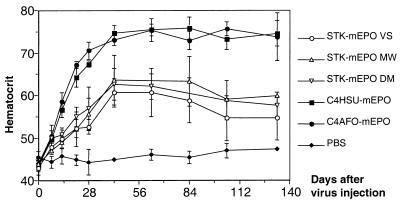
The performance of these vectors was compared in vivo by injecting 106 infectious units in the tail vein of BALB/c mice. This mouse strain was chosen because the mice develop a strong cytotoxic T-lymphocyte response and gene expression from FG vectors declines rapidly (41). Descamps et al. have shown that 5 × 109 infectious units of a FG vector given systemically were necessary to induce a significant increase in the hematocrit level (42). In the experiment shown in Fig. 6, 1 × 106 infectious units of STK120-mEPO caused an increase in the hematocrit level that plateaus at ≈60% and persists for at least 120 days. The result was highly reproducible for three independent stocks prepared by different researchers. At the same dose, C4HSU-mEPO and C4AFO-mEPO gave rise to a hematocrit above 70%, which was also stable for the duration of the experiment. The increased hematocrit level reflects a significantly higher potency for C4HSU-mEPO and C4AFO-mEPO. Our results are in agreement with earlier reports of highly efficient and stable gene transfer with helper dependent vectors (19, 20). All HD vectors used in those studies contain only stuffer DNA of mammalian origin. In contrast, incorporation of prokaryotic DNA (phage lambda) resulted in short-lived gene expression and lambda-specific cytotoxic T-lymphocyte response (37). These findings suggest that tolerance to the vector is as much required for stability as tolerance to the transgene (43). The response may not be limited to elimination of the transduced cells by cytotoxic T-lymphocytes. Intracellular mechanisms may lead to specific shut off in gene expression or elimination of the vector genome once the transduced cell is activated as described for hepatitis B virus in transgenic mice (44). Some of these mechanisms may have a stronger influence in the liver as compared with the heart or muscle, which in some cases can support long term transgene expression from FG vectors (43, 45). Because minimal vectors that lack stuffer sequences completely are also unable to direct stable gene transfer (46), stuffer DNA also seems to play an active role in vector stabilization. We have studied vectors containing identical expression units (promoter and transgene) and different human stuffer sequences in vivo. All vectors maintained expression in immunocompetent mice, but significant differences in expression levels were found between the vectors tested. These differences could be caused by factors associated with the nature of the backbone, such as the level of condensation, partial methylation, or the association with matrix proteins. The presence of a known MAR element in STK120 did not confer any advantage to the vector. In summary, a careful selection of sequences present in helper-dependent vectors is important to match requirements of production with high potency in vivo.
Figure 6.
In vivo comparison between HD vectors C4HSU-mEPO, C4AFO-mEPO, and STK120-mEPO. Groups of female BALB/c mice (n = 5) were infused in the tail vein with 1 × 106 infectious units of different HD-mEPO vectors. Blood samples were taken at the time points indicated and were analyzed for hematocrit levels. The results obtained for three independent stocks of STK120-mEPO prepared by different researchers are shown.
Acknowledgments
We thank Robin Parks and Frank Graham for providing us with the cre-expressing cell line cre4 as well as the helper virus AdLC8cluc. Plasmid pSTK120 was a generous gift of Stefan Kochanek, and STK120-gfp was provided by Fabio Palombo. We thank Paul Olson for helpful suggestions in the experimental design.
Abbreviations
- HD
helper-dependent
- FG
first generation
- CMV
cytomegalovirus
- ITR
inverted terminal repeat
- MAR
matrix attachment region
References
- 1.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickham T J, Tzeng E, Shears L L, II, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon R H, Engelhardt J F, Yang Y, Zepeda M, Weber-Pendleton S, Grossman M, Wilson J M. Hum Gene Ther. 1993;4:771–780. doi: 10.1089/hum.1993.4.6-771. [DOI] [PubMed] [Google Scholar]
- 5.Morral N, O'Neal W, Zhou H, Langston C, Beaudet A. Hum Gene Ther. 1997;8:1275–1286. doi: 10.1089/hum.1997.8.10-1275. [DOI] [PubMed] [Google Scholar]
- 6.Engelhardt J F, Ye X, Doranz B, Wilson J M. Proc Natl Acad Sci USA. 1994;91:6196–6200. doi: 10.1073/pnas.91.13.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armentano D, Sookdeo C C, Hehir K M, Gregory R J, St. George J A, Prince G A, Wadsworth S C, Smith A E. Hum Gene Ther. 1995;6:1343–1353. doi: 10.1089/hum.1995.6.10-1343. [DOI] [PubMed] [Google Scholar]
- 8.Gorziglia M I, Lapcevich C, Roy S, Kang Q, Kadan M, Wu V, Pechan P, Kaleko M. J Virol. 1999;73:6048–6055. doi: 10.1128/jvi.73.7.6048-6055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedieu J F, Vigne E, Torrent C, Jullien C, Mahfouz I, Caillaud J M, Aubailly N, Orsini C, Guillaume J M, Opolon P, et al. J Virol. 1997;71:4626–4637. doi: 10.1128/jvi.71.6.4626-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Neal W K, Zhou H, Morral N, Aguilar-Cordova E, Pestaner J, Langston C, Mull B, Wang Y, Beaudet A L, Lee B. Hum Gene Ther. 1998;9:1587–1598. doi: 10.1089/hum.1998.9.11-1587. [DOI] [PubMed] [Google Scholar]
- 11.Lusky M, Christ M, Rittner K, Dieterle A, Dreyer D, Mourot B, Schultz H, Stoeckel F, Pavirani A, Mehtali M. J Virol. 1998;72:2022–2032. doi: 10.1128/jvi.72.3.2022-2032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks R J, Chen L, Anton M, Sankar U, Rudnicki M A, Graham F L. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitani K, Graham F L, Caskey C T, Kochanek S. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher K J, Choi H, Burda J, Chen S J, Wilson J M. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 15.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar-Singh R, Chamberlain J S. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 18.Chen H H, Mack L M, Kelly R, Ontell M, Kochanek S, Clemens P R. Proc Natl Acad Sci USA. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 20.Morsy M A, Gu M, Motzel S, Zhao J, Lin J, Su Q, Allen H, Franlin L, Parks R J, Graham F L, et al. Proc Natl Acad Sci USA. 1998;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morral N, Parks R J, Zhou H, Langstron C, Schiedner G, Quinones J, Graham F L, Kochanek S, Beaudet A L. Hum Gene Ther. 1998;10:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- 22.Morral N, O'Neal W, Rice K, Leland M, Kaplan J, Piedra P A, Zhou H, Parks R J, Velji R, Aguilar C R E, et al. Proc Natl Acad Sci USA. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alemany R, Dai Y, Lou Y C, Sethi E, Prokopenko E, Josephs S F, Zhang W W. J Virol Methods. 1997;68:147–159. [PubMed] [Google Scholar]
- 24.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Anton M, Graham F L. Somatic Cell Mol Genet. 1996;22:477–488. doi: 10.1007/BF02369439. [DOI] [PubMed] [Google Scholar]
- 26.Lochmuller H, Jani A, Huard J, Prescott S, Simoneau M, Massie B, Karpati G, Acsadi G. Hum Gene Ther. 1994;5:1485–1491. doi: 10.1089/hum.1994.5.12-1485. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Grace M, Casale J, Chang A T, Musco M L, Bordens R, Greenberg R, Schaefer E, Indelicato S R. Hum Gene Ther. 1999;10:113–121. doi: 10.1089/10430349950019246. [DOI] [PubMed] [Google Scholar]
- 28.Young C S, Cachianes G, Munz P, Silverstein S. J Virol. 1984;51:571–577. doi: 10.1128/jvi.51.3.571-577.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahern K G, Wang K, Xu F Y, Mathews C Z, Pearson G D. Proc Natl Acad Sci USA. 1991;88:105–109. doi: 10.1073/pnas.88.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid S I, Hearing P. J Virol. 1997;71:3375–3384. doi: 10.1128/jvi.71.5.3375-3384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittal S K, Bett A J, Prevec L, Graham F L. Virology. 1995;210:226–230. doi: 10.1006/viro.1995.1337. [DOI] [PubMed] [Google Scholar]
- 32.Dix I, Leppard K N. J Gen Virol. 1992;73:2975–2976. doi: 10.1099/0022-1317-73-11-2975. [DOI] [PubMed] [Google Scholar]
- 33.Daniell E. J Virol. 1976;19:685–708. doi: 10.1128/jvi.19.2.685-708.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasson T B, Ornelles D A, Shenk T. J Virol. 1992;66:6133–6142. doi: 10.1128/jvi.66.10.6133-6142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid S I, Hearing P. J Virol. 1998;72:6339–6347. doi: 10.1128/jvi.72.8.6339-6347.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks R J, Graham F L. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks R J, Bramson J L, Wan Y, Addison C L, Graham F L. J Virol. 1999;73:8027–8034. doi: 10.1128/jvi.73.10.8027-8034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fredman J N, Engler J A. J Virol. 1993;67:3384–3395. doi: 10.1128/jvi.67.6.3384-3395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angeletti P C, Engler J A. J Virol. 1996;70:3060–3067. doi: 10.1128/jvi.70.5.3060-3067.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster A, Leith I R, Nicholson J, Hounsell J, Hay R T. J Virol. 1997;71:6381–6389. doi: 10.1128/jvi.71.9.6381-6389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barr D, Tubb J, Ferguson D, Scaria A, Lieber A, Wilson C, Perkins J, Kay M A. Gene Ther. 1995;2:151–155. [PubMed] [Google Scholar]
- 42.Descamps V, Blumenfeld N, Villeval J-L, Vainchenker W, Perricaudet M, Beuzard Y. Hum Gene Ther. 1994;5:979–985. doi: 10.1089/hum.1994.5.8-979. [DOI] [PubMed] [Google Scholar]
- 43.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 44.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 45.Chan S Y, Li K, Piccotti J R, Louie M C, Judge T A, Turka L A, Eichwald E J, Bishop D K. Nat Med. 1999;5:1143–1149. doi: 10.1038/13467. [DOI] [PubMed] [Google Scholar]
- 46.Lieber A, He C Y, Kirillova I, Kay M A. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]