Abstract
Jaagsiekte sheep retrovirus (JSRV) is the etiological agent of a contagious lung cancer of sheep known as ovine pulmonary adenocarcinoma (OPA). Expression of the JSRV envelope protein (Env) is sufficient to transform immortalized and primary fibroblasts, but the precise mechanisms of this process are not known. The cellular receptor for JSRV is hyaluronidase 2 (Hyal-2), the product of a putative tumor suppressor gene that in humans maps to a chromosomal region frequently deleted in the development of lung and breast cancers. Here we report studies to determine whether the Hyal-2-JSRV Env interaction plays a role in virus-induced transformation of rodent fibroblasts. Chimeric Env proteins between JSRV and the unrelated murine retroviruses Moloney murine leukemia virus (MMuLV) and mouse mammary tumor virus (MMTV) showed cell surface expression comparable to that of wild-type MMuLV Env and rescued infection of MMuLV particle pseudotypes. Interestingly, an MMuLV-JSRV chimera in which the putative receptor binding domain (RBD) and proline-rich region (PRR) of JSRV Env were replaced by the RBD and PRR of MMuLV induced transformation of 208F, a rodent fibroblast line. Cell lines derived from foci of MMuLV-JSRV chimera-transformed 208F cells grew in soft agar and showed Akt activation, a hallmark of JSRV-transformed rodent fibroblasts. Transformation assays performed using proteins with amino-terminal deletion mutations showed that the carboxy-terminal 141 amino acids of the transmembrane subunit (TM) were sufficient to induce cell transformation when targeted to the membrane with a myristoylation signal. Thus, the JSRV TM is necessary and sufficient to transform rodent fibroblasts. Taken together these results indicate that the interaction with Hyal-2 at least is not an essential determinant of JSRV-induced transformation of fibroblasts and that the viral TM functions essentially as an oncoprotein.
Jaagsiekte sheep retrovirus (JSRV) is the etiological agent of ovine pulmonary adenocarcinoma (OPA), a naturally occurring lung cancer of sheep (22, 25). OPA is one of the major infectious diseases of sheep (9) and in the last few years has attracted increasing interest as an animal model for lung cancer (22). In vivo, JSRV transforms the differentiated epithelial cells of the distal portion of the respiratory tract: type II pneumocytes and Clara cells, which also represent the major sites of viral expression in OPA-affected animals (21, 26). Low levels of viral expression are detected also in lymphoid tissues, but no evidence of transformation has been detected in these cell types (12, 23).
Of particular interest are the mechanisms by which JSRV induces cell transformation that appear to be unique among the oncogenic retroviruses. For example, expression of the JSRV Env is sufficient to induce transformation of immortalized rodent fibroblasts (18, 29), and the cytoplasmic tail (CT) of the JSRV envelope is a major determinant of viral transformation (24). No other Env of a replication-competent retrovirus has these properties, with the exception of the highly related enzootic nasal tumor virus (1, 10).
The exact mechanisms of JSRV transformation in vitro and in vivo are unclear. A putative docking site for the phosphatidylinositol 3-kinase (PI3-K) in the CT of the viral Env is critical for transformation of rodent fibroblasts (mouse NIH 3T3 and rat 208F cells), and the PI3-K/Akt pathway is activated in JSRV-transformed rodent cells. However, the residues in the PI3-K docking site are not required to transform the chicken DF1 cell line (4), and both DF1 and primary chicken embryo fibroblasts (CEF) can be transformed by the JSRV Env independently of Akt activation (40). On the other hand, CEF transformed by the wild-type JSRV Env grow more readily in soft agar than CEF transformed by Env mutants lacking the PI3-K docking site. In the latter cells, no Akt activation was detected (40). Similarly, Akt phosphorylation was not detected in tumor cells of naturally occurring OPA cases. These previous studies suggested that other determinants of the JSRV Env, besides the CT, may be involved in cell transformation.
The cellular receptor for JSRV is the product of a putative tumor suppressor gene, hyaluronidase 2 (Hyal-2) (28, 29). In humans, the Hyal-2 gene maps to 3p21.3, a 120-kb region that is frequently deleted in lung and breast cancers (37, 38). Intratumoral inoculation of adenovirus vectors expressing Hyal-2 suppresses the growth of lung cancer cell xenografts (13). The attractive possibility that interaction between the JSRV Env and its receptor interferes with the tumor suppressor function of Hyal-2 has been proposed (29, 31).
Here we report studies addressing the potential role of Hyal-2 in JSRV-induced cell transformation. We initially constructed chimeric envelopes in which the putative receptor binding domain (RBD) of JSRV was replaced by the RBD of the unrelated retroviruses mouse mammary tumor virus (MMTV) and Moloney murine leukemia virus (MMuLV). The resulting chimeras were infectious and expressed at the cell surface. One of the chimeras induced transformation of the rodent fibroblast cell line 208F, suggesting that the RBD, and thus likely Hyal-2, are not essential for transformation. Furthermore, a JSRV Env deletion mutant lacking the putative RBD also induced some degree of cell transformation. Finally, expression of an amino-terminal truncation of the JSRV Env transmembrane subunit (TM) was sufficient for virus-induced transformation of rodent fibroblasts. These results establish that the carboxy-terminal 141 amino acids (aa) of the JSRV TM possess the transforming potential of the viral Env and provide strong indirect evidence that suppression of Hyal-2 does not play an essential role in transformation of rodent fibroblasts.
MATERIALS AND METHODS
Plasmids.
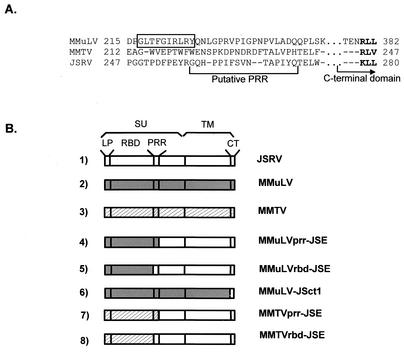
The pCMV3JS21ΔGP plasmid expressing the JSRV21 Env (25) has been described previously (18, 24). pCEE+ (16), encoding the Env of MMuLV driven by the cytomegalovirus immediate-early promoter was provided by Paula Cannon. pENV(Q61) (11) contains the coding sequence for the envelope of MMTV (kindly provided by Susan Ross). The pCMV3JS21ΔGP plasmid has been used as a backbone to insert various portions of the MMuLV or MMTV Env. Coding sequences for chimeric Envs were derived by overlapping PCR and cloned into the BsmI site and the StuI site of pCMV3JS21ΔGP unless otherwise specified. The BsmI site overlaps the first ATG of the JSRV Env gene, while StuI is at the junction between the coding sequences for the surface subunit (SU) and TM domains. The chimeric Env constructs between the JSRV and the MMuLV or the MMTV envelope proteins are schematically represented in Fig. 1. The sequences of the oligonucleotide primers employed in the overlapping PCRs are shown in Table 1. The exact positions of the junctions of each chimera are described in Results. Chimera MMuLVprr-JSE consists of the leader peptide (LP), the RBD, and the proline-rich region (PRR) of MMuLV fused to the putative carboxy-terminal region of the SU and the TM of JSRV Env. The MMuLV Env portion of the MMuLVprr-JSE chimera was amplified with PCR primers MMLVE(BsmI)F (sense primer) and MMLVrbd-prrR (antisense primer). The JSRV portion of MMuLVprr-JSE was amplified with primers JSEΔprr-MMLVF (sense primer) and JSE(StuI)R (antisense primer). PCR products were used in the final overlapping reaction product, which was amplified by using MMLVE(BsmI)F and JSE(StuI)R and cloned in the BsmI and StuI sites of pCMV3JS21ΔGP. MMuLVrbd-JSE consists of the MMuLV LP and RBD fused to the putative PRR and the remaining portions of SU and TM of JSRV Env. The MMuLV Env portion of the chimera MMuLVrbd-JSE was amplified by using PCR primers MMLVE(BsmI)F (sense primer) and MMLVrbdR (antisense primer), while the JSRV portion was amplified with JSEprrF (sense primer) and JSE(StuI)R (antisense primer). The overlapping PCR was analogous to that described for chimera MMuLVprr-JSE. Chimera MMuLV-JSct1 consists of the SU and the ectodomain of the TM of MMuLV and the membrane-spanning domain and the CT of the JSRV TM. The MMuLV portion of the MMuLV-JSct1 chimera was amplified by using primers MMLVE(BsmI)F (sense primer) and MMLVTM-JSER (antisense primer), while the JSRV portion was amplified with primers JSETMF (sense primer) and JSEend(HindIII)R. Overlapping PCR products were amplified by using primers MMLVE(BsmI) and JSEend(HindIII)R, and the final product was cloned into BsmI and HindIII sites of pCMV3JS21ΔGP. MMTVprr-JSE consists of the LP, the putative RBD, and the putative PRR of MMTV fused to the putative carboxy-terminal region of the SU and the TM of JSRV Env. The MMTV Env portion of the MMTVprr-JSE chimera was amplified by using primers MMTVE(BsmI)F (sense primer) and MMTVrbd-prrR (antisense primer), while the JSRV Env portion was amplified by using JSEΔprr-MMTVF (sense primer) and JSE(StuI)R (antisense primer). Overlapping PCR products were amplified with primers MMTVE(BsmI)F and JSE(StuI)R and cloned into the BsmI and StuI sites of pCMV3JS21ΔGP. MMTVrbd-JSE consists of the LP and putative RBD of MMTV fused to the PRR and the remaining portion of the Env of JSRV. The MMTV Env portion of MMTVrbd-JSE was derived by using primers MMTVE(BsmI)F (sense primer) and MMTVrbdR (antisense), while the JSRV Env was amplified with JSErbd-MMTVF (sense primer) and JSE(StuI)R. Overlapping PCR products were amplified with MMTVE(BsmI)F and JSE(StuI)R and inserted into the BsmI and StuI sites of pCMV3JS21ΔGP. Plasmid JSEΔrbd expresses the JSRV Env lacking its putative RBD and was obtained by overlapping PCR. In JSEΔrbd the LP of JSRV was obtained by using primers JSE(BsmI)F (sense primer) and JSEsignalR (antisense primer), while the carboxy-terminal portion of the JSRV SU was amplified with JSEsignal-prrF (sense primer) and JSE(StuI)R. The overlapping PCR product was amplified with JSE(BsmI)F and JSE(StuI)R and inserted into BsmI and StuI sites of pCMV3JS21ΔGP. To express only portions of the TM domain of the JSRV Env, the myristoylation leader signal sequence (MGQSLTTHM) derived from the Rasheed sarcoma virus Gag protein (7) was fused to the amino termini of a series of deletion mutants. Plasmid pMyr-JSE7061 expresses only the CT of the TM, and pMyr-JSE6992 expresses the CT and the membrane-spanning domain of the TM. pMyr-JSE6818 and pMyr-JSE6770 express 58 and 74 aa, respectively, of the TM ectodomain in addition to the membrane-spanning domain and the CT. pΔMyr-JSE6770 is identical to pMyr-JSE6770 with the exception of a glycine-to-alanine mutation in the myristoylation signal. The glycine in the myristoylation signal (after the methionine at position 2) is required for myristoylation to occur (34). Numbers 7061, 6992, 6818, and 6770 in the plasmids described above indicate the position of the first codon of the JSRV TM expressed in each construct with respect to the nucleotide sequence of JSRV21 (25). Thus, the first amino-terminal residue of the JSRV TM expressed in pMyr-JSE6770 is at position 475 with respect to the JSRV Env sequence (KKPYNT…), that expressed in pMyr-JSE6818 is at position 491 (HLQGIW…), that expressed in pMyr-JSE6992 is at position 549 (TLIGVG…), and that expressed in pMyr-JSE7061 is at position 572 (RGMVRD…).
FIG. 1.
Envelope chimeras. (A) Alignment of JSRV Env sequences with those of MMuLV and MMTV in the region of the junctions of the chimeric Envs. The sequences of the last beta strand of MMuLV RBD is boxed. Dashes indicate MMuLV positions missing from MMTV or JSRV. For simplicity, residues 250 to 274 in the PRR of MMuLV, which are missing from MMTV and JSRV, were omitted (dots). The single-letter amino acid code is used to indicate individual residues. The MMTV strain was C3H. Numbers indicate the positions of the first and last residues of each segment in the precursor Env (JSRV) or in mature SU (MMuLV and MMTV). The bracket indicates the putative PRR of JSRV. The first three amino acids in the conserved portion of the C-terminal domain are in boldface type. (B) Schematic representation of the Env chimeras used in this study.
TABLE 1.
Oligonucleotide primers employed in this study to derive the MMuLV-JSRV and MMTV-JSRV Env chimeras
| Primer | Sequence (5′-3′) |
|---|---|
| JSE(Bsml)F | CACAGAATGCCGAAGCGCCGCGCTGGATTCCGG |
| JSEsignalR | GAAAATAGGCGGCCAAAAAGCCGCAGCTGCCCCATT |
| JSEsignal-prrF | GCTTTTTGGCCGCCTATTTTCTCTGTAAAT |
| JSE(Stul)R | CAGGCTCAGGCCTCTCTTAGGGCGGCTGAG |
| MMLVE(Bsml)F | ACTGGAATGCCGCGTTCAACGCTCTCAAAA |
| MMLVrbd-prrR | AGCCAAAAGTTTATTTTCCGTTCCCGCCGGTGGAAG |
| JSEΔprr-MMLVF | ACGGAAAATAAACTTTTGGCTGCTTTTGGT |
| MMLVrbdR | GAAAATAGGCGGGGGTCCTAGATTTTGGTATCTGAG |
| JSEprrF | CTAGGACCCCCGCCTATTTTCTCTGTAAAT |
| MMLVTM-JSER | TACACCAATCAGGGTGGTAAACCAAGGGGATCTGTT |
| JSETMF | TTTACCACCCTGATTGGTGTAGGAATACTTGTG |
| JSEend(HindIII)R | AAAAAGCTTCTACGGGTCGTCCCCCGCATCTCCCCT |
| MMTVE(Bsml)F | AACGAATGCCGAATCACCAATCTGGGTCC |
| MMTVrbd-prrR | AGCCAAAAGTTTAAACAATTCTGTATGGGGAACTAG |
| JSEΔprr-MMTVF | GAATTGTTTAAACTTTTGGCTGCTTTTGGT |
| MMTVrbdR | AGAGAAAATAGGCGGCTTAGGAGAATTTTCCCA |
| JSErbd-MMTVF | CCTAAGCCGCCTATTTTCTCTGTAAATACCGCT |
pMyr-JSE7061, pMyr-JSE6992, pMyr-JSE6818, pMyr-JSE6770, and pΔMyr-JSE6770 were also used to express green fluorescent protein (GFP) fusion proteins to control for relative expression by fluorescence-activated cell sorter (FACS) analysis (see below). Briefly, the coding sequences for the myristoylation signal and the truncated TM from each construct were amplified by PCR and inserted into phrGFP (Stratagene) to obtain pMyr-JSE7061GFP, pMyr-JSE6992GFP, pMyr-JSE6818GFP, pMyr-JSE6770GFP, and pΔMyr-JSE6770GFP. These constructs are under the control of the immediate-early cytomegalovirus promoter and express the myristoylated TM deletion mutants fused at their carboxy termini to the GFP protein.
Cells.
All the cells used in this study were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C, 5% CO2, and 95% humidity unless otherwise specified. 208F cells are an immortalized rat fibroblast cell line (27) (kindly provided by Dusty Miller). Polyclonal lines of 208F cells transformed by pCMV3JS21ΔGP (208F-JE) or by MMuLVprr-JSE (208F-23) or by pMyr-JSE6770 (208F-6770) were derived by isolation and expansion of individual foci. 293-GP and 293-GP-luc are 293-based cell lines expressing MMuLV Gag and Pol (Clontech). 293-GP-AP cells were obtained by selection of 293-GP cells transfected with the retroviral vector pLAPSN (20) (kindly provided by Dusty Miller), expressing the human placental alkaline phosphatase. 293T (15) cells were obtained from Hung Fan. NIH 3T3 cells were grown in DMEM supplemented with 10% calf serum and were originally provided by Robert Weinberg. NIH 3T3-Hyal-2 cells stably express human Hyal-2 and are susceptible to JSRV infection (24). oST, a cell line derived from sheep uterine stroma, was a gift from Tom Spencer (14).
Entry assays.
Cell entry mediated by chimeric envelopes was assessed by determining the ability of the wild-type, chimeric, and truncated Env proteins to pseudotype MMuLV-based vectors (28). 293-GP-AP cells were transfected with the expression plasmids for the envelope protein of MMuLV or JSRV or the MMuLV-JSRV or MMTV-JSRV chimera. Supernatants were collected and stored at −70°C. Subsequently, naive NIH 3T3, NIH 3T3-Hyal-2, and oST cells were exposed to 10-fold serial dilutions of the vector supernatants. Two days postinfection cells were fixed and stained for alkaline phosphatase-positive foci. The viral titer is expressed as alkaline phosphatase foci (APF) per milliliter. Experiments were performed at least two times with two replicates tested for each dilution.
Transformation assays.
Transformation assays with 208F cells were performed essentially as previously described (19). Cells (5 × 105 per 6-cm-diameter dish) were transfected with 10 μg of plasmids by using the CalPhos transfection kit (Clontech) as instructed by the manufacturer. One day after transfection, the cells were trypsinized and reseeded in a 10-cm-diameter dish. After the cells became confluent, the medium (with the addition of 1 μM dexamethasone) was replaced every 3 days, and foci of transformed cells were counted after approximately 2 weeks. Transformation assays were performed at least three times independently with two different plasmid DNA preparations.
Colony assays.
Growth assays in soft agar to assess anchorage independence were performed with 6-cm-diameter petri dishes essentially as described previously (17). Each dish was coated with 2 ml of a base layer formed by 0.5% agar, 1× DMEM, 20% FBS, and 10% tryptose phosphate buffer (TPB). Cells (2 × 104) suspended in 2 ml of incubation medium containing DMEM, 20% FBS, 10% TPB, and 0.3% agar were plated over the base layer. Cells were incubated for 2 weeks at 37°C, 5% CO2, and 95% humidity, and 2 ml of incubation medium was added to the culture every 3 to 4 days. Eleven days after plating, the sizes of 100 randomly selected colonies were measured with Image-Pro-Express software (Media Cybernetics). Representative images were captured with an Olympus IX-70 microscope interfaced with a Magnafire digital camera. Each experiment was done in duplicate and performed independently two times.
Western blotting.
For the detection of Akt, cells were transfected with Env expression plasmids as described for the transformation assay except that they were grown at 37°C and 5% CO2 in DMEM-10% FBS until they reached approximately 80% confluence, washed twice with phosphate-buffered saline, and grown for another 24 h in medium lacking FBS. Cells were then lysed in buffer containing 0.5% NP-40, 50 mM HEPES buffer (pH 7.8), 100 mM sodium fluoride, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and one tablet of protease inhibitor cocktail (Roche) per 50 ml of lysis buffer. Following standard procedures, 5 to 10 μg of cell lysates was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting (5). Filters were incubated with polyclonal rabbit antisera to either Akt (Cell Signaling) or Akt phosphorylated at serine 473 (Cell Signaling), and detection was achieved with donkey anti-rabbit immunoglobulin labeled with horseradish peroxidase (Amersham) followed by an enhanced chemiluminescence detection system (Supersignal; Pierce) as recommended by the manufacturers.
FACS.
Cell surface expression of the MMuLV-JSRV Env chimeras was measured by FACS analysis. 293T cells were transfected with the vector alone (pCDNA3.1; Invitrogen), pCEE+, MMuLVprr-JSE, MMuLVrbd-JSE, pCMV3JS21ΔGP, and MMuLV-JSct1. Thirty-six hours after transfection cells were removed from the dish, washed with 10% FBS-phosphate-buffered saline, and then incubated for 1 h at 4°C with a polyclonal goat anti-Rauscher MuLV gp70 serum (Quality Biotech, Camden, N.J.) or with normal goat serum to measure background staining. Cells were then washed and stained with fluorescein isothiocyanate-conjugated antibodies directed against goat immunoglobulins. Surface expression was measured as the mean fluorescence intensity of cells gated for a specific signal, i.e., fluorescence intensity above that observed in cells transfected with vector alone. Relative expression levels of pMyr-JSE7061GFP,pMyr-JSE6992GFP, pMyr-JSE6818GFP, pMyr-JSE6770GFP, and pΔMyr-JSE6770GFP were also measured by FACS analysis. 293T cells were transfected with pCDNA3.1, phrGFP (expressing GFP alone), pMyr-JSE7061GFP, pMyr-JSE6992GFP, pMyr-JSE6818GFP, pMyr-JSE6770GFP, and pΔMyr-JSE6770GFP. Forty-eight hours after transfection cells were washed and expression was measured directly by measuring the mean fluorescence intensity of cells gated for a specific signal.
Akt kinase assay.
Akt kinase activity was detected with a nonradioactive Akt kinase assay kit (Cell Signaling). Briefly, decreasing amounts of cell lysates from 208F and 208F-JE cells were immunoprecipitated with an Akt antibody cross-linked to hydrazide-agarose beads. The resulting immunoprecipitate was incubated in the presence of ATP with a recombinant GSK-3 fusion protein. Phosphorylation of GSK-3 was measured by Western blotting with a phospho-GSK-3β (Ser 21/9) antibody.
RESULTS
Chimeric envelope proteins.
Hyal-2, the cellular receptor for JSRV and enzootic nasal tumor virus (1, 10, 28, 29), is the product of a putative tumor suppressor gene. To test the hypothesis that JSRV transforms cells by blocking or interfering with the tumor suppressor function of Hyal-2, we constructed chimeric Env between JSRV and the murine retroviruses MMuLV and MMTV (Fig. 1). Typically, the amino-terminal portion of the retroviral SU is the domain that interacts with the cellular receptor. In MMuLV and MMTV the RBD has been identified (6, 8; L. M. Albritton and S. Ross, personal communication). The receptors for MMuLV and MMTV are not associated with cell transformation and are unrelated to Hyal-2. MMuLV uses a cationic amino acid transporter as the cellular receptor (2, 3), while mouse transferrin receptor 1 is the high-affinity entry receptor for MMTV (32).
Dusty Miller and coworkers have reported evidence that the amino-terminal 210 residues of the JSRV SU is important for interaction with Hyal-2 (10), suggesting that the RBD extends at least to that residue. In MMuLV, the RBD is located in the amino terminus of SU and is followed by a short region, termed the PRR, that separates the RBD from the carboxy-terminal domain of SU. These regions have not yet been identified in JSRV. We hypothesized that the domain structure of JSRV SU is similar to that of the better-characterized gammaretroviruses and, based on that hypothesis, generated an alignment of the Env sequences of MMuLV, MMTV, and JSRV from a previously constructed alignment of the MMuLV and MMTV SU sequences (L. M. Albritton and S. Ross, unpublished data). The alignment in the region of the putative PRR is shown in Fig. 1A. Notably, MMTV and JSRV have fewer proline residues in their putative PRRs than MMuLV, but this is true also for some gammaretroviruses such as amphotropic 4070A MuLV (6). The positions of the junctions of chimeras constructed for these studies were based on the alignment shown.
Since it is not known if the PRR folds as a part of the putative RBD, as part of the carboxy-terminal domain, or independently, we used two strategies to increase the possibility that at least one set of chimeras would give stable expression. The first set of chimeras (MMuLVrbd-JSE and MMTVrbd-JSE) joined the carboxy-terminal residue of RBD of MMuLV (aa 1 to 264) or MMTV (aa 1 to 326) to the amino-terminal residue of the PRR of JSRV (aa 242 to 615). The second set of chimeras (MMuLVprr-JSE and MMTVprr-JSE) joined the carboxy-terminal residue of the PRR of the murine retrovirus envelope protein (aa 312 for MMuLV and 343 for MMTV) to the corresponding residue of JSRV (aa 259). In addition, we constructed another chimera formed by the SU and TM of MMuLV joined to the membrane-spanning domain and the CT of JSRV (MMuLV-JSct1). In MMuLV-JSct1 the MMuLV Env sequences terminate at aa 609 and the JSRV Env sequences begin at aa 550 and continue until the end of the JSRV Env.
Characterization of Env chimeras.
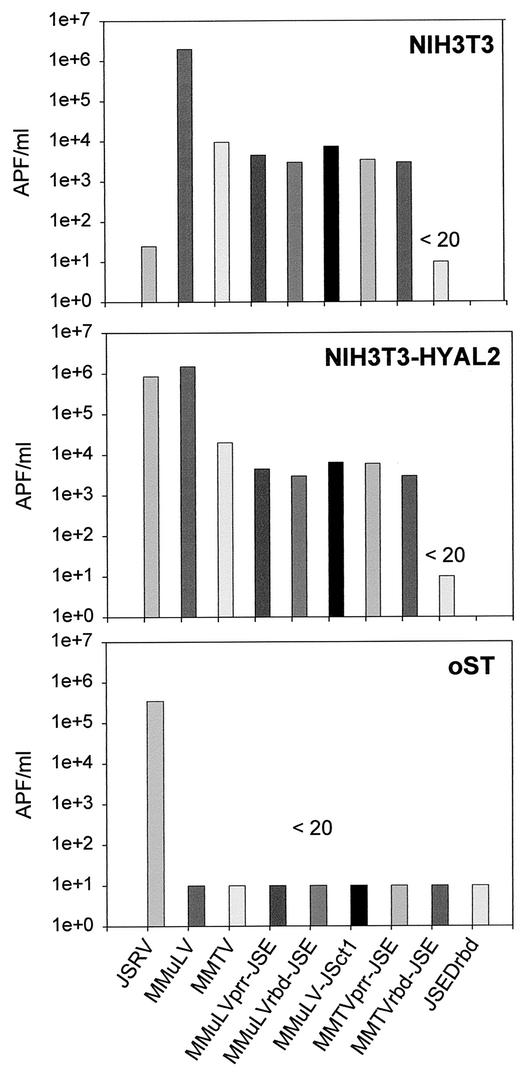
To assess the capacity of the constructed chimeras to mediate viral entry, we quantified the transduction of an alkaline phosphatase-expressing vector (pLAPSN) (20). Pseudotype viruses were obtained by transfecting 293-GP-AP cells (expressing MMuLV Gag, Pol, and pLAPSN) with the chimeric envelope expression plasmids. The resulting pseudotypes were used to infect NIH 3T3, NIH 3T3-Hyal-2, and sheep oST cells. All chimeras were infectious, but the deletion mutant JSEΔrbd was not infectious even on JSRV-susceptible cells, e.g., NIH 3T3-Hyal-2, and oST cells. Chimeras between JSRV and MMTV showed titers within 10-fold of those of the parental wild-type MMTV. In contrast, MMuLV chimeras showed titers approximately 1,000-fold lower than those of wild-type MMuLV or JSRV (Fig. 2). As previously published (1, 10, 28, 29), vectors bearing the JSRV Env efficiently infected NIH 3T3-Hyal-2 cells (titers in the order of 105 to 106 APF/ml) but gave poor infection of mouse NIH 3T3 cells (titers ranging between 101 and 102 APF/ml). Conversely, the infectious titer of the MMuLV and MMTV chimeras in NIH 3T3 cells were comparable to those in NIH 3T3-Hyal-2 cells. These results suggest that the chimeras use the MMuLV and MMTV receptors and not Hyal-2 as their entry receptor. In agreement with this interpretation, only the vector pseudotyped with the JSRV Env entered efficiently into the sheep oST cells. However, we cannot exclude the possibility that the chimeras still interact with Hyal-2 with a lower avidity.
FIG. 2.
Entry assays. Viral entry assays were performed with NIH 3T3, NIH 3T3-Hyal-2, and sheep oST cells and retroviral vectors expressing alkaline phosphatase and pseudotyped with the chimeric Envs. Results are denoted <20 when the titer was less than 20 APF/ml.
Surface expression of MMuLV-JSRV chimeras.
The drop in the infectious titer of the MMuLV-based chimeric Env pseudotypes with respect to the MMuLV or JSRV Env pseudotype may have been due to poor expression of the chimeras indicative of misfolding and degradation. To determine if this was the case, we performed FACS analysis on 293T cells transfected with the MMuLV-JSRV chimeras and compared their cell surface expression to that of parental MMuLV. The results obtained showed that the surface expression levels of MMuLVrbd-JSE and MMuLV-JSct1 were comparable to that of wild type MMuLV Env while MMuLVprr-JSE expression was typically one-half of that level (Table 2). Due to the lack of specific antisera recognizing either JSRV or MMTV Env, the surface expression of MMTV-JSRV chimeras was not quantified.
TABLE 2.
Surface expression of MMuLV-JSRV chimeras
| Envelope | Expt 1
|
Expt 2
|
Expt 3
|
|||
|---|---|---|---|---|---|---|
| % Env+ cellsa | Relative surface expressionb | % Env+ cells | Relative surface expression | % Env+ cells | Relative surface expression | |
| MMuLV | 24.4 (0.91) | 100 | 19.9 (0.67) | 100 | 46.9 (3.99) | 100 |
| MMuLVrbd-JSE | 20.4 (2.27) | 85 | 20.4 (0.6) | 107 | 34.8 (6.43) | 160 |
| MMuLVprr-JSE | 16.8 (1.55) | 52 | 9.8 (0.87) | 59 | 40 (10) | 180 |
| MMuLV-JSct1 | 16 (2.05) | 85 | 10.4 (2.94) | 105 | 48.4 (12.7) | 257 |
% Env+ cells, the percentage of cells emitting fluorescence above that observed in negative control cells transfected with vector pcDNA3.1 alone and incubated with primary goat anti-MuLV SU antiserum and a secondary mouse anti-goat antibody conjugated to fluorescein isothiocyanate. The mean fluorescence intensities (MFI) of negative control cells were 0.321, 0.169, and 0.214 in experiments 1, 2, and 3, respectively. Values in parentheses were obtained by incubating the transfected cells with a normal goat antiserum instead of the goat anti-MuLV SU.
Relative surface expression was calculated as (MFI of chimeric Env+ cells/MFI of MMuLV Env+ cells) × 100. The MFI of MMuLV Env+ cells were 6.33, 5.25, and 5.79 in experiments 1, 2, and 3, respectively.
Transformation of 208F cells by chimeric Envs.
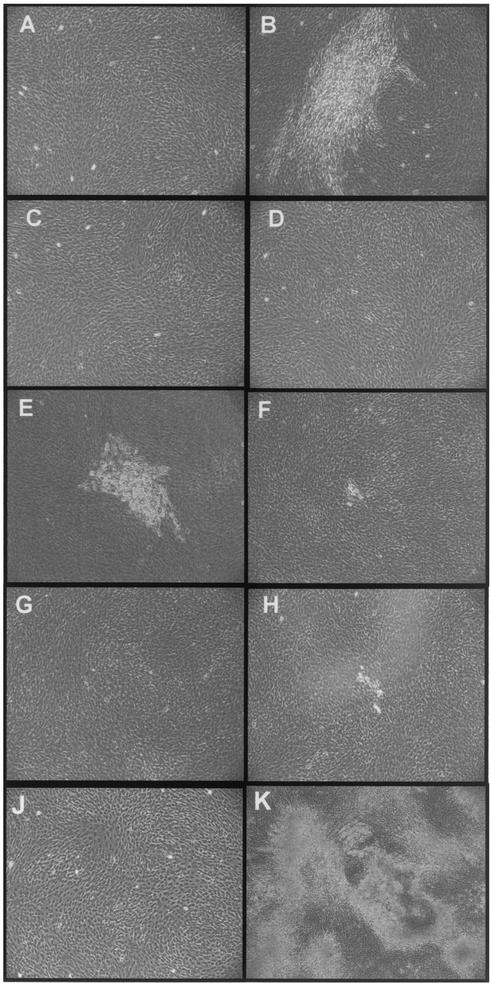
Transformation assays used the rat fibroblast cell line 208F (27), a cell line that is transformed efficiently by the JSRV Env (1, 29). Plasmids expressing the wild-type JSRV, MMuLV, and MMTV Envs and the Env chimeras were transfected into 208F cells, and foci of transformed cells were counted 2 weeks posttransfection. As expected the plasmid expressing the JSRV Env induced numerous foci of transformed cells (Fig. 3B) while foci were absent from the mock-transfected 208F cells or cells transfected with expression plasmids for MMuLV and MMTV Envs (Fig. 3A, C, and D). Typically, pCMV3JS21ΔGP (the expression plasmid for the JSRV Env) induced 100 to 130 foci per 5 × 105 transfected 208F cells.
FIG. 3.
Transformation assays of 208F cells with the Env chimeras. 208F cells were transfected with chimeric and wild-type Env expression plasmids as described in the text. Transfected cells were examined by light microscopy for morphological alterations at 2 weeks postinfection. Cell transformation, represented by the formation of foci of refractive cells, was achieved with the expression plasmids for the JSRV Env (B) and MMuLVprr-JSE (E). Some morphological changes were observed with MMuLVrbd-JSE (F) and MMTVprr-JSE (H). No signs of cell transformation were observed in mock-transfected cells (A) or in cells transfected with pCEE+ (C) (expressing the MMuLV Env), pENV (Q61) (D) (expressing the MMTV Env), MMTVrbd-JSE (G), or MMuLV-JSct1 (J). (K) The 208F-23 cell line, derived from the isolation and expansion of MMuLVprr-JSE-transformed cells. 208F-23 cells have lost the contact inhibition typical of 208F cells.
pMMuLVprr-JSE, MMuLVrbd-JSE, and MMTVprr-JSE induced morphological changes in transfected cells (Fig. 3E, F, and H). However, only MMuLVprr-JSE-transfected cells showed clear evidence of foci at the end point of the experiment (Fig. 3E), and they were less numerous and smaller than the foci induced by wild-type JSRV Env. In three independent experiments, the number of foci induced by MMuLVprr-JSE was 33.2% ± 3.9% of the number of foci induced by the JSRV Env (taken as 100%).
MMuLVrbd-JSE and MMTVprr-JSE induced some morphological changes, represented by a small number of refractive cells (Fig. 3F and H), not observed in control plates (Fig. 3A, C, and D). pMMTVrbd-JSE and pMMuLV-JSct1 did not induce foci or morphological changes (Fig. 3G and J). To confirm cell transformation by MMuLVprr-JSE, we isolated and expanded several foci of 208F transformed cells and we obtained a polyclonal cell line (208F-23). 208F-23 cells lost the contact inhibition characteristic of the parental cells and appeared markedly different in morphology (Fig. 3K).
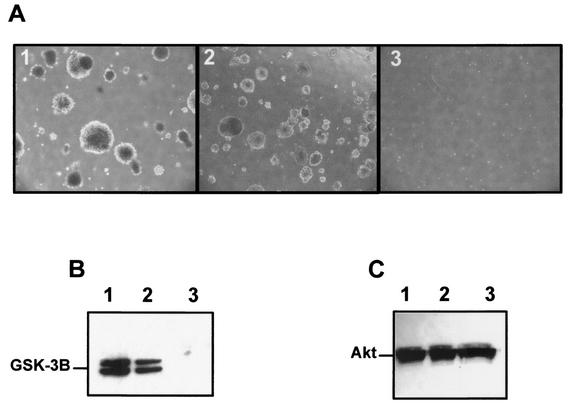
We performed assays of colonies in soft agar and compared 208F-23 with parental 208F and 208F-JE, a cell line derived from a focus of 208F cells transformed by the wild-type JSRV Env. 208F-23 cells grew readily in soft agar at levels comparable to those for 208F-JSFlag, while 208F cells did not form colonies, as was expected (Fig. 4A). Notably, 208F-JE cells grew faster than 208F-23 cells. At 11 days postseeding the average diameters of 100 colonies in randomly selected fields were 429 ± 90 μm for 208F-JSFlag and 256.5 ± 26 μm for 208F-23. These results were repeated twice in two independent experiments. In addition 208F-23 cells showed constitutive activation of Akt (Fig. 4B), a hallmark of rodent fibroblasts transformed by the JSRV Env (1, 24).
FIG. 4.
Colony formation and Akt assays of MMuLVprr-JSE-transformed cells. (A) Anchorage independence of the 208F-JSFlag (1), 208F-23 (2), and parental 208F (3) cell lines was assessed by growth in soft agar as described in Materials and Methods. Shown is a representative example of the morphology and size of the colonies at 11 days after seeding (magnification, ×40). Virtually no background was seen in 208F cells. (B) Akt activation was assessed in cell lysates of serum-starved 208F-JE, 208F-23, and 208F cells by using an Akt kinase kit (Cell Signaling) as described in Materials and Methods. Shown is the phosphorylation of GSK-3β in 208F-JE cells (lane 1) and 208F-23 cells (lane 2) but not in 208F cells (lane 3). The same cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting (C) by using an antiserum to Akt as a loading control.
Transformation of rodent fibroblasts by the JSRV Env is receptor independent.
The results described above indirectly suggest that an interaction with Hyal-2 is not a primary factor in JSRV-induced cell transformation and, by inference, that the putative tumor suppressor function of Hyal-2 is not absolutely required for this process to occur. However, two alternative explanations could not be ruled out. First, the binding of the JSRV RBD to its receptor may induce conformational changes that are important to the transforming activity of the viral envelope. The binding of the MMuLVprr-JSE chimera to the MMuLV receptor might induce similar structural changes. Second, the MMuLV RBD portion of the chimera might interact with molecules other than the entry receptor and in this manner compensate for the role of the JSRV RBD in cell transformation. For example, the MMuLV and MMTV Envs were recently shown to activate B cells via an interaction with the toll-like receptor 4 (TLR-4) (30). Since JSRV Env has greater homology to MMTV Env than does MMuLV Env (39), JSRV Env may also interact with TLR-4.
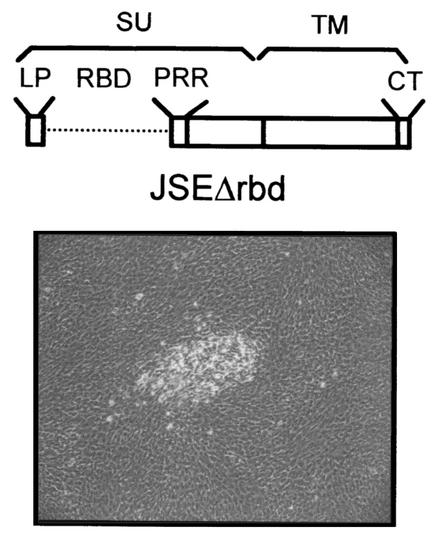
To rule out a possible role for the MMuLV RBD, we constructed a JSRV envelope protein (JSEΔrbd) with the putative RBD (aa 82 to 241) deleted. JSEΔrbd did not rescue the infectivity of a retroviral vector expressed in 293-GP-AP cells (Fig. 2), indirect evidence that this deletion mutant does not interact with Hyal-2. However when transfected into 208F cells, JSEΔrbd induced foci of transformed cells similarly to MMuLVprr-JSE (Fig. 5). In three separate experiments, JSEΔrbd induced between 31 and 36% of the number of foci induced by the wild-type JSRV Env. The ability of JSEΔrbd to induce cell transformation further supports the hypothesis that the JSRV Env induces transformation of rodent fibroblasts in an entry receptor-independent manner.
FIG. 5.
JSEΔrbd induces cell transformation. (Top) Schematic of JSEΔrbd. (Bottom) Typical focus of transformed cells at 14 days posttransfection.
Envelope-induced transformation of 208F cells can occur in the absence of the SU domain.
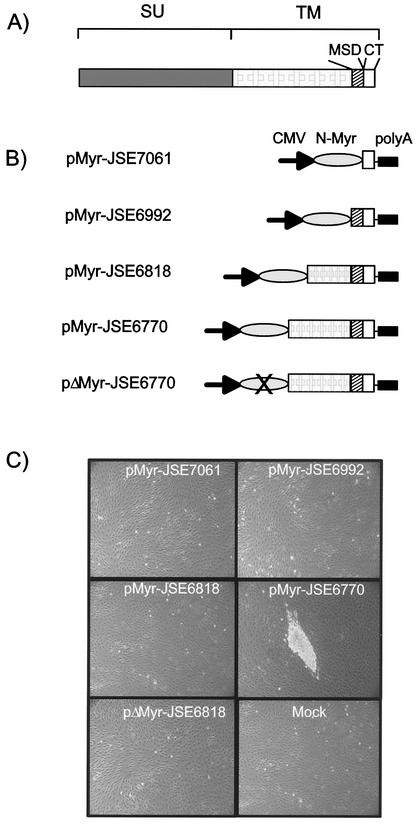
The results shown so far indirectly demonstrate that the determinants of JSRV Env transformation are localized either at the carboxy-terminal domain of SU or in TM. We previously showed that the CT of the TM domain is a major determinant of JSRV transformation (1, 24, 40). Consequently, the sequences of the CT alone might be sufficient to transform rat 208F cells provided they associate with cell membranes. In this study, a myristoylation signal was placed on the amino terminus of a series of TM truncation mutants in order to target them to the cell membrane (Fig. 6). The plasmid expressing only the CT of the viral TM (pMyr-JSE7061) did not transform 208F cells. Plasmids pMyr-JSE6818 and pMyr-JSE6992 also did not induce any morphological changes in 208F cells. The last two plasmids express longer carboxy-terminal portions of the JSRV TM domain. In contrast, pMyr-JSE6770, which expresses the carboxy-terminal 141 aa of the JSRV TM, induced between 33 and 67% as many foci as the full-length JSRV Env (Fig. 6). A cell line derived from foci of 208F cells transformed by pMyr-JSE6770 also showed constitutive activation of Akt and anchorage independence characterized by its ability to grow in soft agar (data not shown). These results shows that the JSRV TM is the oncogenic determinant of the envelope protein at least in 208F cells and that cell transformation can occur in the absence of the SU domain.
FIG. 6.
Determinants of JSRV TM transformation. (A) Schematic organization of the JSRV Env. MSD, membrane-spanning domain. (B) Schematic organization of the plasmids expressing different portions of the JSRV TM with an NH2 myristoylation signal. In plasmid pΔMyr-JSE6772 the codon for the glycine in position 2 of the myristoylation signal has been mutated into an alanine codon. (C) Transfection of 208F cells with the plasmids shown in panel B results in the induction of transformed foci only with pMyr-JSE6770. The myristoylation signal seems to be essential for transformation to occur, as pΔMyr-JSE6770 failed to transform 208F cells.
We also checked for the relative expression of the TM deletion mutant constructs by FACS analysis of GFP-tagged constructs (Table 3). The level of expression of pMyr-JSE6770GFP is comparable to (or lower than) the levels of expression of pMyr-JSE7061GFP, pMyr-JSE6818GFP, and pMyr-JSE6992GFP. This indirectly suggests that the lack of transforming activity exhibited by pMyr-JSE7061, pMyr-JSE6818, and pMyr-JSE6992 is not due to low expression levels of these constructs. Thus, residues of the JSRV Env between aa 475 and 491 in the TM ectodomain might be essential for transformation or might merely be crucial for the functional structural conformation of the pMyr-JSE6770 construct. However, in a previously published report (24), mutation of tyrosine 478 (into phenylalanine) greatly reduced the transforming potential of the resulting JSRV Env mutant in NIH 3T3 cells. These results support the notion that residues 475 to 491 are critical for Env-induced cell transformation.
TABLE 3.
Relative expression of GFP-tagged TM deletion mutants
| Plasmid | Expt 1
|
Expt 2
|
Expt 3
|
|||
|---|---|---|---|---|---|---|
| % GFP+ cellsa | Relative expressionb | % GFP+ cells | Relative expression | % GFP+ cells | Relative expression | |
| pMyr-JSE6770GFP | 11.9 | 100 | 19.7 | 100 | 9.1 | 100 |
| pΔMyr-JSE6770GFP | 33.2 | 93.5 | 27 | 77.5 | 23.1 | 69.1 |
| pMyr-JSE6818GFP | 7.2 | 92.7 | 7.9 | 119.7 | 23.2 | 102.1 |
| pMyr-JSE6992GFP | 31 | 695 | 40.1 | 732 | 57.8 | 317 |
| pMyr-JSE7061GFP | 16.1 | 430 | 19.1 | 575 | 22.3 | 207 |
% GFP+ cells, the percentage of cells emitting fluorescence above that observed in negative control cells transfected with vector pcDNA3.1 alone. The mean fluorescence intensities (MFI) of negative control cells were 0.522, 0.516, and 0.466 in experiments 1, 2, and 3, respectively.
Relative expression was calculated as (MFI of test plasmid/MFI of pMyr-JSE6770GFP+ cells) × 100. The MFI of pMyr-JSE6770 GFP+ cells were 17.25, 20.65, and 20.47 in experiments 1, 2, and 3, respectively.
Myristoylation in pMyr-JSE6770 also appears to be essential for transformation. pΔMyr-JSE6770 was derived by introducing a critical mutation in the glycine of the myristoylation signal of pMyr-JSE6770. pΔMyr-JSE6770 was not able to induce foci of transformed cells.
DISCUSSION
JSRV is a unique replication-competent oncogenic retrovirus that exhibits some properties of the acutely transforming retroviruses. In vivo, JSRV induces rapid tumorigenesis when inoculated intratracheally into newborn lambs (33, 36). In vitro, expression of the JSRV Env is sufficient to transform immortalized rodent and chicken fibroblasts (4, 18, 29). Thus, studies of JSRV-induced cell transformation can give insights into novel mechanisms of retroviral transformation.
Prior to this study, the CT of the JSRV TM had been identified as a determinant of viral transformation (24) but there was evidence suggesting that other domains of Env are involved. Activation of the PI3-K/Akt pathway was found to be critical in JSRV-induced cell transformation of rodent fibroblasts, and this activation correlated with the presence of a Y-X-X-M motif in the CT of TM (24). However, transformation of immortalized and primary chicken fibroblasts occurs also by an Akt-independent pathway, and the Y-X-X-M motif is not absolutely required in these cells (4, 40). Thus, the mechanisms of JSRV Env-induced cell transformation are not completely clarified. It has been speculated that JSRV might induce cell transformation by interfering with the tumor suppressor function of its entry receptor, Hyal-2 (29, 31), implying that SU might also be an important determinant of viral transformation.
In this study we evaluated the role of SU in JSRV-induced cell transformation and further defined the determinants of viral transformation. Expression of JSEΔrbd, containing a deletion of the putative JSRV RBD, or of MMuLVprr-JSE, a chimera containing the RBD and PRR of MMuLV in place of JSRV sequences, induced transformation of 208F cells although not as efficiently as the wild-type JSRV Env. Although not overtly transforming, chimeras MMuLVrbd-JSE and MMTVrbd-JSE induced some morphological changes that were absent in the mock-transfected 208F cells at the end point of the experiment (2 weeks). The reasons for the difference in the transforming potentials of the various chimeras are not known, but FACS analysis showed that this is not due to differences in cell surface expression of the chimeric Envs, as MMuLVprr-JSE was expressed even less efficiently than MMuLVrbd-JSE in some experiments. Deletion of the putative JSRV RBD in JSEΔrbd also induced transformation of 208F cells but abolished infection, suggesting that an interaction with the entry receptor was lost and that this interaction was not required for fibroblast transformation.
However, we could not exclude with certainty the possibility that JSEΔrbd and MMuLVprr-JSE are still able to interact with Hyal-2 and that this interaction, although not sufficient to mediate viral entry, might have a role in cell transformation. In some retroviruses, e.g., feline leukemia virus subgroup B, portions outside the RBD can contribute to receptor specificity (35). Thus, a series of deletion mutants were characterized. These mutants lacked SU as well as increasing amounts of the amino-terminal sequences of TM. The results indicate that the TM of the JSRV Env is the major determinant of rodent fibroblast transformation and essentially functions as the viral oncogene. This notion is in agreement with the previous observations that mutation of tyrosine 478 in the ectodomain of the TM (included in pMyr6770-JSE) markedly reduced the transforming potential of the JSRV Env (24), and truncation of the CT or its replacement by the CT of the nontransforming, endogenous enJS56A1 sequence resulted in loss of the ability to transform DF1 cells and primary CEF (4, 40).
The smallest fragment of Env that induced transformation of rodent fibroblasts consisted of the carboxy-terminal 75 aa of the TM ectodomain in addition to the membrane-spanning domain and the CT of TM (encoded by pMyr6770-JSE). Almost nothing is known about the domain structure of JSRV TM. However, alignment of its sequences with those of MMuLV TM shows that most of the key leucine residues in the heptad repeat of MMuLV TM are not conserved in JSRV (L. M. Albritton, unpublished observation). Instead, JSRV has a leucine-rich motif downstream of its cysteine residues (L. M. Albritton, unpublished observation). The presence of this putative coiled-coil region in the smallest fragment of JSRV Env that exhibited transforming potential raises the interesting possibility that oligomerization may be relevant to the process of virus-induced transformation.
It is possible that the JSRV Env uses different mechanisms to transform cell lines derived from different tissues and/or species. However, we favor the hypothesis that TM contains the main determinants of JSRV Env transformation of all cell types and that cell type-specific transformation involves the use of TM in possibly cell type-specific pathways. In vivo, oncogenesis by JSRV is most likely a multistep process. Expression of the JSRV Env might not be sufficient to transform type II pneumocytes and Clara cells in vivo because additional steps are required to achieve carcinogenesis. Comparison of the mechanisms of cell transformation in vitro and in vivo will give a full understanding of JSRV oncogenesis.
Acknowledgments
We thank Susan Ross, Paula Cannon, and Dusty Miller for providing useful reagents mentioned in the text. We thank Hung Fan for reagents and continuous discussions and encouragement, Don Evans for help with the FACS analysis, and Paula Cannon for useful suggestions.
This work was funded by grants CA95706 (M.P.) from the National Cancer Institute and AI33410 (L.M.A.) from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health and the Georgia Cancer Coalition (M.P.). M.P. is a member of the Biomedical Health and Science Institute and the Comparative Oncology Program of the University of Georgia.
REFERENCES
- 1.Alberti, A., C. Murgia, S.-L. Liu, M. Mura, C. Cousens, M. Sharp, A. D. Miller, and M. Palmarini. 2002. Envelope-induced cell transformation by ovine betaretroviruses. J. Virol. 76:5387-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton, L. M., J. W. Kim, L. Tseng, and J. M. Cunningham. 1993. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J. Virol. 67:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 4.Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The Jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 83:2733-2742. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buss, J. E., P. A. Solski, J. P. Schaeffer, M. J. MacDonald, and C. J. Der. 1989. Activation of the cellular proto-oncogene product p21Ras by addition of a myristylation signal. Science 243:1600-1603. [DOI] [PubMed] [Google Scholar]
- 8.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMartini, J. C., and D. F. York. 1997. Retrovirus-associated neoplasms of the respiratory system of sheep and goats. Ovine pulmonary carcinoma and enzootic nasal tumor. Vet. Clin. N. Am. Food Anim. Pract. 13:55-70. [DOI] [PubMed] [Google Scholar]
- 10.Dirks, C., F. M. Duh, S. K. Rai, M. I. Lerman, and A. D. Miller. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 76:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golovkina, T. V., J. Dzuris, B. van den Hoogen, A. B. Jaffe, P. C. Wright, S. M. Cofer, and S. R. Ross. 1998. A novel membrane protein is a mouse mammary tumor virus receptor. J. Virol. 72:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland, M. J., M. Palmarini, M. Garcia-Goti, L. Gonzalez, I. Mckendrick, M. De las Heras, and J. M. Sharp. 1999. Jaagsiekte retrovirus is widely distributed in both T and B lymphocytes and mononuclear phagocytes of sheep with naturally and experimentally acquired pulmonary adenomatosis. J. Virol. 73:4004-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, L., M. Nishizaki, B. Gao, D. Burbee, M. Kondo, C. Kamibayashi, K. Xu, N. Yen, E. N. Atkinson, B. Fang, M. I. Lerman, J. A. Roth, and J. D. Minna. 2002. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 62:2715-2720. [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, G. A., R. C. Burghardt, G. R. Newton, F. W. Bazer, and T. E. Spencer. 1999. Development and characterization of immortalized ovine endometrial cell lines. Biol. Reprod. 61:1324-1330. [DOI] [PubMed] [Google Scholar]
- 15.Lebkowsky, J. S., S. Clancy, and M. P. Calos. 1985. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature 317:169-171. [DOI] [PubMed] [Google Scholar]
- 16.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacPherson, I., and L. Montagnier. 1964. Agar suspension culture for the selective assay of cells transformed by polyoma virus. Virology 23:291-294. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by Jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, A. D., T. Curran, and I. M. Verma. 1984. c-fos protein can induce cellular transformation: a novel mechanism of activation of a cellular oncogene. Cell 36:51-60. [DOI] [PubMed] [Google Scholar]
- 20.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmarini, M., P. Dewar, M. De las Heras, N. F. Inglis, R. G. Dalziel, and J. M. Sharp. 1995. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J. Gen. Virol. 76:2731-2737. [DOI] [PubMed] [Google Scholar]
- 22.Palmarini, M., and H. Fan. 2001. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J. Natl. Cancer Inst. 93:1603-1614. [DOI] [PubMed] [Google Scholar]
- 23.Palmarini, M., M. J. Holland, C. Cousens, R. G. Dalziel, and J. M. Sharp. 1996. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J. Gen. Virol. 77:2991-2998. [DOI] [PubMed] [Google Scholar]
- 24.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol-3-kinase (PI-3K) docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 75:11002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmarini, M., J. M. Sharp, M. De las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt, J. A., N. Kraipowich, F. Villafane, and J. C. DeMartini. 2002. Alveolar type II cells expressing Jaagsiekte sheep retrovirus capsid protein and surfactant proteins are the predominant neoplastic cell type in ovine pulmonary adenocarcinoma. Vet. Pathol. 39:341-352. [DOI] [PubMed] [Google Scholar]
- 27.Quade, K. 1979. Transformation of mammalian cells by avian myelocytomatosis virus and avian erythroblastosis virus. Virology 98:461-465. [DOI] [PubMed] [Google Scholar]
- 28.Rai, S. K., J. C. DeMartini, and A. D. Miller. 2000. Retrovirus vectors bearing Jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J. Virol. 74:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for Jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rassa, J. C., J. L. Meyers, Y. Zhang, R. Kudaravalli, and S. R. Ross. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. USA 99:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg, N. 2001. New transformation tricks from a barnyard retrovirus: implications for human lung cancer. Proc. Natl. Acad. Sci. USA 98:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, S. R., J. Schofield, C. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp, J. M., K. W. Angus, E. W. Gray, and F. M. Scott. 1983. Rapid transmission of sheep pulmonary adenomatosis (Jaagsiekte) in young lambs. Brief report. Arch. Virol. 78:89-95. [DOI] [PubMed] [Google Scholar]
- 34.Solski, P. A., L. A. Quilliam, S. G. Coats, C. J. Der, and J. E. Buss. 1995. Targeting proteins to membranes using signal sequences for lipid modification. Methods Enzymol. 250:435-454. [DOI] [PubMed] [Google Scholar]
- 35.Sugai, J., M. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verwoerd, D. W., E. M. De Villiers, and R. C. Tustin. 1980. Aetiology of Jaagsiekte: experimental transmission to lambs by means of cultured cells and cell homogenates. Onderstepoort J. Vet. Res. 47:13-18. [PubMed] [Google Scholar]
- 37.Wistuba, I. I., C. Behrens, S. Milchgrub, D. Bryant, J. Hung, J. D. Minna, and A. F. Gazdar. 1999. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene 18:643-650. [DOI] [PubMed] [Google Scholar]
- 38.Wistuba, I. I., C. Behrens, A. K. Virmani, G. Mele, S. Milchgrub, L. Girard, J. W. Fondon III, H. R. Garner, B. McKay, F. Latif, M. I. Lerman, S. Lam, A. F. Gazdar, and J. D. Minna. 2000. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 60:1949-1960. [PubMed] [Google Scholar]
- 39.York, D. F., R. Vigne, D. W. Verwoerd, and G. Querat. 1992. Nucleotide sequence of the Jaaksiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 66:4930-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zavala, G., C. Pretto, J. Y.-H. Chow, L. Jones, A. Alberti, E. Grego, M. De las Heras, and M. Palmarini. Relevance of Akt phosphorylation in cell transformation induced by Jaagsiekte sheep retrovirus. Virology, in press. [DOI] [PubMed]