Abstract
We have characterized a conformational epitope on capsids of hepatitis B virus (HBV) by cryo-electron microscopy and three-dimensional image reconstruction of Fab-labeled capsids to ∼10-Å resolution, combined with molecular modeling. The epitope straddles the interface between two adjacent subunits and is discontinuous, consisting of five peptides—two on one subunit and three on its neighbor. Together, the two icosahedral forms of the HBV capsid—T=3 and T=4 particles—present seven quasiequivalent variants of the epitope. Of these, only three bind this Fab. Occupancy ranges from ∼100 to ∼0%, reflecting conformational variations in the epitope and steric blocking effects. In the former, small shifts of the component peptides have large effects on binding affinity. This approach appears to hold general promise for elucidating conformational epitopes of HBV and other viruses, including those of neutralizing and diagnostic significance.
Antibody-combining sites on proteins are of two types, called linear and conformational epitopes, respectively (37). Linear epitopes typically consist of 6 to 12 consecutive amino acid residues: antibodies that recognize them also bind to the peptides in question and to the denatured protein, for example, in Western blots. However, some linear peptides are not recognized on the native protein because they occupy sites that are inaccessible to antibodies (cryptic epitopes). Conformational epitopes, on the other hand, are presented only when the antigen assumes its native conformation and are not recognized on peptides or denatured proteins. Most such epitopes, also called discontinuous epitopes (1), consist of two or more peptides from different parts of the polypeptide chain spatially juxtaposed by the protein's three-dimensional fold.
Complex antigens such as viral capsids potentially contain many epitopes. In the course of a natural infection, however, the number of immunodominant epitopes tends to be limited. The factors involved in selecting them are incompletely understood. The majority of epitopes on native (i.e., folded) protein antigens are thought to be conformational (4). These include many clinically important viral epitopes, such as the capsid-associated core antigen of hepatitis B virus (HBcAg) (30) and surface antigen (sAg), its envelope glycoprotein (12). An important goal for vaccine development is to be able to characterize immunodominant conformational epitopes, both in terms of the contributing peptides and structurally, in order to ultimately engineer their presentation on alternative platforms as synthetic antigens.
Linear epitopes may be mapped on the antigen's amino acid sequence by testing the reactivity of defined fragments with the antibody in question. This operation may be done systematically with the Pepscan technique, using a set of overlapping peptides that spans the entire sequence (18). Conformational epitopes cannot be characterized in this way, and mapping inferences tend to be indirect, based on binding competitions with other antibodies that have known linear epitopes. However, this approach is subject to pitfalls: antibodies are large molecules, and binding of an antibody to one site can block access of a second antibody to other sites considerably removed in space and greatly displaced along the antigen's primary sequence or even located on another subunit.
Conformational epitopes may be identified from crystal structures of the antigen complexed with Fab fragments of a monoclonal antibody, and such identifications have been achieved in a few cases (e.g., see references 24 and 32). However, this approach imposes daunting requirements in terms of the amount of material, crystallinity, and data analysis. Here we demonstrate an alternative approach based on cryo-electron microscopy (cryo-EM) of Fab-decorated antigens (31, 38), which requires less material by >3 orders of magnitude and has no need for crystals. The basic idea is as follows: provided that the antigen structure is known to high resolution, a cryo-EM structure of the antigen-Fab complex at moderate resolution, probed by molecular modeling with a generic Fab structure from the protein database, contains sufficient information to allow identification of the peptides that make up the epitope. We demonstrate proof of principle by using this approach to dissect the binding of monoclonal antibody 3120, which is specific for a hitherto unidentified conformational epitope on hepatitis B virus (HBV) capsids (35).
MATERIALS AND METHODS
Preparation of HBcAg capsids.
Capsids were prepared essentially as described (39). Construct Cp149.3CA consists of residues 1 to 149 in which the cysteines at positions 48, 61, and 107 have been changed to alanines. Capsids recovered from bacterial extracts by gel filtration were dissociated with 1.5 M urea at pH 9.5 into dimeric protein and further purified by gel filtration. Reassembly was induced by either dilution or dialysis into 100 mM HEPES-350 mM NaCl, pH 7.0. Capsids were freed of unassembled protein and buffer exchanged by gel filtration with 50 mM HEPES, 100 mM NaCl, pH 8.0. The particles were concentrated by ultrafiltration to 2 mg/ml. Protein concentration was determined by absorbance at 280 nm corrected for light scattering (ɛ280 = 29,500 M−1 · cm−1).
Generation of Fabs and decoration of capsids.
Monoclonal antibody (MAb 3120) was purchased from the Institute of Immunology, Tokyo, Japan. To produce Fab fragments, MAbs at 0.5 mg/ml were first reduced by adding EDTA to 1 mM and TCEP (Tris[2-carboxyethylphosphine] hydrochloride) (Pierce, Rockford, Ill.) to 3.5 mM and incubating for 0.5 h at room temperature. Immobilized papain resin (Pierce) was equilibrated with 10 mM EDTA-3.5 mM TCEP · HCl-20 mM PO4, pH 7.0. Antibody (55 μl) and centrifugally drained resin (15 μl) were mixed and incubated for 1 h at 37°C with tumbling. The resin was removed by centrifugal filtration. Fabs were mixed with capsids at a 2:1 ratio of Fab:dimer and incubated overnight at room temperature. Fc fragments were removed by adding 0.1 volume of settled immobilized protein G resin (Pierce), incubating for 0.5 h at room temperature, pelleting the resin in a microcentrifuge, and recovering the supernatant. The volume of the sample was reduced under a stream of nitrogen gas until a particle density satisfactory for cryo-electron microcopy was attained.
Cryo-EM.
Data were collected according to Cheng et al. (13). In brief, specimens were vitrified in thin films suspended over holey carbon films, and micrographs were recorded at a magnification of ×38,000 on a CM200-FEG microscope (FEI, Eindhoven, Netherlands) fitted with a Gatan 626 cryoholder, operating at 120 keV. Focal pairs of micrographs were recorded at ∼10 e−/Å2 per exposure. The first exposure was defocused such that the first zero of the contrast transfer function was at a frequency of (17 Å)−1 − (20 Å)−1. For the second exposure, the defocus was increased by 0.4 μm.
Image reconstruction.
Six focal pairs were digitized with a SCAI scanner (Z/I Imaging, Huntsville, Ala.), giving 7-μm pixels (1.8 Å at the specimen). Density maps were calculated with the PFT algorithm (2) as described previously (16), using preexisting maps of Cp147 capsids (T=4[15] and T=3[13]) as starting models. The reconstructions included all particles with correlation coefficients above a threshold of (mean − 1 standard deviation) for each of the three parameters calculated by PFT. Specifically, the threshold values were as shown in Table 1. Thus, the T=4 map used 1,710 out of 2,758 originally picked particles and had a resolution of 10.1 Å (Fourier shell correlation threshold, 0.3). The T=3 map used 479 particles out of 839 and had a resolution of 13.8 Å.
TABLE 1.
Threshold values for each of three correlation coefficients calculated by PFT
| Icosahedral form | Threshold value
|
||
|---|---|---|---|
| ccPFT | ccPRJ | ccCMP | |
| T = 4 | 0.416 | 0.458 | 0.372 |
| T = 3 | 0.428 | 0.386 | 0.341 |
Molecular modeling.
Coordinates of HBV capsid protein (PDB 1QGT [43]) were fitted into cryo-EM envelopes of T=3 and T=4 reconstructions by visual criteria (39), using the program O (21). The coordinates of an immunoglobulin G2a (IgG2a) Fab-peptide antigen complex (PDB 1FPT [40]) were then fitted into the Fab portion of the T=4 HBV-Fab 3120 density map at the site overlying the C-D interface (see Fig. 1 for designation of A, B, C, D—the four quasiequivalent subunits per T=4 capsid). Colores (11) was then used to refine the fits of the HBV subunits into both capsid reconstructions and of Fab 1FPT into the capsid-Fab 3120 reconstruction. The Fab coordinates were split into antigen-binding and non-antigen-binding domains to allow variation in the elbow angle. To save computation time and avoid redundant solutions, only the portion of the reconstruction immediately surrounding each subunit being fitted (2 to 3 Å beyond the surface of the volume determined by visual fitting) was included in the Colores fitting. The resulting T=4 coordinates differed by root-mean-square deviations of 1.0 and 3.2 Å from the “by-eye” and 1QGT coordinates, respectively. To test the robustness of our 1FPT fit, additional IgG2a structures—1FOR (23), 1AIF (3), and 1BOG (22) were fitted into the Fab density via Colores. The root mean square (RMS) differences between the three additional fits and the 1FPT fit were 0.9 to 1.5 Å as measured from the Cα positions of all residues in β-strands of the antigen-binding-domain, or 1.1 to 1.8 Å from the Cα positions of all residues not in variable-length loops.
FIG. 1.
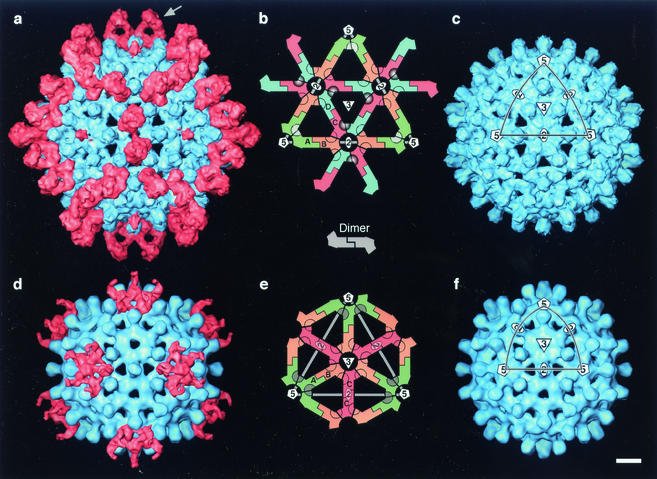
Cryo-EM reconstructions of HBV capsids labeled with Fab 3120. (a) T=4 capsid; (d) T=3 capsid. Capsid protein is blue; Fab density is pink. The capsids are viewed along a twofold symmetry axis. Control (unlabeled) capsids are shown (c and f) (from reference 15). (b and e) Lattice diagrams of single triangular facets, as marked (on panels c and f): they show the packing of dimers; the positions of symmetry axes; and the placement of quasiequivalent subunits (A, B, C, and D). Rings marking the positions of the epitopes are shaded according to occupancy: the C-D site on the T=4 capsid is dark, 100% occupancy; the A-A sites around the fivefold vertex are gray, 20 to 40% occupancy; the unoccupied D-B and B-C sites on T=4 and B-C and C-B sites on T=3 have empty rings. Bar = 50 Å.
To compare epitopes at different quasiequivalent sites, a pair of adjacent C-D subunits was aligned visually with corresponding pairs of A-A, B-C, and D-B subunits on the T=4 capsid and with pairs of A-A, B-C, and C-B subunits on the T=3 capsid. The C25-29 residues were used as a primary reference in these alignments. The positions of the Fabs bound at the A=A sites on both the T=4 and T=3 capsids were assigned by applying the same rotations and translations that moved the C and D capsid subunits to the A=A sites to the Fab overlying the C-D epitope. To allow a second Fab to bind at the same fivefold axis on the T=3 capsid, the first Fab was rotated a few degrees about its long axis, and a second Fab placed at an A=A site situated 144° around the fivefold axis from the first site. Density was then computed according to procedures and programs previously described (6, 20).
RESULTS
Visualization of Fab-labeled capsids.
HBV capsid protein consists of a core domain (residues 1 to 149) and a nucleic acid-binding protamine domain (residues 150 to 183). Core domains self-assemble into capsids (7). HBV capsids are dimorphic, the two variants having triangulation numbers of T=4 (120 icosahedrally coordinated dimers) and T=3 (90 dimers), respectively (17, 42). For the present experiments, capsids were prepared by expressing a core domain construct in Escherichia coli, isolating the capsids, dissociating them and further purifying the protein, and then reassembling capsids. The majority of capsids in such preparations are T=4 (44).
The immunogen used to generate MAb 3120 was HBV nucleocapsids isolated from enveloped virions (35). We found this antibody to be negative in Western blots (data not shown), indicating that it recognizes a conformational epitope (see also reference 25). A positive immunoprecipitation reaction was elicited only with capsids, this assay being negative with dissociated but folded dimers (data not shown).
Fab fragments were prepared and incubated with capsids. Negative staining EM showed that the capsids were densely covered with Fabs: accordingly, cryo-micrographs were also recorded. Three-dimensional density maps were calculated for both capsids (Fig. 1a and d and 2a and d). The T=4 map has a resolution of ∼10 Å, and the T=3 map, based on fewer particles, is at ∼14 Å. Fab-related density (pink in Fig. 1 and 2) is associated with the surfaces of both capsids, but its distribution differs markedly between them.
FIG. 2.
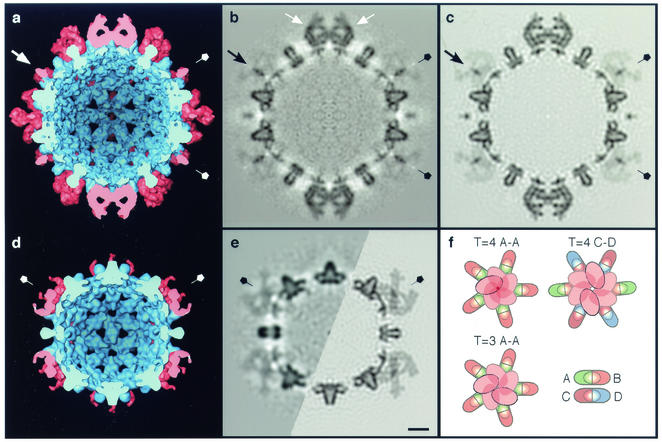
Interior views of cryo-EM density maps of HBV capsids labeled with Fab 3120, as represented by surface rendering (a and d) and in central sections (b and e [left half]). (a and b) T=4 capsid; (d and e) T=3 capsid. The capsids are viewed along a twofold axis of symmetry. (a and d) Capsid protein is blue, and Fab-derived density is pink. In the sections, protein density is dark. The corresponding sections from the quasi-atomic models, as limited to the same resolution as the T=4 reconstruction, are also shown (c and e [right half]). Note the close match between the cryo-EM density and the modeled density (cf. panels b and c and the left and right halves of panel e). (b) The two Fabs bound at C-D epitope sites on the T=4 capsid are marked in with white arrows. (a to e) Fivefold symmetry axes are labeled with pentagons. One example of the axial pencil of density on the T=4 fivefold axis is marked with a black arrow (b and c) or a white arrow in (a). (f) Schematic diagrams of the steric exclusion patterns at the fivefold vertices of both capsids (T=4 and T=3 A-A sites) and at the twofold axis of the T=4 capsid (T=4 C-D site) are shown. Bar = 50 Å (a to e).
Locations of Fab binding sites on the capsid surface.
There are four quasiequivalent subunits on the T=4 capsid and three on the T=3 capsid, each present in 60 copies. The T=4 subunits are designated A, B, C, and D, and the T = 3 subunits are A, B, and C (44), although the latter do not necessarily have the same conformations and intralattice interactions as the similarly named T = 4 subunits. The building blocks are dimers and their placement on both surface lattices is shown schematically in Fig. 1b and e. Together, the two capsids present seven quasiequivalent variants of any epitope.
The 25-Å-long spikes that protrude from the capsid surface are four-helix bundles (44) formed by two α-helical hairpins at the dimer interface (9, 15). Two copies of the so-called immunodominant loop, covering approximately residues 76 to 84 (28), are present at the tip of each spike (14, 43). However, MAb 3120 does not bind to the spike: rather, its epitope lies on the intervening “floor” region. This property is consistent with the observation that MAb 3120 bound to capsids in which the immunodominant loop was substituted with an exogenous peptide (29).
On the T=4 capsid, only one of the four quasiequivalent sites is occupied by density corresponding in size and shape to a Fab molecule (Fig. 1a, arrow). Elsewhere, and on the T=3 capsid, the observed distribution of bound density represents the superposition of Fabs bound at adjacent sites which cannot be simultaneously occupied because of steric interference (Fig. 2f). Consequently, their molecular envelopes merge in the density maps, so that features seen in surface renderings do not have the shapes of individual Fabs. Nevertheless, the maps are interpretable (see below).
The Fabs' footprints are marked on lattice diagrams in Fig. 1b and 1e, shaded according to occupancy, where the darkest shades indicate full occupancy and the lightest shades indicate zero occupancy. On the T=4 capsid, the fully occupied epitope overlies the interface between the C and D subunits. Interestingly, there is zero occupancy of the D-B and B-C sites around the same axis of twofold symmetry (which is also a quasi-sixfold axis). It follows that the avidity of Fab 3120's binding to the C-D epitope is much higher than to the D-B and B-C epitopes, presumably reflecting local structural variations. A consequence of this dominance by the C-D sites is that Fabs bound there do not become merged in the density map with Fabs at the D-B and B-C sites, which are unoccupied. This serendipitous property facilitated quasi-atomic modeling of the interaction between the Fab and the T=4 capsid, and identification of the peptides that make up the epitope (see below).
Differential binding of Fabs to quasiequivalent epitopes.
A thin pencil of density extends along the fivefold axis of the T=4 capsid (Fig. 1a and 2a and b). Although noise levels in icosahedral reconstructions tend to be elevated close to fivefold axes, this density is at the same level as the capsid shell (representing 100% occupancy) and far exceeds the noise level. It is contributed by Fabs bound to the A-A sites around this axis. The low value of other Fab-related density in this region (Fig. 2b) implies that only one of the five A-A sites is occupied at any given vertex, where a bound Fab occludes the other four sites slightly but enough to repel Fabs from them. This exclusion pattern is shown schematically in Fig. 2f. Thus, the pencil of axial density represents the overlap volume common to Fabs bound at all five sites, one of which is randomly occupied on each vertex. Accordingly, there are ∼72 Fabs bound per T=4 capsid: ∼60 at C-D sites at ∼100% occupancy and ∼12 at A-A sites at ∼20% occupancy.
The T=3 capsid exhibits less binding: in particular, there is zero occupancy of any of the six potential sites (three B-C and three C-B) around the threefold axis (cf. Fig. 1d and e). There is density associated with the A-A sites around the fivefold axis but it is distributed differently from the corresponding region of the T=4 capsid (cf. Fig. 2e and b, as well as Fig. 2d and a). We infer that a slight alteration in the binding aspect of Fabs at the T=3 A-A sites allows two molecules to bind per vertex instead of one, i.e., a bound Fab occludes the two adjacent sites but not the sites on the other side of the fivefold axis, where a second Fab may bind (Fig. 2f). This small change in binding aspect results in a qualitatively distinct pattern of density in the reconstruction.
In this context, we note that the two Fabs bound at apposing C-D sites on the T=4 capsid are almost touching (Fig. 2b, white arrows; Fig. 3b), and slight pivoting of one bound Fab toward the twofold axis that lies between them would occlude the other site. At the T=4 A-A sites, such impedance does occur and limits binding to one Fab per vertex. At the T=3 A-A sites, a slightly different binding aspect allows a second Fab to attach on the other side of the same vertex. When portrayed by surface rendering (Fig. 1d), the bound Fabs produce a five-fingered ring of coalescent density whose fingers represent the overlap volumes of pairs of adjacent subunits (Fig. 2f). Thus, maximum occupancy of accessible A-A sites corresponds to 24 Fabs per T=3 capsid.
FIG. 3.
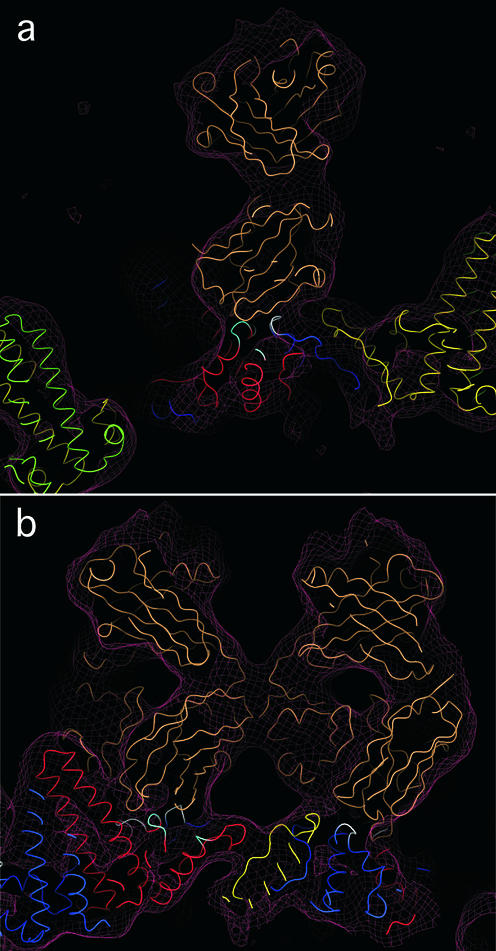
Quasi-atomic model of the interaction of Fab 3120 with the T=4 capsid in the vicinity of the C-D epitope site. Atomic coordinates of a Fab (PDB—1FPT [40] shown in tan) were fitted into the cryo-EM reconstruction of the capsid-Fab 3120 complex (maroon mesh), which already housed a quasi-atomic model of the capsid: subunit A (green), B (yellow), C (red), and D (blue). An excellent fit was obtained. Capsid protein residues at the capsid-Fab interface are colored white or cyan. (a) This slab of density, showing a side-view of one Fab, contains three of the five peptide segments that make up the epitope, marked in cyan and white. (b) Face-on view of two Fabs related by a twofold symmetry axis. This slab contains four of the five the epitope peptides, marked cyan and white (see Fig. 4). The figure was produced with DINO (http://www.dino3d.org).
Characterization of the epitope.
To explore the interaction of Fab 3120 with its epitope, we used our cryo-EM density maps of decorated capsids to construct quasi-atomic models. For the capsids, we used models derived from the crystal structure of the T=4 capsid at 3.3 Å resolution (43) in which the dimers were positioned so as to optimally fit our cryo-EM density maps. For the Fab, we used four crystal structures of molecules of the same subtype (IgG2a). The Fabs were docked into the density overlying the C-D site by an automatic procedure (11). All four probes fitted snugly into the density map (e.g., Fig. 3). To test the robustness of the fit, we calculated pairwise RMS deviations between them for the antigen-binding domains, omitting the variable-length loops from these comparisons. The deviations ranged from 1.1 to 1.8 Å. We conclude that although the structure of MAb 3120 has not been determined, it is likely to be closely similar to the Fabs analyzed and therefore its position is specified by the cryo-EM map to within ∼1 Å in each dimension.
To further appraise the verisimilitude of the quasi-atomic model of the Fab-labeled capsid, we excised its central section after band-limiting the model to the same resolution as the cryo-EM map and compared them (cf. Fig. 2c and b). An excellent match is observed not only with respect to the capsid shell but also with respect to the detailed substructure of the Fabs bound at the C-D sites (white arrows, Fig. 2b). To complete the labeling, a Fab was also assigned to the interface between adjacent A-A subunits at the same position relative to them as it occupies relative to the C-D interface (see Materials and Methods). Each of the five A-A sites per vertex was labeled at 20% occupancy. This procedure generated almost perfectly the ′pencil of density' that represents the overlap region along the 5-fold axis, and also reproduced the salient features of the faint surrounding density (cf. Fig. 2c and b). The same procedure was applied at the A-A sites on the T=3 capsid, in this case rotating the Fab by a few degrees and applying 40% occupancy, according to the inferred labeling pattern (Fig. 2f). Again, this procedure closely reproduced the Fab-associated features seen in the cryo-EM density map (cf. the apposing halves of Fig. 2e).
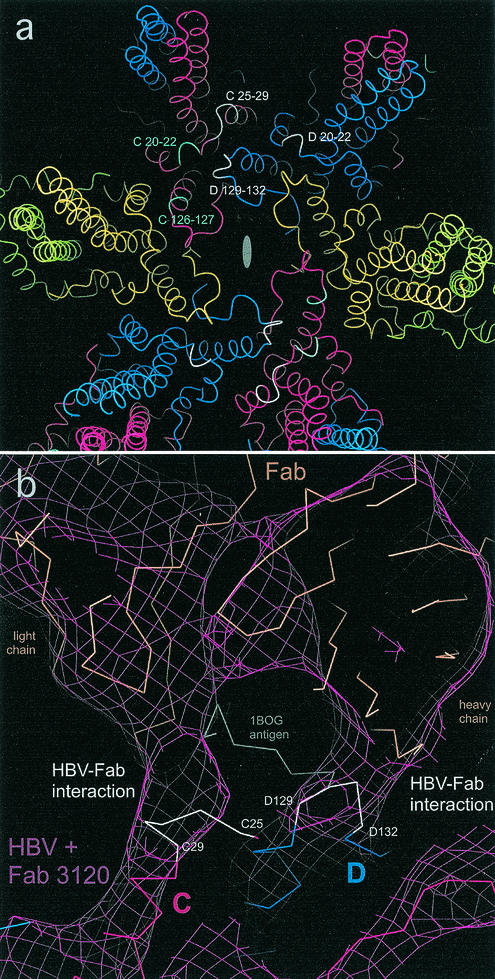
Two of the four Fab crystal structures coopted for this analysis included bound peptides—their respective linear epitopes. We initially thought that once the Fab-peptide complexes were docked into the cryo-EM map, the peptides might serve as markers for the viral epitope, but their positions vary considerably between the two examples. Accordingly, we focused on the regions at which there is strongest continuity of density between Fab 3120 and the capsid at the C-D site as primary indicator of the interaction. The portion of the density map shown in Fig. 4b shows two such interactions. The modeling suggested that the first complementarity-determining loop on the light chain of MAb 3120 and the second complementarity-determining loop on its heavy chain are, respectively, engaged. On the basis of this analysis, we identified the capsid protein peptides that come closest to the Fab, concluding that five peptides on two adjacent subunits contribute to the epitope. D20-D22, D129-D132, and C25-C29 (white in Fig. 4a) are in the region of highest interfacial density and therefore are likely to have the strongest interactions: C126-C127 and C20-C22 (cyan in Fig. 4a) are also involved, but in lower density regions. The positions of the five peptides are marked in Fig. 4a.
FIG. 4.
(a) Model of region around a twofold axis on the HBV T=4 capsid, showing the residues that bind to Fab 3120 at the C-D interface. Subunits are colored as follows: A, green; B, yellow; C, red; D, blue. An oval marks the twofold axis. The major interactions involve peptides D20-D22, D129-D132, and C25-C29 (white), where continuity of density is seen between the capsid and the Fab when the map is contoured at 2.3 sigma above background. Secondary interactions involve C126-C127 and C20-C22 (cyan), where continuity of density is seen between the capsid and the Fab when the map is contoured at 1.8σ above background. (b) Very high magnification detail of the interface region between Fab 3120 and the C-D epitope. Cryo-EM density is contoured at 2.3σ (maroon mesh) with quasi-atomic models showing interaction of Fab 3120 with capsid residues C25-C29 (left) and D129-D132 (right). A Fab (tan) (PDB-1BOG; [22]), fitted into the cryo-EM density (maroon mesh), is shown along with its peptide antigen (gray). Subunits A and B and the interactions of D20-D22, C20-C22, and C126-C127 with the Fab are not seen in this view. Residues also implicated in the interaction (density = 1.8σ in interface) are cyan. The figure was produced with DINO.
DISCUSSION
Epitopes exposed on folded proteins mainly involve surface loops. It has been argued that all such epitopes should be discontinuous, on grounds that the size of the antigen-combining surface of a Fab is such that more than one peptide loop is required to fully engage it (4, 24). Formally, this argument is persuasive but, in practice, many peptides are sufficiently reactive with antibodies to serve as highly specific reagents in immunochemical assays. In such cases, although several peptides may participate in the native epitope, one of them binds the antibody strongly enough to yield a significant interaction on its own; i.e., it is de facto a linear epitope. On the other hand, strictly conformational antibodies recognize only the native antigen. The present study is concerned with methods to identify the peptides involved in epitopes of the latter kind and with exploring the dependence of binding affinity upon conformation. Whereas conformational antibodies are supposed to bind their native antigens, MAb 3120 binds to only three of seven quasiequivalent variants of its epitope.
Quasiequivalent variations in binding affinity.
The epitope lies on the capsid surface, straddling the interface between adjacent subunits packed around symmetry axes (Fig. 1). The principal binding site involves C and D subunits on the T=4 capsid. There are two such sites on either side of each twofold axis. Two more pairs of quasiequivalent sites around the same axis (D-B and B-C) are unoccupied. Once the C-D sites are occupied, access to the neighboring sites is blocked, so the question of whether they retain some binding affinity is moot.
On both capsids, the Fab binds to A-A sites around the fivefold axes but steric blocking prevents occupation of more than one site per vertex on the T=4 capsid or two sites per vertex on the T=3 capsid.
On the T=3 capsid, both the B-C and the C-B sites (three of each) around the threefold axis are unoccupied (Fig. 1e). Under the conditions of this assay, once binding to the C-D and A-A sites was complete, there was a 3:2 molar ratio of unbound Fabs to unoccupied epitopes, or an 8.4:1 molar ratio of unbound Fabs to threefold axes on T=3 capsids (anticipating steric accessibility of one or two Fabs per such axis). Thus, the affinities of the B-C and C-B sites must be very low. However, it is not straightforward to interpret occupancies observed in cryo-EM experiments in terms of association constants.
Variability in occupancy among quasiequivalent versions of the same epitope appears to be quite common among viral antigens. For example, marked differences in this respect have been observed between the penton and hexon forms of the major capsid protein of herpes simplex virus (36) and cucumber mosaic virus (10) and on L1, the major capsid protein of papillomavirus (8). We suspect that a similar mechanism may underlie these disparities, i.e., small relative shifts of the peptides that make up the epitope, between different quasiequivalent sites.
Conformational nature of the MAb 3120 epitope.
According to our docking experiments, this epitope consists of five peptides on two subunits of neighboring dimers. This observation explains why MAb 3120 does not react with denatured monomeric capsid protein nor with folded but unassembled dimers.
We tried challenging the binding of MAb 3120 to capsids with a 10-fold molar excess of two peptides, covering residues 21 to 29 and 126 to 131, respectively; together, they present all of the five shorter peptides that we infer to constitute the epitope. However, essentially no interference was registered in a Biacore assay (data not shown), whence we conclude that the association constants of the peptides are too low for them to compete effectively with the composite epitope, appropriately configured on the capsid surface.
We also compared the modeled structures of the seven quasiequivalent versions of the epitope. RMS differences of ∼2 Å were detected among them, but these differences did not correlate intelligibly with occupancy. Differences of the same order were encountered between the crystal structure of the T=4 capsid and the quasi-atomic model, reflecting either limitations of our cryo-EM model at 8-Å resolution (15) (subsequently refined from 9 Å to 8 Å resolution [J. F. Conway, unpublished results]), or that there are small but real differences between the structures studied, arising from the mutant used, crystal packing effects, or the preparation histories of the capsids. We conclude that subtle shifts in the relative positions of the five peptides—which are below our threshold of reliable detection—account for the observed distinctions in Fab binding affinity.
Characterizing conformational epitopes on other antigens.
The first cryo-EM investigations of capsids decorated with antibodies were performed over ten years ago (27, 38a, 38). When feasible, molecular modeling enhances the interpretability of the resulting density maps (38). This area of research, recently reviewed by Smith (31), now covers numerous applications, many of which have addressed mechanisms of antibody-based neutralization (e.g., see references 19, 26, and 41), while others have identified the locations of specific proteins (27) or individual peptides (14, 33, 34). In the present study, the relatively high resolution of the cryo-EM analysis (10 Å) made it possible to identify the peptides present in a discontinuous, strictly conformational, epitope.
We are optimistic that this approach will prove useful in other systems. In virological applications, it should help to elucidate antigenicities and thus aid in vaccine design. The conditions for applicability are as follows: first, a high resolution structure of the antigen—or at least the component that the antibody of interest recognizes—should be available. Second, the antigen should be large enough to be amenable to cryo-EM analysis, i.e., at least 250 kDa or so (5) (all capsids qualify). Fab binding facilitates such analysis because it augments the size and irregularity of the structure to be determined. A third consideration is the quality of the cryo-EM map. Although much the same conclusion with respect to epitope placement could have been reached at lower resolution, the 10-Å resolution of our T=4 map made it possible to specify the interactions at the Fab-capsid interface. In summary, the signal-to-noise ratio and occupancy should be high enough to clearly delineate the Fab; the resolution should suffice to specify its orientation relative to the point of contact; and the hand of the EM map should have been established.
One potential complication with antigens that have many copies of the same epitope is steric interference between neighboring binding sites, which results in the distribution of bound density having the shape not of a single Fab but rather, of an amalgam of overlapping Fabs—e.g., see references 8 and 10 and our A-A sites in Fig. 1. However, this problem may be tackled by first identifying the footprint of the Fab on the capsid surface and then pivoting the model structure about this contact point to optimize the fit in the proximal portion of the density map (D. M. Belnap et al., unpublished data).
Acknowledgments
We thank D. Milich (Scripps) and R. Tedder and B. Ferns (Middlesex Hospital) for helpful correspondence, P. Ober (NIAMS) for programming help, W. Wriggers (Scripps) for help with Colores, and B. Heymann (LSBR) for software.
REFERENCES
- 1.Atassi, M. Z., and J. A. Smith. 1978. A proposal for the nomenclature of antigenic sites in peptides and proteins. Immunochemistry 15:609-610. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 3.Ban, N., C. Escobar, K. W. Hasel, J. Day, A. Greenwood, and A. McPherson. 1995. Structure of an anti-idiotypic Fab against feline peritonitis virus-neutralizing antibody and a comparison with the complexed Fab. FASEB J. 9:107-114. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, D. J., M. S. Edwards, and J. M. Thornton. 1986. Continuous and discontinuous protein antigenic determinants. Nature 322:747-748. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister, W., and A. C. Steven. 2000. Macromolecular electron microscopy in the era of structural genomics. Trends Biochem. Sci. 25:624-631. [DOI] [PubMed] [Google Scholar]
- 6.Belnap, D. M., A. Kumar, J. T. Folk, T. J. Smith, and T. S. Baker. 1999. Low-resolution density maps from atomic models: how stepping “back” can be a step “forward.” J. Struct. Biol. 125:166-175. [DOI] [PubMed] [Google Scholar]
- 7.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 64:3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booy, F. P., R. B. Roden, H. L. Greenstone, J. T. Schiller, and B. L. Trus. 1998. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 Å resolution. J. Mol. Biol. 281:95-106. [DOI] [PubMed] [Google Scholar]
- 9.Böttcher, B., S. A. Wynne, and R. A. Crowther. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88-91. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, V. D., E. S. Chase, A. W. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacon, P., and W. Wriggers. 2002. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317:375-384. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. C., K. Delbrook, C. Dealwis, L. Mimms, I. K. Mushahwar, and W. Mandecki. 1996. Discontinuous epitopes of hepatitis B surface antigen derived from a filamentous phage peptide library. Proc. Natl. Acad. Sci. USA 93:1997-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, N., J. F. Conway, N. R. Watts, J. F. Hainfeld, V. Joshi, R. D. Powell, S. J. Stahl, P. E. Wingfield, and A. C. Steven. 1999. Tetrairidium, a four-atom cluster, is readily visible as a density label in three-dimensional cryo-EM maps of proteins at 10-25 Å resolution. J. Struct. Biol. 127:169-176. [DOI] [PubMed] [Google Scholar]
- 14.Conway, J. F., N. Cheng, A. Zlotnick, S. J. Stahl, P. T. Wingfield, D. M. Belnap, U. Kanngiesser, M. Noah, and A. C. Steven. 1998. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J. Mol. Biol. 279:1111-1121. [DOI] [PubMed] [Google Scholar]
- 15.Conway, J. F., N. Cheng, A. Zlotnick, P. T. Wingfield, S. J. Stahl, and A. C. Steven. 1997. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386:91-94. [DOI] [PubMed] [Google Scholar]
- 16.Conway, J. F., and A. C. Steven. 1999. Methods for reconstructing density maps of “single particles”from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106-118. [DOI] [PubMed] [Google Scholar]
- 17.Crowther, R. A., N. A. Kiselev, B. Böttcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 18.Geysen, H. M., S. J. Rodda, T. J. Mason, G. Tribbick, and P. G. Schoofs. 1987. Strategies for epitope analysis using peptide synthesis. J. Immunol. Methods 102:259-274. [DOI] [PubMed] [Google Scholar]
- 19.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 20.Heymann, J. B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156-169. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 22.Keitel, T., A. Kramer, H. Wessner, C. Scholz, J. Schneider-Mergener, and W. Hohne. 1997. Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity. Cell 91:811-820. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H., T. J. Smith, W. M. Lee, A. G. Mosser, R. R. Rueckert, N. H. Olson, R. H. Cheng, and T. S. Baker. 1994. Structure determination of an Fab fragment that neutralizes human rhinovirus 14 and analysis of the Fab-virus complex. J. Mol. Biol. 240:127-137. [DOI] [PubMed] [Google Scholar]
- 24.Mariuzza, R. A., S. E. Phillips, and R. J. Poljak. 1987. The structural basis of antigen-antibody recognition. Annu. Rev. Biophys. Biophys. Chem. 16:139-159. [DOI] [PubMed] [Google Scholar]
- 25.Milich, D. R., J. Hughes, J. Jones, M. Sallberg, and T. R. Phillips. 2001. Conversion of poorly immunogenic malaria repeat sequences into a highly immunogenic vaccine candidate. Vaccine 20:771-788. [DOI] [PubMed] [Google Scholar]
- 26.Porta, C., G. Wang, H. Cheng, Z. Chen, T. S. Baker, and J. E. Johnson. 1994. Direct imaging of interactions between an icosahedral virus and conjugate F(ab) fragments by cryoelectron microscopy and X-ray crystallography. Virology 204:777-788. [DOI] [PubMed] [Google Scholar]
- 27.Prasad, B. V., J. W. Burns, E. Marietta, M. K. Estes, and W. Chiu. 1990. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature 343:476-479. [DOI] [PubMed] [Google Scholar]
- 28.Sällberg, M., U. Ruden, L. O. Magnius, H. P. Harthus, M. Noah, and B. Wahren. 1991. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. J. Med. Virol. 33:248-252. [DOI] [PubMed] [Google Scholar]
- 29.Schödel, F., A. M. Moriarty, D. L. Peterson, J. A. Zheng, J. L. Hughes, H. Will, D. J. Leturcq, J. S. McGee, and D. R. Milich. 1992. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J. Virol. 66:106-114. (Erratum, 66:3977.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifer, M., and D. N. Standring. 1995. Assembly and antigenicity of hepatitis B virus core particles. Intervirology 38:47-62. [DOI] [PubMed] [Google Scholar]
- 31.Smith, T. J. Structure of antibody/virus complexes. In W. Chiu and J. E. Johnson (ed.), Virus structure, in press. Academic Press, San Diego, Calif.
- 32.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer, J. V., B. L. Trus, F. P. Booy, A. C. Steven, W. W. Newcomb, and J. C. Brown. 1997. Structure of the herpes simplex capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology 228:229-235. [DOI] [PubMed] [Google Scholar]
- 33a.Steven A. C., W W. Newcomb, F. P. Booy, J. C. Brown, and B. L. Trus. 1992. Immuno-electron miroscopy at sub-nanometer resolution. Proc. 50th Ann. Mtg. EMSA 1:522-523. [Google Scholar]
- 34.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, K., A. Machida, G. Funatsu, M. Nomura, S. Usuda, S. Aoyagi, K. Tachibana, H. Miyamoto, M. Imai, T. Nakamura, Y. Miyakawa, and M. Mayumi. 1983. Immunochemical structure of hepatitis B e-antigen in the serum. J. Immunol. 130:2903-2907. [PubMed] [Google Scholar]
- 36.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Regenmortel, M. V. H. 1992. Molecular dissection of protein antigens, p. 1-29. In M. V. H. van Regenmortel (ed.), Structure of antigens, vol. 1. CRC Press, Boca Raton, Fla.
- 38.Wang, G. J., C. Porta, Z. G. Chen, T. S. Baker, and J. E. Johnson. 1992. Identification of a Fab interaction footprint site on an icosahedral virus by cryoelectron microscopy and X-ray crystallography. Nature 355:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watts, N. R., J. F. Conway, N. Cheng, S. J. Stahl, D. M. Belnap, A. C. Steven, and P. T. Wingfield. 2002. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 21:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wien, M. W., D. J. Filman, E. A. Stura, S. Guillot, F. Delpeyroux, R. Crainic, and J. M. Hogle. 1995. Structure of the complex between the Fab fragment of a neutralizing antibody for type 1 poliovirus and its viral epitope. Nat. Struct. Biol. 2:232-243. [DOI] [PubMed] [Google Scholar]
- 41.Wikoff, W. R., G. Wang, C. R. Parrish, R. H. Cheng, M. L. Strassheim, T. S. Baker, and M. G. Rossmann. 1994. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure 2:595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingfield, P. T., S. J. Stahl, R. W. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 43.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 44.Zlotnick, A., N. Cheng, J. F. Conway, F. P. Booy, A. C. Steven, S. J. Stahl, and P. T. Wingfield. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35:7412-7421. [DOI] [PubMed] [Google Scholar]