Abstract
Chromosome 18q is frequently deleted in lung cancers, and a common region of 18q deletions was mapped to chromosome 18q21. Since the DCC candidate tumor suppressor gene has been mapped in this region, mutation and expression of the DCC gene were examined in 46 lung cancer cell lines, consisting of 14 small cell lung carcinomas (SCLCs) and 32 non-small cell lung carcinomas (NSCLCs), to elucidate the pathogenetic significance of DCC alterations in human lung carcinogenesis. A heterozygous missense mutation was detected in a NSCLC cell line, Ma26, while homozygous deletion was not detected in any of the cell lines. The DCC gene was expressed in 11 (24%) of the 46 cell lines, and the incidence of DCC expression was significantly higher in SCLCs (7/14, 50%) than in NSCLCs (4/32, 13%) (P = .01, Fisher's exact test). Therefore, genetic alterations of DCC are infrequent; however, the levels of DCC expression vary among lung cancer cells, in particular, between SCLCs and NSCLCs. The present result does not implicate DCC as a specific mutational target of 18q deletions in human lung cancer; however, it suggests that DCC is a potential target of inactivation by genetic defects including intron or promoter mutations and/or epigenetic alterations. The present result also suggests that DCC expression is associated with some properties of SCLCs, such as a neuroendocrine (NE) feature.
Keywords: DCC, lung cancer, chromosome 18, tumor suppressor gene, mutation
Introduction
We previously reported that loss of heterozygosity (LOH) on chromosome 18q occurs frequently (>60%) in advanced non-small cell lung carcinomas (NSCLCs). The incidence of LOH in brain metastases was significantly higher than that in stage I NSCLCs [1,2]. Frequent occurrence of 18q deletions in NSCLC was also shown by cytogenetic and comparative genomic hybridization analyses [3,4]. LOH on 18q was also observed in 30% of small cell lung carcinomas (SCLCs) [5]. These results indicate that the tumor suppressor gene(s) on 18q plays an important role in the acquisition of malignant phenotypes in lung cancers. Recently, we performed a deletion mapping of chromosome 18q in 62 surgical specimens of lung cancer, and mapped a commonly deleted region at chromosome 18q21 [6].
The chromosome 18q21 region contains three candidates for tumor suppressor genes, SMAD4/DPC4, SMAD2/MADR2/JV18-1 and DCC [7–10]. SMAD4 and SMAD2 proteins are known to reside in a pathway of transforming growth factor-β signaling. SMAD4 was identified as a target gene for homozygous 18q21 deletions in pancreatic cancer [7], and up to the present, inactivation of the SMAD4 gene by homozygous deletions and/or mutations have been reported in several human cancers including pancreatic cancer, colorectal cancer, hepatocellular cancer and ovarian cancer [7,11–13]. SMAD2 is also mutated in a small subset of colorectal cancer and hepatocellular cancer [8,9,12]. However, recent studies revealed that alterations of these two genes are rare in lung cancer, suggesting that genes other than SMAD4 and SMAD2 at 18q21 function as tumor suppressors in human lung carcinogenesis [14,15].
The DCC gene was isolated from the region commonly deleted in colorectal cancer [10], and it encodes a 1447-amino-acid transmembrane protein with four immunoglobulin-like and six fibronectin III-like domains [16]. Recently, it was shown that DCC is also a target gene for homozygous 18q21 deletions in pancreatic cancer [17]. DCC is expressed in most epithelial cells, including lung, and is abundantly expressed in the central and peripheral nervous system [18]. Mutations of the DCC gene have been detected in a subset of several human cancers, including colorectal cancer, esophageal cancer and neuroblastoma [19–21], while reduction or loss of DCC expression is frequently observed in a variety of human cancers [21–27]. DCC has an activity to induce apoptosis and cell cycle arrest in mammalian cells [28,29], and suppress the tumorigenicity of cancer cells [30–32]. It was also shown that DCC is a key factor for the differentiation of neural cells [33,34]. It is well known that a large portion of SCLCs and a subset of NSCLCs have features of neuroendocrine (NE) differentiation [35]. Based on these results, we hypothesized that DCC can be a target tumor suppressor gene for 18q deletions in human lung cancer.
In the present study, we examined the genetic and expression status of the DCC gene in 46 lung cancer cell lines (14 SCLCs and 32 NSCLCs) to elucidate the pathogenetic significance of DCC gene alterations in human lung carcinogenesis. Genetic alterations, such as mutations and homozygous deletions, were infrequent in these cell lines, indicating that DCC is not a target tumor suppressor gene for 18q deletions in human lung carcinogenesis. DCC transcripts were detected in 12 (26%) of the 46 cell lines, and the incidence of the DCC expression was significantly higher in SCLCs than in NSCLCs. The pathogenetic significance for differential expression of the DCC gene in lung cancer cells will be discussed.
Materials and Methods
Samples
Forty-six lung cancer cell lines (14 SCLCs and 32 NSCLCs) were used in this study. The SCLC cell lines were Lu24, Lu134, Lu135, Lu139, H69, H82, N417, LCMA, SBC5, H526, H841, MS18, H209 and H774, while the NSCLC cell lines were A427, A549, PC3, PC7, PC9, PC10, PC13, PC14, Lu99, LC1-Sq, RERF-LCOK, RERF-LCD, RERF-LCMS, ABC1, EBC1, Lu65, H596, H23, H157, H322, H441, H520, H1155, Ma1, Ma2, Ma10, Ma12, Ma17, Ma24, Ma25, Ma26 and Ma29 [36]. The Ma26 cell line is derived from an adenocarcinoma (stage IIIb) of the lung of a 79-year-old male who was admitted to the Osaka Prefectural Habikino Hospital, Osaka, Japan. A primary culture of normal human small airway epithelial cells, Cryo-SAEC (strain 5336), a bronchial epithelial cell line, tpc16B, and a lung fibroblast cell line, WI38, were also used. Cryo-SAEC was purchased from Clonetics (Walkersville, USA). Detailed information on the cell lines used in the present study can be obtained upon request. High-molecular-weight DNA was prepared from the cell lines by proteinase K digestion and phenol-chloroform extraction as described previously [36]. High-molecular-weight DNA prepared from non-cancerous tissues of patients with lung cancers and other cancers were also used in this study [36]. Poly(A) RNA was prepared from cell lines by the Fast Track mRNA isolation kit (Invitrogen, Carlsbad, CA). Poly (A)+ RNA from human normal lung tissues was purchased from Clontech Laboratories Japan Ltd. (Tokyo, Japan).
Polymerase Chain Reaction (PCR) Single-Strand Conformation Polymorphism (SSCP) Analysis
PCR was carried out by using 50 ng of genomic DNA as a template in a 20-µl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 125 ng of each primer, 250 µM each deoxynucleotide triphosphate and 0.1 unit of Taq DNA polymerase (TaKaRa, Tokyo, Japan). DNA fragments for each primer were amplified for 30 cycles of 94°C for 60 seconds, 55°C for 60 seconds, 72°C for 90 seconds followed by a final extension for 10 minutes at 72°C after the initial denaturation step for 5 minutes at 94°C. The primer pairs used for the analysis of exons 1–13, 15–19 and 23–29 of DCC were previously reported [21]. Primers used for the analysis of exons 14, 20 and 22 (5′-AGTAACTTTCTTCCCCCCTG-3′ and 5′-TCAGAGTGATAGTAACTGGGA-3′ for exon 14, 5′-TAGGACACCAAATTAAGTCACA-3′ and 5′-CCCACAGGAAAAGAAAAGCAT-3′ for exon 20, and 5′-GACATTGTGACATGCTCTCC-3′ and 5′-ATATACTTACCATTTAGGGTGC-3′ for exon 22) were set based on the information of Cho et al. [17]. Exon 21, 33 bp in size, was not analyzed due to the lack of sequence information of the exon-intron boundary for the exon [Ref. [17]; Fearon E, personal communication]. SSCP analysis was performed in a low pH buffer system that showed improved separation of long mutant fragments of up to 800 bp [37]. Gels were dried and exposed to Kodak XAR films for 24 to 48 hours at -80°C.
Sequence Analysis
PCR products showing different mobilities were purified by using a QIA quick-spin PCR purification kit (Qiagen Inc., Tokyo, Japan), and directly sequenced in both directions with Thermo-Sequence dye terminator cycle sequencing pre-mix kits (Amersham Pharmacia Biotech, Tokyo, Japan) and ABI 373S DNA Sequence System (PE Biosystems Japan, Tokyo, Japan). Sequencing primers were the same as PCR primers.
Reverse Transcription PCR (RT-PCR) Analysis
Randomly primed cDNAs were reverse-transcribed from 0.5 µg mRNAs by using SuperScriptII Reverse Transcriptase (Gibco BRL, Grand Island, NY) according to the supplier's protocol. One microliter of the cDNA conversion mixture was amplified under the following conditions; 60 seconds at 95°C, 60 seconds at 63°C (for DCC) or 55°C (for GAPDH), and 60 seconds at 72°C for 32 cycles (for DCC) or 25 cycles (for GAPDH), followed by 10 minutes at 72°C. Two sets of primers used in previous studies [10,21,27,38] — 5′-TTCCGCCATGGTTTTTAAATCA-3′ and 5′-AGCCTCATTTTCAGCCACACA-3′, and 5′-GCCTGAGCATTCTAGCAGC-3′ and 5′-AAAATTCACCTCATCCATTCAG-3′ — were used for the amplification of DCC cDNA (nucleotides 987–1218 and 3951–4375 of Genbank NM 005215), and a set of primers — 5′-AAGGTCATCCATGACAAC-3′ and 5′-CACCCTGTTGCTGTAGCCA-3′ — was used for the GAPDH gene (nucleotides 544–1032 of Genbank M33197). The DCC and GAPDH primer pairs were designed to amplify cDNA fragments encompassing two adjacent exons. The PCR products were electrophoresed on 3.0% agarose gel and stained with ethidium bromide.
Results
We examined 46 lung cancer cell lines for mutations in 28 exons of the DCC gene by genomic PCR-SSCP analysis using 28 sets of primers. All the exons analyzed were amplified in these 46 cell lines indicating a lack of homozygous deletions of DCC exons. DNA samples showing band shifts in the SSCP analysis were subjected to direct sequencing. A known non-synonymous (associated with amino acid substitutions) single nucleotide polymorphism (SNP), C601G leading to Arg201Gly [20,21], was observed with allele frequency of 0.55/0.45 in lung cancer cell lines (Table 1). In addition, eight nucleotide substitutions were detected in exons 3, 15, 17, 19, 20, 23, and 24. Two of them, T2277G and G2494A, were non-synonymous. T2277G substitution, which was detected in five cell lines as heterozygotes, was also detected in normal DNA obtained from 80 unrelated individuals (160 alleles); therefore, it was a genetic polymorphism. In contrast, G2494A, which was detected as a heterozygous substitution in a NSCLC cell line, Ma26, was not observed in these 80 normal DNA samples; therefore, this variant was likely to be a somatic mutation. However, since the corresponding normal tissue DNA for this cell line was not available, we could not exclude the possibility that it was a rare genetic polymorphism. The remaining six substitutions were synonymous (not associated with amino acid substitutions). They consisted of a common polymorphism, T3108C, and five rare polymorphisms, A651T, G291OA, T3276A, C3540T and A/G in intron 23.
Table 1.
Nucleotide Substitutions of the DCC Gene in Human Lung Cancer Cell Lines.
| Nucleotide | Amino acid change | Allele frequency | Cell line* | |
| Cell line | Normal lung | |||
| C601G | Arg201Gly | 0.55/0.45 | ND | |
| A651T | Silent | 0.99/0.01 | ND | HI57 |
| T2277G | lle759Met | 0.95/0.05 | 0.97/0.03 | Lu24 |
| Lu134 | ||||
| SBC5 | ||||
| LC1-Sq | ||||
| RERF-LCMS | ||||
| G2494A | Asp832Asn | 0.99/0.01 | 1.00/0.00 | Ma26 |
| C2910A | Silent | 0.99/0.01 | ND | H157 |
| T3108C | Silent | 0.67/0.33 | ND | |
| T3276A | Silent | 0.99/0.01 | ND | H157 |
| C3540T | Silent | 0.98/0.02 | ND | H520 |
| A/G (+3 bp from exon 23) | - | 0.99/0.01 | ND | H774 |
ND, not done.
Cell lines with minor alleles of the frequency less than 10% are listed.
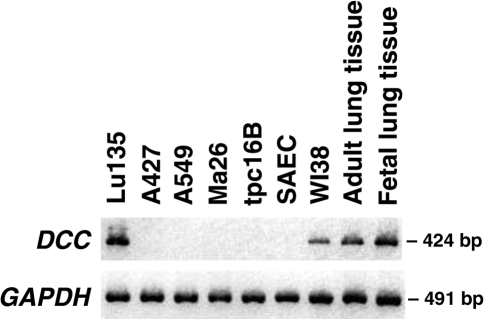
We next examined the expression of the DCC gene by RT-PCR analysis using two sets of primers, which amplify cDNA fragments encompassing exons 6–7 and 27–29, respectively. Among 46 cell lines analyzed in total, DCC gene expression was detected in 14 (30%) cell lines (Table 2). However, the levels of DCC expression in three of them, SBC5, PC7, and Ma10, seemed to be much lower than these in the remaining 11 cell lines, since only faint bands were detected by one of the two sets of primers. Therefore, we concluded that 11 (24%) of the 46 cell lines had the readily detectable levels of DCC expression. DCC expression was more frequently observed in SCLC cell lines (7/14, 50%) than in NSCLC cell lines (4/32, 13%), and the difference in the incidence between SCLC and NSCLC was statistically significant (P = .01, Fisher's exact test) (Table 3). DCC was not expressed in the Ma26 cell line carrying a heterozygous missense DCC mutation (Figure 1). To further investigate the DCC expression in human non-cancerous lung cells, RT-PCR analysis was performed using mRNAs from fetal and adult lung tissues, a primary culture of bronchial epithelial cells, SAEC, a bronchial epithelial cell line, tpc16B, and a lung fibroblast cell line, WI38. DCC expression was detected in fetal and adult lung tissues and the lung fibroblast cell line, but not in the primary culture of bronchial epithelial cells and the bronchial epithelial cell line (Figure 1).
Table 2.
Expression of the DCC Gene in Human Lung Cancer Cell Lines.
| Cell line | Histological subtype | mRNA expression | ||
| nt 986–1218 | nt 3951–4375 | |||
| 1 | Lu24 | SCC | - | - |
| 2 | Lu134 | SCC | + | + |
| 3 | Lu135 | SCC | + | + |
| 4 | Lu139 | SCC | + | + |
| 5 | H69 | SCC | + | + |
| 6 | H82 | SCC | + | + |
| 7 | N417 | SCC | - | + |
| 8 | H209 | SCC | - | - |
| 9 | H526 | SCC | - | - |
| 10 | SBC5 | SCC | ± | - |
| 11 | H774 | SCC | - | - |
| 12 | MS18 | SCC | + | + |
| 13 | H841 | SCC | - | - |
| 14 | LCMA | SCC | - | - |
| 15 | PC3 | ADC | - | - |
| 16 | PC7 | ADC | ± | - |
| 17 | PC9 | ADC | - | - |
| 18 | PC14 | ADC | - | - |
| 19 | A427 | ADC | - | - |
| 20 | A549 | ADC | - | - |
| 21 | H23 | ADC | - | - |
| 22 | H322 | ADC | - | - |
| 23 | H441 | ADC | - | - |
| 24 | LCD | ADC | - | - |
| 25 | LCOK | ADC | - | - |
| 26 | LCMS | ADC | - | - |
| 27 | ABC1 | ADC | - | - |
| 28 | Ma1 | ADC | - | - |
| 29 | Ma10 | ADC | ± | - |
| 30 | Ma12 | ADC | - | - |
| 31 | Ma17 | ADC | - | - |
| 32 | Ma24 | ADC | - | - |
| 33 | Ma26 | ADC | - | - |
| 34 | Ma29 | ADC | - | - |
| 35 | H596 | ADSQC | - | - |
| 36 | LC1-Sq | SQC | + | + |
| 37 | EBC1 | SQC | - | - |
| 38 | H520 | SQC | - | + |
| 39 | PC10 | SQC | + | + |
| 40 | H157 | SQC | - | - |
| 41 | H1155 | LCC | + | + |
| 42 | PC13 | LCC | - | - |
| 43 | Lu65 | LCC | - | - |
| 44 | Lu99 | LCC | - | - |
| 45 | Ma2 | LCC | - | - |
| 46 | Ma25 | LCC | - | - |
Table 3.
Incidence of DCC Expression in Human Lung Cancer Cell Lines.
| Type of tumor | Expression (%) |
| NSCLC | 4/32(13)* |
| Adenocarcinoma | 0/20 (0) |
| Squamous cell carcinoma | 3/5 (60) |
| Large cell carcinoma | 1/6(17) |
| Adenosquamous cell carcinoma | 0/1 (0) |
| SCLC | 7/14 (50)* |
| Total | 11/46(24) |
The difference in the incidence between NSCLC and SCLC was statistically significant (P = .01, Fisher's exact test).
Figure 1.
Expression of the DCC gene detected by RT-PCR analysis. Primers designed to amplify the 3′-portion of DCC cDNA (nucleotides 3951–4375 of Genbank NM 005215) were used GAPDH was analyzed to standardize the RNA amount of each sample.
Discussion
Forty-six lung cancer cell lines were examined for genetic alterations and expression of the DCC gene. Nine different types of nucleotide substitutions were identified, and three of them were non-synonymous. However, only one of the three non-synonymous substitutions, which was detected in the Ma26 cell line, was considered as a somatic mutation because it was not observed in 80 unrelated individuals. The substitution was heterozygous and was associated with the change of an evolutionally conserved amino acid located between the fibronectin III-like domains 4 and 5 [16,19,39,40]. There was no case of homozygous deletion in the DCC exons. Therefore, genetic alterations, including nucleotide substitutions and homozygous deletions of the coding region, in the DCC gene are infrequent in human lung cancer. In contrast to the low frequency of genetic alterations, loss of DCC expression was observed in a large portion of lung cancer cell lines, including the Ma26 cell line with heterozygous missense mutation of DCC. At present, molecular mechanisms for the loss or reduction of DCC expression are unclear. DCC was not expressed in lung epithelial cells, but expressed in a fibroblast cell line. Therefore, the status of DCC expression in lung cancer cell lines may reflect that in their precursor cells. However, only two kinds of cultured bronchial epithelial cells were examined in this study, and precursor cells for lung cancer are still obscure at present; therefore, more detailed analysis of DCC expression in cancerous and non-cancerous lung tissues should be necessary to draw a conclusion. Alternatively, the loss or reduction of DCC expression could be caused by genetic alterations in the region not examined, such as the promoter region and intron sequences, since DCC is a large gene and the coding regions comprise only a small portion of the gene's total size [10,19]. It is also possible that DCC transcription is silenced by epigenetic changes, such as hypermethylation of the CpG island as in the cases of the p16 and VHL genes [41]. To investigate this possibility, we preliminarily examined DCC expression in several lung cancer cell lines without DCC expression after culturing for 3 days with 1 µM 5-aza-deoxycytidine; however, re-expression of the DCC gene was not observed in these cell lines (data not shown). Therefore, hypermethylation could not be a cause for the loss of DCC expression. We previously examined the allelic status of 11 microsatellite loci on chromosome 18q in the 46 cell lines used in this study, and 25 (54%) of them were inferred as having 18q LOH since significant contiguous homozygosity of these loci was observed [6]. DCC was expressed in 4 (19%) of the 25 cell lines inferred as having 18q LOH and in 8 (38%) of the remaining 21 cell lines. Therefore, it is unlikely that the loss of DCC expression is linked to 18q LOH. The present result does not implicate DCC as a specific mutational target of 18qLOH in human lung cancer; however, it does not definitively exclude DCC as a potential target of inactivation by both genetic and epigenetic mechanisms. Genetic defects including intron or promoter mutations or unknown epigenetic mechanisms could inactivate both DCC alleles in cases that lacked 18q LOH. Further investigation should be performed to clarify the mechanisms regulating DCC expression in human lung cancer cells.
Studies on several cancers indicated that DCC expression is associated with several properties of cancer cells, such as differentiation, progression, and/or malignancy [21–27]. In the present study, DCC expression was observed with the higher incidence in SCLCs than in NSCLCs, raising the possibility that DCC expression is associated with a unique property of SCLC. It is known that a large portion of SCLCs, but a small subset of NSCLCs, show NE differentiation and express NE markers such as neuron-specific enolase [35]. Since DCC is involved in neural differentiation [33,34], we further investigated the association of DCC expression with the properties of NE differentiation in lung cancer cells. To our knowledge, 17 of the 46 cell lines used in the present study have been examined for expression of NE markers, and seven (six SCLCs and a NSCLC) were shown as having some properties of NE differentiation, while 10 (four SCLCs and six NSCLCs) were not [42,43]. DCC expression was detected in six of the seven cell lines with NE property, but 2 of the 10 cell lines without NE property, and the difference was statistically significant (P = .01, Fisher's exact test). Therefore, it is possible that DCC expression is associated with the NE differentiation, a common feature of SCLCs, in lung cancer cells. It is also possible that the DCC expression has some effects on differentiation of lung cancer cells. Thus, the status of DCC expression should be further examined in primary lung tumors of various stages and histological subtypes in order to elucidate the biological significance of differential DCC expression in lung cancer.
In the course of the mutation study of DCC, a novel non-synonymous SNP at codon 759 was identified in addition to the known non-synonymous one at codon 201 [20,21]. These two SNPs lead to the substitution of evolutionally conserved amino acids located in the immunoglobulin-like C2 domain 2 and fibronectin III-like domain 4, respectively [39,40]; therefore, it is possible that they have some effects on the physiological activities of DCC protein. Interestingly, the mouse Dcc gene is reported to be a candidate gene for Par2 (pulmonary adenoma resistance 2), a putative gene involved in the susceptibility to lung tumorigenesis [44,45]. Therefore, it would be worth examining the allele distribution of these two non-synonymous SNPs in the DCC gene in a case-control study of lung cancer in order to investigate the involvement of this gene in human lung cancer susceptibility.
The low frequency of genetic alterations of DCC, SMAD2 and SMAD4 [this study, Refs. [14,15] in human lung cancer indicates that the 18q21 region harbors unknown tumor suppressor gene(s) involved in lung carcinogenesis. Recently, we identified a minimum overlapping region of 18q deletions in lung cancer at the 18q21 region between the SMAD4 and DCC loci. The size of the region was estimated as being less than 300 kb [Kohno T, unpublished result], and the region did not include coding exons of DCC, SMAD4 and SMAD2 [6]. Thus, it is possible that a tumor suppressor gene involved in lung carcinogenesis is located in this region. In an effort to identify the unknown tumor suppressor gene at 18q21, construction of a BAC contig covering the minimum overlapping region and a search for genes in the region are in progress in our laboratory.
Acknowledgements
We thank the following scientists for providing lung cancer cell lines: Y. Hayata of Tokyo Medical College; T. Terasaki and S. Hirohashi of the National Cancer Center Research Institute, Japan; M. Takada of Izumisano Municipal Hospital; and A. F. Gazdar and J. D. Minna of the University of Texas Southwestern Medical Center. Cell lines were also obtained from the American Type Culture Collection. We are grateful to M. Noguchi of University of Tsukuba for providing us with a human bronchial epithelial cell line, tpc16B. S. T., K. T., K. I. and M. N. are recipients of the Research Resident Fellowship from the Foundation for Promotion of Cancer Research.
Abbreviations
- SCLC
small cell lung carcinoma
- NSCLC
non-small cell lung carcinoma
- SSCP
single-strand conformation polymorphism
- SNP
single nucleotide polymorphism
- LOH
loss of heterozygosity
- AI
allelic imbalance
- RT-PCR
reverse transcription polymerase chain reaction
- NE
neuroendocrine
Footnotes
This work was supported, in part, by a Grant-in-Aid from the Ministry of Health and Welfare for the second term Comprehensive 10-Year Strategy for Cancer Control, a Grant-in-Aid from the Ministry of Health and Welfare for Research on Human Genome and Gene Therapy, Grants-in-Aid from the Ministry of Health and Welfare and from the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.Shiseki M, Kohno T, Nishikawa R, Sameshima Y, Mizoguchi H, Yokota J. Frequent allelic losses on chromosome 2q, 18q, and 22q in advanced non-small cell lung carcinoma. Cancer Res. 1994;54:5643–5648. [PubMed] [Google Scholar]
- 2.Shiseki M, Kohno T, Adachi J, Okazaki T, Otsuka T, Mizoguchi H, Noguchi M, Hirohashi S, Yokota J. Comparative allelotype of early and advanced stage non-small cell lung carcinomas. Gene Chromosomes Cancer. 1996;17:71–77. doi: 10.1002/(SICI)1098-2264(199610)17:2<71::AID-GCC1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.Testa JR, Siegfried JM, Liu Z, Hunt JD, Feder MM, Litwin S, Zhou JY, Taguchi T, Keller SM. Cytogenetic analysis of 63 non-small cell lung carcinomas: recurrent chromosome alterations amid frequent and widespread genomic upheaval. Genes Chromosomes Cancer. 1994;11:178–194. doi: 10.1002/gcc.2870110307. [DOI] [PubMed] [Google Scholar]
- 4.Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, Langreck H, Gellert K, Reichel M, Just K, du Manoir S, Cremer T, Dietel M, Ried T. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997;57:2331–2335. [PubMed] [Google Scholar]
- 5.Kawanishi M, Kohno T, Otsuka T, Adachi J, Sone S, Noguchi M, Hirohashi S, Yokota J. Allelotype and replication error phenotype of small cell lung carcinoma. Carcinogenesis. 1997;18:2057–2062. doi: 10.1093/carcin/18.11.2057. [DOI] [PubMed] [Google Scholar]
- 6.Takei K, Kohno T, Hamada K, Takita J, Noguchi M, Matsuno Y, Hirohashi S, Uezato H, Yokota J. A novel tumor suppressor locus on chromosome 18q involved in the development of human lung cancer. Cancer Res. 1998;58:3700–3705. [PubMed] [Google Scholar]
- 7.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 8.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGF-β-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 9.Riggins GJ, Thiagalingam S, Rozenblum E, Weinstein CL, Kern SE, Hamilton SR, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mad-related genes in the human. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 10.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, Vogelstein B. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science (Washington, DC) 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 11.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero RA, Jr, Meltzer PS, Hahn SA, Kern SE. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 12.Yakicier MC, Irmak MB, Romano A, Kew M, Ozturk M. Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene. 1999;18:4879–4883. doi: 10.1038/sj.onc.1202866. [DOI] [PubMed] [Google Scholar]
- 13.Takakura S, Okamoto A, Saito M, Yasuhara T, Shinozaki H, Isonishi S, Yoshimura T, Ohtake Y, Ochiai K, Tanaka T. Allelic imbalance in chromosome band 18q21 and SMAD4 mutations in ovarian cancers. Genes Chromosomes Cancer. 1999;24:264–271. doi: 10.1002/(sici)1098-2264(199903)24:3<264::aid-gcc12>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 14.Nagatake M, Takagi Y, Osada H, Uchida K, Mitsudomi T, Saji S, Shimokata K, Takahashi T, Takahashi T. Somatic in vivo alterations of the DPC4 gene at 18q21 in human lung cancers. Cancer Res. 1996;56:2718–2720. [PubMed] [Google Scholar]
- 15.Uchida K, Nagatake M, Osada H, Yatabe Y, Kondo M, Mitsudomi T, Masuda A, Takahashi T, Takahashi T. Somatic in vivo alterations of the JV18-1 gene at 18q21 in human lung cancers. Cancer Res. 1996;56:5583–5585. [PubMed] [Google Scholar]
- 16.Hedrick L, Cho KR, Fearon ER, Wu TC, Kinzler KW, Vogelstein B. The DCC gene product in cellular differentiation and colorectal tumorigenesis. Genes Dev. 1994;8:1174–1183. doi: 10.1101/gad.8.10.1174. [DOI] [PubMed] [Google Scholar]
- 17.Hilgers W, Song JJ, Hayes M, Hruban RR, Kern SE, Fearon ER. Homozygous deletions inactivate DCC, but not DPC4, in a subset of pancreatic and biliary cancers. Genes Chromosomes Cancer. 2000;27:353–357. [PubMed] [Google Scholar]
- 18.Reale MA, Hu G, Zafar AI, Getzenberg RH, Levine SM, Fearon ER. Expression and alternative splicing of the deleted in colorectal cancer (DCC) gene in normal and malignant tissues. Cancer Res. 1994;54:4493–4501. [PubMed] [Google Scholar]
- 19.Cho KR, Oliner JD, Simons JW, Hedrick L, Fearon ER, Preisinger AC, Hedge P, Silverman GA, Vogelstein B. The DCC gene: structural analysis and mutations in colorectal carcinomas. Genomics. 1994;19:525–531. doi: 10.1006/geno.1994.1102. [DOI] [PubMed] [Google Scholar]
- 20.Miyake S, Nagai K, Yoshino K, Oto M, Endo M, Yuasa Y. Point mutations and allelic deletion of tumor suppressor gene DCC in human esophageal squamous cell carcinomas and their relation to metastasis. Cancer Res. 1994;54:3007–3010. [PubMed] [Google Scholar]
- 21.Kong XT, Choi SH, Inoue A, Xu F, Chen T, Takita J, Yokota J, Bessho F, Yanagisawa M, Hanada R, Yamamoto K, Hayashi Y. Expression and mutational analysis of the DCC, DPC4, and MADR2/JV18-1 genes in neuroblastoma. Cancer Res. 1997;57:3772–3778. [PubMed] [Google Scholar]
- 22.Scheck AC, Coons SW. Expression of the tumor suppressor gene DCC in human gliomas. Cancer Res. 1993;53:5605–5609. [PubMed] [Google Scholar]
- 23.Ekstrand BC, Mansfield TA, Bigner SH, Fearon ER. DCC expression is altered by multiple mechanisms in brain tumours. Oncogene. 1995;11:2393–2402. [PubMed] [Google Scholar]
- 24.Reale MA, Reyes-Mugica M, Pierceall WE, Rubinstein MC, Hedrick L, Cohn SL, Nakagawara A, Brodeur GM, Fearon ER. Loss of DCC expression in neuroblastoma is associated with disease dissemination. Clin Cancer Res. 1996;2:1097–1102. [PubMed] [Google Scholar]
- 25.Shibata D, Reale MA, Lavin P, Silverman M, Fearon ER, Steele G, Jr, Jessup JM, Loda M, Summerhayes IC. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727–1732. doi: 10.1056/NEJM199612053352303. [DOI] [PubMed] [Google Scholar]
- 26.Reyes-Mugica M, Rieger-Christ K, Ohgaki H, Ekstrand BC, Helie M, Kleinman G, Yahanda A, Fearon ER, Kleihues P, Reale MA loss of DCC expression and glioma progression. Cancer Res. 1997;57:382–386. [PubMed] [Google Scholar]
- 27.Yoshida Y, Itoh F, Endo T, Hinoda Y, Imai K. Decreased DCC mRNA expression in human gastric cancers is clinicopathologically significant. Int J Cancer. 1998;79:634–639. doi: 10.1002/(sici)1097-0215(19981218)79:6<634::aid-ijc14>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 29.Chen YQ, Hsieh JT, Yao F, Fang B, Pong RC, Cipriano SC, Krepulat F. Induction of apoptosis and G2/M cell cycle arrest by DCC. Oncogene. 1999;18:2747–2754. doi: 10.1038/sj.onc.1202629. [DOI] [PubMed] [Google Scholar]
- 30.Klingelhutz AJ, Hedrick L, Cho KR, McDougall JK. The DCC gene suppresses the malignant phenotype of transformed human epithelial cells. Oncogene. 1995;10:1581–1586. [PubMed] [Google Scholar]
- 31.Velcich A, Corner G, Palumbo L, Augenlicht L. Altered phenotype of HT29 colonic adenocarcinoma cells following expression of the DCC gene. Oncogene. 1999;18:2599–2606. doi: 10.1038/sj.onc.1202610. [DOI] [PubMed] [Google Scholar]
- 32.Kato H, Zhou Y, Asanoma K, Kondo H, Yoshikawa Y, Watanabe K, Matsuda T, Wake N, Barrett JC. Suppressed tumorigenicity of human endometrial cancer cells by the restored expression of the DCC gene. Br J Cancer. 2000;62:459–466. doi: 10.1054/bjoc.1999.0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawlor KG, Narayanan R. Persistent expression of the tumor suppressor gene DCC is essential for neuronal differentiation. Cell Growth Differ. 1992;3:609–616. [PubMed] [Google Scholar]
- 34.Piercell WE, Cho KR, Getzenberg RH, Reale MA, Hedrick L, Vogelstein B, Fearon E. NIH3T3 cells expressing the deleted in colorectal cancer tumor suppressor gene product stimulate neurite outgrowth in rat PC12 pheochromocytoma cells. J Cell Biol. 1994;124:1017–1027. doi: 10.1083/jcb.124.6.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gazdar AF. Advances in the biology of lung cancer. Clinical significance of neuroendocrine differentiation. Chest. 1989;96:39S–41S. [PubMed] [Google Scholar]
- 36.Hamada K, Kohno T, Takahashi M, Yamazaki M, Tashiro H, Sugawara C, Ohwada S, Sekido Y, Minna JD, Yokota J. Two regions of homozygous deletion clusters at chromosome band 9p21 in human lung cancer. Genes Chromosomes Cancer. 2000;27:308–318. [PubMed] [Google Scholar]
- 37.Kukita Y, Tahira T, Sommer SS, Hayashi K. SSCP analysis of long DNA fragments in low pH gel. Hum Mutat. 1997;10:400–407. doi: 10.1002/(SICI)1098-1004(1997)10:5<400::AID-HUMU11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 39.Masu K-K, Masu M, Hinck L, Leonardo ED, Chan SS-Y, Culotti JG, Lavigne MT. Deleted in colorectal cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 40.Cooper HM, Armes P, Britto J, Gad J, Wilks AF. Cloning of the mouse homologue of the deleted in colorectal cancer gene (mDCC) and its expression in the developing mouse embryo. Oncogene. 1995;11:2243–2254. [PubMed] [Google Scholar]
- 41.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 42.Terasaki T, Shimosato Y, Nakajima T, Tsumuraya M, Morinaga S, Hirohashi S, Yamaguchi K, Kato K, Ichinose H, Nagatsu T. Changes in cell characteristics due to culture conditions in cell lines from human small cell lung cancer. Jpn J Clin Oncol. 1986;16:203–212. [PubMed] [Google Scholar]
- 43.Carbone DP, Koros AM, Linnoila RI, Jewett P, Gazdar AF. Neural cell adhesion molecule expression and messenger RNA splicing patterns in lung cancer cell lines are correlated with neuroendocrine phenotype and growth morphology. Cancer Res. 1991;51:6142–6149. [PubMed] [Google Scholar]
- 44.Obata M, Nishimori H, Ogawa K, Lee GH. Identification of the Par2 (pulmonary adenoma resistance) locus on mouse chromosome 18, a major genetic determinant for lung carcinogen resistance in BALB/cByJ mice. Oncogene. 1996;13:1599–1604. [PubMed] [Google Scholar]
- 45.Devereux TR, Anna CH, Patel AC, White CM, Festing MF, You M. Smad4 (homolog of human DPC4) and Smad2 (homolog of human JV18-1): candidates for murine lung tumor resistance and suppressor genes. Carcinogenesis. 1997;18:1751–1755. doi: 10.1093/carcin/18.9.1751. [DOI] [PubMed] [Google Scholar]