Abstract
In multicellular organisms, cell proliferation and death must be regulated to maintain tissue homeostasis. Many observations suggest that this regulation may be achieved, in part, by coupling the process of cell cycle progression and programmed cell death by using and controlling a shared set of factors. An argument in favor of a link between the cell cycle and apoptosis arises from the accumulated evidence that manipulation of the cell cycle may either prevent or induce an apoptotic response. This linkage has been recognized for tumor suppressor genes such as p53 and RB, the dominant oncogene, c-Myc, and several cyclin-dependent kinases (Cdks) and their regulators. These proteins that function in proliferative pathways may also act to sensitize cells to apoptosis. Indeed, unregulated cell proliferation can result in pathologic conditions including neoplasias if it is not countered by the appropriate cell death. Translating the knowledge gained by studying the connection between cell death and cell proliferation may aid in identifying novel therapies to circumvent disease progression or improve clinical outcome.
Keywords: apoptosis, cell cycle, c-myc, p53, pRb, Cdks
Introduction
Apoptosis is a highly conserved mechanism by which eucaryotic cells commit suicide. It enables an organism to eliminate unwanted and defective cells through an orderly process of cellular disintegration that has the advantage of not inducing an undesirable inflammatory response [1]. Apoptotic elimination of cells occurs during normal development and turnover, as well as in a variety of pathological conditions. Indeed, improper regulation of apoptosis contributes to disorders such as cancer, viral infection, autoimmune diseases, neurodegenerative disorders, stroke, anemia and AIDS [2].
Apoptosis can be triggered by a wide variety of signals. These include Fas ligand, tumor necrosis factor, growth factor withdrawal, viral or bacterial infection, oncogenes, irradiation, ceramide, and chemotherapeutic drugs [2]. Often, these signals are cell-type-specific. Even if these agents vary from cell to cell, there is basic biochemical machinery underlying the process of regulated cell death. The morphological changes characteristic of the apoptotic process are mainly due to caspases, a family of cysteine proteases that act as effectors of the cell death pathway [3]. Their activation leads to the cleavage of specific proteins that include lamins, topoisomerases, DNA-dependent protein kinase (DNA-PK), poly (ADP-ribose) polymerase (PARP) and some cell cycle regulators.
The cell cycle is a conserved mechanism by which eucaryotic cells replicate themselves. In metazoans, the process of cell loss and cell gain must be homeostatically balanced in order to generate and maintain the complex architecture of tissues, and also to allow adaptation to changing circumstances. One way in which this connection may be achieved is through the coupling of the cell cycle and programmed cell death, perhaps by using or controlling a shared a set of factors [4].
A direct link between apoptosis and the cell cycle may be supposed from noting that mitosis and apoptosis display very similar morphological features. During both processes, cells lose substrate attachment, become rounded, shrink, condense their chromatin and display membrane blebbing. Although mitosis and apoptosis share some features, of course there are critical differences. For example, in apoptotic cells, DNA is degraded at internucleosomal linker sites, yielding DNA fragments in multiples of 180 bp resulting in a nucleosomal ladder. In addition, during apoptosis, cell membrane proteins are cross-linked, making the membrane more rigid [2], and apoptotic cells are typically phagocytosed by adjacent cells or macrophages. In contrast, during mitosis, DNA is segregated and the cell is divided by cytokinesis, resulting in two healthy, viable daughter cells.
A more solid argument in favor of a link between the cell cycle and apoptosis is based on several instances in which apoptosis is regulated by genes that are involved in cell cycle progression. There is accumulating evidence that manipulation of the cell cycle may prevent or induce an apoptotic response depending upon the cellular context [5]. Because cellular context is crucial, these proteins may serve to tie cell death to proliferative signals even though they may not be part of the cell's apoptotic machinery. It is possible that these proliferative proteins act to sensitize cells to apoptosis.
This review, after briefly describing cell cycle regulation, summarizes the role of cell proliferation regulators in apoptosis. Specifically, we will discuss p53, Myc and pRb, players with important relevance in tumor progression. We will also describe the involvement of cyclin-dependent kinases (Cdks) and their regulators in apoptosis.
Cell Cycle Machinery
The cell cycle is a set of events responsible for cell duplication [6]. Transmission of genetic information from one cell generation to the next requires genome replication during the S-phase, and its segregation to the two new daughter cells during mitosis or M-phase. S- and M-phases are crucial events rigorously ordered in a cyclic process that allows for the correct duplication of the cell without accumulating genetic abnormalities. In a normal cell cycle, S-phase is always proceeded by M-phase and M-phase does not occur until S-phase is complete. Between the S- and M-phases, there are two preparatory gaps. G1 separates M from S, and G2 is between S and M. When the cell undergoes differentiation, it exits from the G1 phase of the cell cycle to enter into a quiescent state referred to as G0.
The timing and order of cell cycle events are monitored during cell cycle checkpoints that occur at the G1/S boundary, in S-phase, and during the G2/M-phases [6]. The checkpoints are a series of control systems enabling proliferation only in the presence of stimulatory signals such as growth factors. They also contribute to the fidelity with which genetic information is passed from one generation to the next. The checkpoints also are activated by DNA damage and by mis-aligned chromosomes at the mitotic spindle. In this case, the growth arrest caused by checkpoints allows the cell to repair the damage. After damage repair, progression through the cell cycle resumes. If the damage cannot be repaired, the cell is eliminated through apoptosis.
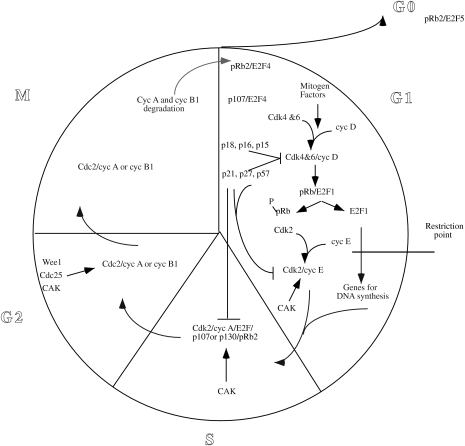
Progression of the eucaryotic cell through the four phases of the cell cycle is mediated by sequential activation and inactivation of Cdks (Figure 1). The Cdks belong to a well-conserved family of serine/threonine protein kinases. Their kinase activity is dependent on the presence of activating subunits called cyclins. The abundance of specific cyclins increases during the phase of the cell cycle in which they are required and decreases during phases in which they are not needed. Cyclin D associates with Cdk4 and Cdk6 during early G1. The primary target of the G1 kinases is the pRb family of proteins. Their phosphorylation permits the transcription of genes necessary for S-phase. The cyclin D/Cdk complexes are crucial to the cell cycle by coupling extracellular signals to the cell cycle [6]. In fact, upon mitogenic stimulation, cyclin D/Cdk complexes are activated and the cells progress from G0 into G1. In late G1, after the restriction point (R-point) is passed, the cell is committed to entering S-phase and cyclin D-Cdk activity is no longer required for cell cycle progression. Cyclin E activates Cdk2 during the G1-to-S-phase transition. Cyclin A binds to Cdk2 during S-phase or to Cdc2 in the G2-to-M-phase transition. The cyclin B/Cdc2 complex also functions during the G2-to-M-phase transition.
Figure 1.
Schematic representation of the relationship between Cdks and Cdk regulators during the cell cycle.
The activity of Cdk complexes is also regulated by other mechanisms. Their activation requires association with their cyclin partner and phosphorylation by Cdk-activating kinase (CAK). In addition, Cdk activity is suppressed by phosphorylation at conserved threonine and tyrosine residues. Dephosphorylation of these residues and consequent Cdk activation are mediated by the Cdc25 phosphatase family. Another mechanism regulating the Cdk activity is mediated by the Cdk-inhibitory subunits (CKIs). In mammalian cells, two classes of CKIs, the Cip/Kip and the Ink4 families, provide a tissue-specific mechanism by which cell cycle progression can be restrained in response to extracellular and intracellular signals [7]. The Cip/Kip family includes p21Cip1, p27Kip1 and p57Kip2 and predominantly inhibits the Cdks of the G1-to-S-phase transition. The Ink4 (inhibitors of Cdk4) family contains four members, p15Ink4b p16Ink4, p18Ink4c and p19Ink4d, several of which are mutated or deleted in certain types of human cancers.
pRb, the retinoblastoma protein, is a negative regulator of cell growth and is a tumor suppressor. It is mutated or deleted in many cancers, such as retinoblastoma and carcinomas of the lung, breast, bladder, bone and prostate [8]. It is the principal of a family of proteins that also encompasses pRb2/p130 and p107 [9]. These proteins have overlapping, but distinct, activities in regulating the cell cycle. In general, hypophosphorylated forms of these three proteins are functionally active in blocking transcription of S-phase genes. This ability depends on their capacity to bind and actively repress the E2F factors [10]. The E2F factors are transcriptional activators composed of an E2F protein and a member of the DP family of proteins [11]. In this review, these complexes are referred simply as E2F. E2F positively regulates the transcription of S-phase genes. Upon mitogen stimulation, G1 Cdks phosphorylate pRb and pRb-like proteins [6], thereby releasing the E2F complexes and allowing expression of genes for DNA synthesis.
c-myc and the Dual Signal Model
The c-myc proto-oncogene has been studied extensively and its product is the best characterized member of the Myc family of short-lived nuclear phosphoproteins [12]. It encodes a protein that functions as a transcription factor that stimulates cell proliferation. Myc's activity depends upon association with Max, and this association is required for mitogenesis [12]. Myc is induced by mitogens in proliferating cells and is absent in quiescent cells. When Myc is ectopically expressed, it drives cells into the cell cycle. Conversely, inhibition of c-myc expression leads to growth arrest [13].
A connection between Myc and apoptosis has also been demonstrated. Myc overexpression promotes apoptosis during serum deprivation or hypoxia [14]. Moreover, anti-apoptotic genes, such as Bcl-2, suppress Myc-induced apoptosis [15]. The mitogenic and pro-apoptotic functions of Myc both require dimerization with Max and sequence specific DNA binding [12]. It is most likely that Myc-Max dimers execute their functions by regulating specific target genes.
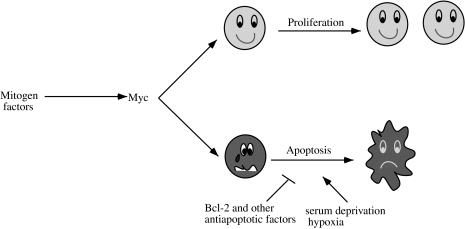
Several models have been proposed to explain the apparently contradictory roles of Myc in both proliferation and apoptosis. It is tempting to speculate that inappropriate Myc expression pushes cells into a cell cycle during serum deprivation or hypoxia for which they were not prepared, thereby sensitizing cells to apoptosis. However, Myc-induced apoptosis is independent of cell cycle position and so this explanation is not satisfying [5]. The “dual signal” model has been postulated in which Myc activates genes involved in both proliferation and apoptotic pathways [5]. Mitogens stimulate Myc's proliferation pathway, while anti-apoptotic factors, such as Bcl-2, may shut down Myc's apoptotic pathway (Figure 2). The fact that Myc-induced apoptosis, but not proliferation, is inhibited by Bcl-2 suggests that there are two distinct sets of genes involved in these pathways that can be modulated by different signals [15]. In addition, Myc apoptosis does not require the synthesis of novel polypeptides, indicating that the Myc-induced apoptotic pathway is “set” in growing cells but that it is being suppressed.
Figure 2.
The “Dual Signal Model”. c-Myc induces proliferation or apoptosis depending on the presence or the absence of anti-apoptotic regulators.
The precise mechanism by which Myc induces apoptosis has not been demonstrated. However, Cdc25A is a well-established transcriptional target of Myc [16]. Cdc25A mediates Myc's effect on the cell cycle by inducing Cdk activity. Cdc25A also induces apoptosis in serum-deprived fibroblasts [16], identical to what has been observed for Myc. Inhibition of Cdc25A expression by antisense also diminishes the ability of Myc to induce apoptosis. Therefore, it is possible that Cdc25A mediates Myc's apoptotic effect; however, additional research is required to determine how Cdc25A, in turn, induces apoptosis.
p53: Growth Arrest or Apoptosis?
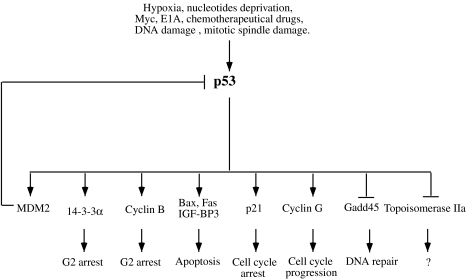
The importance of p53 in normal cell growth is supported by the fact that its function is lost in approximately half of all human cancers. It is involved in several different aspects of cell cycle arrest, apoptosis, control of genome integrity, and DNA repair [17] (Figure 3). It regulates a variety of processes by transactivating genes that are involved in different cellular functions (e.g. p21, Gadd45, Mdm2, Egfr, PCNA, cyclin D1, cyclin G, TGFα, 14-3-3σ, Bax, Bcl-XL, Fas1, FasL, DR5 and lgf-BP3). p53 also can inhibit the expression of specific genes, such as topoisomerase IIa.
Figure 3.
p53 pathways. Several factors induce p53 activity. p53 controls G1 and G2 checkpoints activating or inhibiting genes involved in cell cycle, apoptosis and DNA repair.
p53 is a nuclear DNA-binding phosphoprotein that exists normally as a tetramer able to bind specific DNA sequences. A variety of stimuli causes its activation by increasing the protein's half-life and the rate of translational initiation of its mRNA [18]. Posttranslational modification of the protein, as well as alternative splicing and binding of regulatory proteins, also are involved in activating p53 [19].
p53 influences proliferation by acting predominately in the G1 phase of the cell cycle progression. Oncogenic and hyperproliferative stimuli (e.g. Myc, Ras, E1A), DNA damage by UV, hypoxia, γ-irradiation, nucleotide deprivation or chemotherapeutic drugs activate p53 [17]. Activated p53 causes a G1 arrest by inducing expression of p21 and the consequent inhibition of cyclin D/Cdks. In these conditions, pRb is not phosphorylated and cells do not progress through the G1-to-S-phase transition. Schneider et al. [20] reported that p53 also can regulate CAK activity leading to Cdk2 down regulation. These findings imply a direct involvement of p53 in triggering growth arrest by its interaction with the CAK complex without the need of Cdk inhibitors.
Numerous studies demonstrate that p53 also functions at the G2/M checkpoint [19]. The ability of p53 to induce a G2 arrest is cell-type-specific. Quite frequently in many mouse, rat and human cell lines overexpression of p53 inhibits entry into mitosis [21]. This checkpoint seems to be activated when DNA synthesis is blocked and prevents segregation of damaged or incompletely synthesized DNA. The mechanism behind this G2/M growth arrest still is unknown. Probably, p53 induces a G2 arrest by decreasing cyclin B1 transcription and synthesis [22]. p53 may induce a G2 arrest also thorough 14-3-3σ. 14-3-3σ inactivates Cdc25C and, consequently, Cdc2 activity [23].
p53 is also involved in the spindle checkpoint that blocks re-replication of DNA when the mitotic spindle has been damaged, by inhibiting entry into S-phase [24]. In addition, p53 plays a role in controlling centrosome duplication. In fact, it has been observed that p53 directly associates with centrosomes [25] and prevents mitotic failure regulating the number of centrosomes [26].
p53 plays an important role in triggering apoptosis in certain cell types. p53-dependent apoptosis is induced by DNA damage, hypoxia, withdrawal of growth factors, or expression of Myc or E1A [17]. In most cases, p53-induced apoptosis appears to be independent of its transcriptional function because it occurs in the presence of protein synthesis inhibitors and because p53 mutants unable to transactivate can still induce apoptosis [27]. p53 also represses the transcription of specific genes that inhibit its capability to induce apoptosis such as Bcl-2 [28]. Protein-protein interaction between p53 and factors involved in the DNA repair mechanism can account for additional ways by which p53 induces apoptosis without transcription activation. One example is the interaction of p53 with TFIIH, a transcription factor involved in DNA repair [29].
However, in some systems, the transactivation function of p53 plays an important role in inducing apoptosis. For example, the pro-apoptotic proteins, Bax and lgF-Bp3, are transcriptional targets of p53. p53 also transactivates Fas transcription [30]. Fas is a cell surface protein that triggers apoptosis upon ligand (FasL) binding. Fas belongs to the TNFR family of genes coding for membrane receptors, and are involved in the regulation of cell proliferation. In the presence of DNA damage, p53 transactivates also the DR5 gene [31]. DR5 is another member of TNFR family. Like Fas, the binding of the ligand (TRAIL) to DR5 activates caspase-dependent apoptosis. The induction of Fas and DR5 transcription by p53 indicates that the p53 transcriptional function may assist in modulating the apoptotic response triggered by certain stimuli.
After stimulation, p53 may induce cell death or cell cycle arrest; the different outcome depends upon a variety of variables. The genetic background of the cell can be important. For instance, p21-null cells do not arrest in response to DNA damage but proceed to apoptose in a p53-dependent manner [32]. Impaired cross-talk between p53 and pRb can lead to the same result (see below). The anti-apoptotic effect of Bcl-2 and adenoviral E1B can prevent p53-mediated apoptosis [33,34]. In some hematopoietic cells, the presence of specific interleukins (ILs) can direct the cell towards apoptosis or cell arrest after p53 induction [35]. The extent of DNA damage and p53 protein levels are also factors that contribute to making the choice between life and death. It may be that during p53-induced cell cycle arrest, the cell attempts to repair damage, perhaps with the assistance of its enhanced repair capacity from p53 induction of Gadd45. If the damage is too extensive to be repaired, the cell then is committed to die.
It has been shown that p53 can be regulated by oncogenic and hyperproliferative stimuli. CKI p14 and its murine counterpart p19, coded by the ARF/INK4 locus, are the main players in this mechanism. Mitogenic signals derived by oncogenes such as Myc, E1A or E2F are able to induce p14 synthesis. Subsequently, p14 binds Mdm2 inhibiting its capability to induce p53 degradation [for review, see Ref. [36]]. Indirect stabilization of p53 by p14 results in cell death.
pRb
pRb is not only a growth suppressor but also an anti-apoptotic factor. In vivo studies show that pRb knockout mice die in utero at 14 to 15 days [37]. They have defects in the haematopoietic system and have impaired development of the central and the peripheral nervous system because of massive cell death. Inappropriate cell death also is present in the liver, lens, and skeletal muscle precursors.
In vitro experiments confirm results obtained with transgenic mice. TGF-β1 causes cell death by suppressing pRb expression and phosphorylation [38]. IFNγ-induced apoptosis is blocked in pRb-positive cells [39]. Restoring the function of pRb in a pRb-null osteosarcoma cell line inhibits irradiation-induced apoptosis [40]. The presence of HPV in HeLa cells abolishes functional p53 and pRb. In this cell line, ectopic expression of p53 leads to cell death but the lethal effect of p53 can be reverted by cotransfecting pRb [27]. Additional support for a protective role of pRb against apoptosis comes from the observation that pRb is the target of caspases. pRb is cleaved in different apoptotic systems such as tumor necrosis factor-, staurosporine-, Fas- and cytosine arabinoside-induced apoptosis [41–43]. The cleavage eliminates 42 amino acids at the C-terminal of the protein and it is blocked by caspase tetrapeptide inhibitors [41,42]. The deletion does not affect the growth-suppressive function of the protein but impairs its interaction with Mdm2 [43]. Mdm2, similar to pRb, is a caspase substrate [44]. Mdm2 may function with pRb in inhibiting apoptosis. Its function may be simply to associate with pRB and to maintain pRb protein stability or, in a more complex model, it may cooperate with pRb in directly blocking E2F and p53 apoptotic pathways.
The role of pRb in apoptosis has yet to be clarified. Studies conducted on E2F-1 may help clarify the anti-apoptotic function of pRb. Transgenic mice null for E2F-1 show inappropriate cell proliferation in the thymus and lymph nodes. Considering E2F-1's role in stimulating cell cycle progression, the expected effect of inactivating E2F-1 is hypoproliferation. The explanation for this surprising result may be that E2F-1, in addition to promoting cell proliferation, controls apoptotic pathways. Indeed, E2F-1 mutants unable to interact with pRb show an even greater ability to induce apoptosis. This suggests that pRb has the ability to regulate E2F-1 function during the cell cycle, but also inhibits the apoptotic pathway that E2F-1 stimulates. In fact, E2F-1 cannot induce apoptosis when pRb is coexpressed [38]. The current data indicate that pRb inhibits apoptosis by avoiding improper activation of E2F-1.
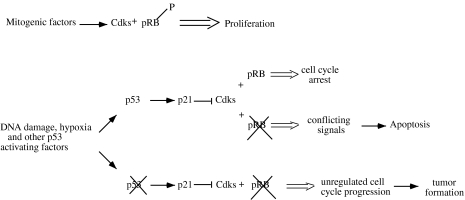
The last argument remaining to be discussed with respect to pRb is the interplay of this molecule with p53 in regulating cell cycle arrest and apoptosis (Figure 4). As discussed above, in some circumstances, p53 can induce cell arrest, but in others, it triggers apoptosis. pRb is among the different cellular parameters that leads to one pathway or to the other. To better clarify the cooperation between pRb and p53, mice heterozygous for pRb and null for p53 were generated [45]. In this case, in addition to the tumors that each individual mutation gene causes, other tumors developed, such as pinealoblastoma and islet tumors. This indicates that p53 normally represses tumorigenesis when pRb is absent. It does so by inducing apoptosis, as studies conducted with viral proteins have demonstrated. During an infection, the virus must accomplish two goals: 1) to make the cell proliferate abnormally and 2) to shut off apoptotic mechanisms induced by uncontrolled proliferation. Adenovirus SV 40 and HPV solve these problems by simultaneously disrupting p53 and pRb. E1A, SV40 Tag and HPV E7 bind and inactivate pRb function. SV 40, HPV E6 and adenoviral E1B inactivate p53. Studies performed on developing mouse lens have been useful for understanding the effect of viral proteins on pRb and p53 functions. Expressing E7 in the developing lens causes apoptosis, because the deadly effect of p53 is not counteracted by pRb. However, if p53 is eliminated by coexpressing E6, apoptosis is inhibited, resulting in tumor formation [46]. A similar effect is caused by SV40 Tag, which inactivates both p53 and pRb [47]. SV40 Tag also has been used to examine the effect of pRb and p53 deficiency in choroid plexus and thymocytes. In these experiments, mutant versions of SV40 Tag, no longer able to bind pRb, cause apoptosis while the wild-type version is able to induce tumorigenesis [48].
Figure 4.
p53 and pRb cooperation. In normal cells, mitogenic factors and antiproliferative signals regulate cell cycle proliferation and influence the phosphorylation state of pRb. In damaged cells, p53 is activated and causes cell cycle arrest by inducing p21 and by inhibiting pRb phosphorylation by Cdks. If pRb is mutated, the cell cycle is not arrested and the conflict between the p53 signal to stop cell growth and the Cdk signal to proliferate leads to apoptosis. If p53 also is mutated, the cell does not receive a signal to arrest the cell cycle and uncontrolled proliferation leads to tumor formation.
Little data are currently available regarding the role of the other two Rb family members, pRb2/p130 and p107, in apoptosis. It is probable that these two proteins have roles in apoptosis because they share other characteristics with the better known pRb. Data supporting a p107 anti-apoptotic role come from a study conducted in mice. The liver and the central nervous system of pRb-/-, p107-/- embryos show more extensive apoptosis than pRb-/- embryos. In addition, the double knock-out animals die 2 days prior to the single mutant [49].
Cdks and Cyclins
Accumulating data show that Cdk/cyclin complexes influence the decision of whether a cell lives or dies. However, it is unclear exactly how Cdks participate in this decision and if their involvement is direct or indirect.
In favor of direct participation of Cdks in apoptosis, there are several studies that illustrate that cyclin A/Cdk complexes positively influence cellular apoptosis. For example, during the effector phase of cell death, cyclin A/Cdk complexes are activated in the nucleus, and their activation depends upon caspases [50]. Another study performed in lymphocytes shows that cyclin A/Cdk2 and cyclin A/Cdc2 activities are necessary during HIV-1 Tat-induced apoptosis [51]. Moreover, dominant negative mutants of Cdc2, Cdk2 and Cdk3 block TNF-induced apoptosis in HeLa cells and can be reverted by coexpressing cyclin A [52]. Additional data for an active role of Cdks in apoptosis come from studies performed on Cdk2 activation in apoptotic thymocytes. In these cells, Cdk2 activation is required for cell death and only occurs subsequent to Bax expression or Bcl-2 repression [53]. These studies support that Cdk activity is modulated by known apoptotic factors working in the induction or execution phases of apoptosis.
However, during apoptosis, Cdk activation can be only a consequence and not the cause. Indeed, the induction of apoptosis does not directly depend on Cdk activity in every case, but on cellular context such as the generation of conflicting signals for proliferation and for cell cycle arrest. In these situations, Cdks may function as signals to sensitize cells to apoptosis. For example, a recent paper by Chen et al. [54] demonstrates that a short peptide that impairs the interaction between Cdk2 and its cyclin partner inhibits the activity of Cdk2 and consequently induces apoptosis. However, the inactivation of Cdk complexes also causes a deregulation of E2F activity in transformed cells that are programmed to proliferate. This results in conflicting signals of growth and arrest, which is resolved with the death of the cell.
Cdc2 activity is necessary for cell death in many systems [55]. For example, a dramatic increase in Cdc2 kinase characterizes Granzyme B-induced apoptosis in lymphoma cells and the inactivation of this kinase blocks also the proapoptotic action of this protease. Cdc2 activity also plays an important role in staurosporine- and in HIV-1 Tat-induced apoptosis. Induction of cyclin B/Cdc2 activity occurs also during apoptosis in taxol-induced apoptosis [56]. However, abolishing Cdc2 function does not impair apoptosis induction in several apoptotic systems such as etoposide, dexamethasone, UV irradiation, serum starvation or Fas-dependent apoptosis [55]. Cdc2 cannot be considered a universal regulator of apoptosis or at least an essential component for all apoptosis. In many cases, in particular when apoptosis is induced in G2 or M-phase, the activation of cdc2 activity could just lead to mitotic catastrophe.
PITSLRE is a Cdk whose role in the cell cycle remains to be established. However, its apoptotic involvement is well demonstrated. Overexpressing PITSLRE induces apoptosis [57] and PITSLRE kinase activity increases during Fas-dependent apoptosis and TNF-mediated apoptosis [58]. Moreover, PITSLRE is cleaved by caspases. The cleavage removes the PITSLRE N-terminus, thereby activating the kinase [57]. During Fas-dependent apoptosis, PITSLRE is phosphorylated and the phosphorylation enhances the N-terminal cleavage [59]. No further information is now available regarding the cellular function of PITSLRE. No cyclins were found to be associated with this kinase. This fact could explain that PITSLRE is a Cdk-related protein involved in events far from the regulation of the cell cycle. It still remains to explain the role of this protein in apoptosis and to find its location in the apoptotic pathway.
Among the cyclins, the involvement of cyclins D in apoptosis has been studied extensively. Cyclin D1 actively promotes apoptosis. Cyclin D1 overexpression causes apoptosis in neuronal differentiated N1E-115 cells and fibroblasts [55]. Apoptosis induced by serum starvation or hydroxyurea is characterized by increased expression of cyclin D1 [60]. Among the cyclin D family of proteins, cyclin D3 also is involved in apoptosis. Janicke et al. [61] found that cyclin D3 promotes TNF-induced apoptosis. As for Cdks, upregulation of cyclin D expression results in pushing cells to cycle. Therefore, the apoptotic effect of cyclins may depend on the concomitant signals of arrest, such as differentiation or serum starvation, and proliferation such as cyclin synthesis. Their own synthesis may function to sensitize cells to undergo apoptosis. Therefore, in these situations, the cyclins may not be principle players in these apoptotic pathways but part of an unbalanced mechanism.
Cdk Regulators
Observations supporting the apoptotic role of Cdks come from studies performed on some Cdk regulators.
Among the CKIs, p21 is one of the most highly studied for its involvement in apoptosis. According to several studies, p21 is able generally to rescue cells from programmed cell death during phenomena such as myocytes differentiation [62] and NGF-induced neuronal differentiation [63]. p21 is able to repress DNA-damage-induced such as X-ray-irradiation- or adriamycin-dependent apoptosis [64]. The involvement of Cdk2 in this process is demonstrated by the fact that a p21 mutant unable to bind Cdk2 cannot repress apoptosis.
In contrast to p21, p27's role in apoptosis is less clear. It has been demonstrated that overexpression of p27 can cause apoptotic death in several human cell lines [65,66]. However, p27 may also have anti-apoptotic effects. Hiromura et al. [67] showed that p27-/- mesangial cells and fibroblasts are susceptible to serum starvation apoptosis. Restoring p27 presence rescues the cells from cell death. In this case, apoptosis is caused by enhanced cyclin A/Cdk2 activity because of a lack of p27.
Recent data demonstrate that p21 and p27 are targets for caspases, further linking these molecules to apoptotic pathways. During serum starvation-induced apoptosis in endothelial cells, the C-terminus of p21 and p27 is eliminated and their association with Cdk2 is reduced. Dominant negative Cdk2 and a mutant p21 that is no longer cleaved by caspases suppress apoptosis [68]. p21 is a caspase target also during γ-irradiation-induced apoptosis [69] and in TNF-induced apoptosis [70]. p27 is cleaved by caspase-3 to generate a p23 form during G1 arrest in mouse hybridoma and in human myeloma cells. The p23 form is localized to the cytoplasm [71].
p57, another CKI, appears to be involved in apoptosis; its ablation in transgenic heterozygous mice causes apoptosis and delayed differentiation [72].
With regard to p16, it has been observed that adenoviral overexpression of p16 causes G1 arrest and prevents drug-induced apoptosis in glioma cells [73]. In contrast with this observation, it was found that p16 and p53 coexpression induces cell death in several cell lines [74]. It is likely that the cellular genetic environment plays a critical role in this activity. p16 overexpression is able to induce apoptosis only in RB-negative cells such as HeLa cells. In pRb-positive cells, p16 induces cell cycle arrest; the two inhibitors, p18 and p27, have similar effects [75].
The above studies suggest that Cdk inhibitors are indirectly involved in apoptosis at best. Their hyperactivation or mis-regulation leads to improper Cdk activity, which could impart conflicting signals for cell division or arrest. No linkage has been found between these molecules and apoptotic regulators that lead us to suppose a direct influence on the induction or execution of apoptosis.
Conclusion
The regulation of cell proliferation and cell death can share common molecules in multicellular organisms. Indeed, several genes involved in cell cycle regulation are also involved in apoptosis.
Myc's role in cell proliferation and apoptosis has been studied extensively. Evan proposed that Myc directly activates genes involved in proliferation and apoptosis. Mitogens stimulate Myc's proliferation pathway, while anti-apoptotic factors, such as Bcl-2, shut down Myc's apoptotic pathway. When other signals inactivate these anti-apoptotic factors, Myc induces cell death.
Half of all human cancers is characterized by a mutated p53. p53 regulates a cell's fate by inducing either death or cell cycle arrest. The specific pathway chosen depends upon a variety of factors such as the extent of DNA damage and on the genetic background of the cell. In addition, the presence of functional p21 and the cross-talk with pRb are critical determinants in p53's role.
pRb not only is a growth suppressor but also has a protective function against apoptosis. Additional support for a protective role for pRb against apoptosis comes from the observation that pRb is the target of caspases. Current data indicate that pRb inhibits apoptosis by properly regulating the activation of E2F.
Cdks also have been implicated in apoptosis. Several studies demonstrate the active participation of cyclin A/Cdk complexes, Cdc2, cyclin D1, and PISTALRE in apoptosis. Further observations supporting the apoptotic role of Cdks come from studies performed on Cdk regulators. While Cdks do not appear to belong to the apoptotic machinery, they do activate several pathways that converge toward apoptosis. They may function to sensitize cells that are receiving inappropriate growth signals to undergo apoptosis. Cdk involvement in apoptosis is cell-type-specific, however, and also depends on environmental conditions and differentiation states.
In conclusion, the interplay of cell cycle regulators and apoptotic factors influences the destiny of a cell in multi-cellular organism in order to regulate tissue homeostasis. It becomes a matter of life and death and the survival of the organism lies in the balance. Deregulation of cell proliferation and/or programmed cell death affects the physiology of an organism. Potentially, this could lead to pathological conditions, including cancer and AIDS [2,76]. Understanding the linkage between cell cycle and apoptosis is of primary importance in the search for new therapeutic strategies to combat these diseases.
Acknowledgements
The authors thank Marco Tafani for critically reading the manuscript and for his comments. M. K. was supported by NCI training grant 5 T32 CA 09137.
Footnotes
This work was supported by grants from the National Institutes of Health awarded to A. G. and in part by The Sbarro Institute.
References
- 1.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Wyllie AH. British Medical Bulletin 53. Dorchester: The Dorset Press; 1997. Apoptosis. [DOI] [PubMed] [Google Scholar]
- 3.Golstein P. Controlling cell death. Science. 1997;275:1081–1082. doi: 10.1126/science.275.5303.1081. [DOI] [PubMed] [Google Scholar]
- 4.King KL, Cidlowsky JA. Cell cycle and apoptosis: common pathways to life and death. J Cell Biochem. 1995;58:175–180. doi: 10.1002/jcb.240580206. [DOI] [PubMed] [Google Scholar]
- 5.Evan GI, Brown L, Whyte M, Harrington E. Apoptosis and the cell cycle. Curr Opin Cell Biol. 1995;7:825–834. doi: 10.1016/0955-0674(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 6.MacLachlan TK, Sang N, Giordano A. Cyclins, cyclin-dependent kinases and Cdk inhibitors: implications in cell cycle control and cancer. Crit Rev Eukaryotic Gene Expression. 1995;5:127–156. doi: 10.1615/critreveukargeneexpr.v5.i2.20. [DOI] [PubMed] [Google Scholar]
- 7.Harper JW, Elledge SJ. Cdk inhibitors in development and cancer. Curr Opin Genet Dev. 1996;6:56–84. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 8.Riley DJ, Lee EY-HP, Lee WH. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 9.Sidle A, Palaty C, Dirks P, Wiggan O, Kiess M, Gill RM, Wong AK, Hamel PA. Activity of the retinoblastoma family proteins, pRb, p107 and p130, during cellular proliferation and differentiation. Crit Rev Biochem Mol Biol. 1996;31:237–271. doi: 10.3109/10409239609106585. [DOI] [PubMed] [Google Scholar]
- 10.Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 11.Lam EW, La Thangue NB. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 12.Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heikkila R, Schwab G, Wichstrom E, Loke SL, Pluznik DH, Watt R, Nackars LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S-phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- 14.Graeber TG, Osmanian C, Jacks T, Houseman DE, Koch CJ, Lowe SW, Giaccia AJ. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumors. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 15.Wagner AJ, Small MB, Hay N. Myc-mediated apoptosis is blocked by ectopic expression of Bcl-2. Mol Cell Biol. 1993;13:2432–2440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galaktionov K, Chen X, Beach D. Cdc25 cell cycle phosphatases as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 17.Levine A. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 18.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 synthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Schneider E, Montenarh M, Wagner P. Regulation of CAK kinase activity by p53. Oncogene. 1998;17:2733–2741. doi: 10.1038/sj.onc.1202504. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innocente SA, Abrahamson JLA, Cogswell JP, Lee JM. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 Sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 24.Di Leonardo A, Khan SH, Linke SP, Greco V, Seidita G, Wahl GM. DNA re-replication in presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 25.Brown CR, Doxsey SJ, White, Welch WJ. Both viral (adenovirus E1B) and cellular (hsp70, p53) components interact with centrosomes. J Cell Physiol. 1994;160:47–60. doi: 10.1002/jcp.1041600107. [DOI] [PubMed] [Google Scholar]
- 26.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 27.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by transactivation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita T, Harigai M, Hanada M, Reed JC. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
- 29.Wang XW, Vermeulen W, Coursen JD, Gibson M, Lupold SE, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers JH, Harris CC. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 30.Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, Roth JA, Deisseroth AB, Zhang WW, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu GS, Burns TF, McDonald ER, III, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, El-Deiry WS. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 32.Polyak K, Waldman T, He TC, Kinzlre KW, Vogestein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 33.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 34.Chiou SK, Rao L, White E. Bcl-2 blocks p53-dependent apoptosis. Mol Cell Biol. 1994;14:2556–2563. doi: 10.1128/mcb.14.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrel M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 36.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 37.Jacks T, Fazeli A, Schmitt E, Branson R, Goodell M, Weinberg R. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 38.Fan G, Ma X, Kren BT, Steer CJ. The retinoblastoma gene product inhibits TGF-β1 induced apoptosis in primary rat hepatocytes and in human HuH-7 hepatoma cells. Oncogene. 1996;12:1909–1919. [PubMed] [Google Scholar]
- 39.Berry DE, Lu Y, Schmidt B, Fallon PG, O'Connell, Hu SX, Xu HJ, Blanck G. Retinoblastoma protein inhibits IFNγ-induced apoptosis. Oncogene. 1996;12:1809–1819. [PubMed] [Google Scholar]
- 40.Haas-Kogan DA, Kogan SC, Levi D, Dazin P, T'ang A, Fung YK, Israel MA. Inhibition of apoptosis by the retinoblastoma gene product. EMBO J. 1995;14:461–472. doi: 10.1002/j.1460-2075.1995.tb07022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An B, Dou QP. Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as a candidate. Cancer Res. 1996;56:438–442. [PubMed] [Google Scholar]
- 42.Dou QP, An B, Antoku K, Johnson DE. Fas stimulation induces RB dephosphorylation and proteolysis that is blocked by inhibitors of the ICE protease family. J Cell Biochem. 1997;64:586–594. [PubMed] [Google Scholar]
- 43.Janicke RU, Walker PA, Lin XY, Porter AG. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- 44.Erhardt P, Tomaselli KJ, Cooper GM. Identification of the MDM2 oncoprotein as a substrate for CPP32-like apoptotic proteases. J Biol Chem. 1997;272:15049–15052. doi: 10.1074/jbc.272.24.15049. [DOI] [PubMed] [Google Scholar]
- 45.Williams BO, Remington L, Albert DM, Mukai S, Branson RT, Jacks T. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 46.Pan H, Griep E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implication for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 47.Fromm L, Shawlot W, Gunning K, Butel JS, Overbeek PA. The retinoblastoma binding region of simian virus 40 large T antigen alters cell cycle regulation in lenses of transgenic mice. Mol Cell Biol. 1994;14:6743–6754. doi: 10.1128/mcb.14.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy SA, Symonds HS, Van Dyke T. Regulation of apoptosis in transgenic mice by simian virus 40 T antigen-mediated inactivation of p53. Proc Natl Acad Sci USA. 1994;92:3979–3983. doi: 10.1073/pnas.91.9.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MH, Williams BO, Mulligan G, Mukai S, Branson RT, Kyson N, Harlow E, Jacks T. Target disruption of p107: functional overlap between p107 and pRb. Genes Dev. 1996;16:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 50.Harvey KJ, Blomquist JF, Ucker DS. Commitment and effector phases of physiological cell death pathway elucidated with respect to Bcl-2, caspase and cyclin-dependent kinase activities. Mol Cell Biol. 1998;18:2912–2922. doi: 10.1128/mcb.18.5.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li CJ, Friedman DJ, Wang C, Metelev V, Pardee AB. Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 52.Meikrantz W, Gisselbrecht S, Tam S, Schlegel R. Activation of cyclin A-dependent protein kinases during apoptosis. Proc Natl Acad Sci USA. 1994;91:3754–3758. doi: 10.1073/pnas.91.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gil-Gomez G, Berns A, Brady HJM. A link between cell cycle and cell death: Bax and Bcl-2 modulate Cdk2 activation during thymocyte apoptosis. EMBO J. 1998;17:7209–7218. doi: 10.1093/emboj/17.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen YN, Sharma SK, Ramsey TM, Jiang L, Martin MS, Baker K, Adams PD, Bair KW, Kaelin WG. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonist. Proc Natl Acad Sci USA. 1999;13:4221–4223. doi: 10.1073/pnas.96.8.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasten MM, Giordano A. pRb and the cdks in apoptosis and the cell cycle. Cell Death Differ. 1998;5 doi: 10.1038/sj.cdd.4400323. 1342-140. [DOI] [PubMed] [Google Scholar]
- 56.Ling YH, Consoli U, Tornos C, Andreef M, Perez-Soler R. Accumulation of cyclin B, activation of cyclin B1-dependent kinase and induction of programmed cell death in human epidermoid carcinoma KB cells treated with taxol. Int J Cancer. 1998;75:925–932. doi: 10.1002/(sici)1097-0215(19980316)75:6<925::aid-ijc16>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 57.Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Mol Cell Biol. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahiti JM, Gururajan R, Vandenabeele P, Fiers W. Cleavage of PITSLRE kinases by ICE/CASP1 and CPP32/CASP3 during apoptosis induced by tumor necrosis factor. J Biol Chem. 1997;272:11694–11697. doi: 10.1074/jbc.272.18.11694. [DOI] [PubMed] [Google Scholar]
- 59.Tang D, Gururajan R, Kidd VJ. Phosphorylation of PITSLRE p110 isoforms accompanies their processing by caspases during Fas-meditated cell death. J Biol Chem. 1998;273:16601–16607. doi: 10.1074/jbc.273.26.16601. [DOI] [PubMed] [Google Scholar]
- 60.Han EK, Begemann M, Sgambato A, Soh JW, Doki Y, Xing WQ, Liu W, Weinsten IB. Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27Kip1, inhibits growth and enhanced apoptosis. Cell Growth Differ. 1996;7:699–710. [PubMed] [Google Scholar]
- 61.Janicke RU, Lin XY, Porter AG. Cyclin D3 sensitizes tumor cells to TNF-induced, c-Myc-dependent apoptosis. Mol Cell Biol. 1996;16:5245–5253. doi: 10.1128/mcb.16.10.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Walsh K. Resistance to apoptosis conferred by CDK inhibitors during myocyte differentiation. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poluha W, Poluha D, Chang B, Crosbie NE, Schonhoff CM, Kilpatrick DL, Ross AH. The cyclin-dependent kinase inhibitor p21WAF1 is required for survival of differentiating neuroblastoma cells. Mol Cell Biol. 1996;16:1335–1341. doi: 10.1128/mcb.16.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Y, Yamagishi N, Yagi T, Takebe H. Mutated p21(WAF1/CIP1/SDI1) lacking CDK-inhibitory activity fails to prevent apoptosis in human colorectal carcinoma cells. Oncogene. 1998;16:705–712. doi: 10.1038/sj.onc.1201585. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Gorospe M, Huang Y, Holbrook NJ. p27Kip1 overexpression causes apoptotic death of mammalian cells. Oncogene. 1997;15:2991–2997. doi: 10.1038/sj.onc.1201450. [DOI] [PubMed] [Google Scholar]
- 66.Scott DW, Donjerkovic D, Maddox B, Ezhevsky S, Grdina T. Role of c-myc and p27 in anti-IgM-induced B-lymphoma apoptosis. Curr Topics Microbiol Immunol. 1997;224:102–112. doi: 10.1007/978-3-642-60801-8_10. [DOI] [PubMed] [Google Scholar]
- 67.Hiromura K, Pippin JW, Fero ML, Roberts JM, Shankland SJ. Modulation of apoptosis by the cyclin-dependent kinase inhibitor p27(Kip1) J Clin Invest. 1999;103:597–604. doi: 10.1172/JCI5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levkau B, Koyama H, Raines EW, Clurman BE, Herren B, Orth K, Roberts JM, Russel R. Cleavage of p21CIP1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 69.Gervais JLM, Seth P, Zhang H. Cleavage of CDK inhibitor p21Cip/Waf1 by caspases is an early event during DNA damage-induced apoptosis. J Biol Chem. 1998;273:19207–19212. doi: 10.1074/jbc.273.30.19207. [DOI] [PubMed] [Google Scholar]
- 70.Donato N, Perez M. Tumor necrosis factor-induced apoptosis stimulates p53 accumulation and p21WAF1 proteolysis in ME-180 cells. J Biol Chem. 1998;273:5067–5072. doi: 10.1074/jbc.273.9.5067. [DOI] [PubMed] [Google Scholar]
- 71.Loubat A, Rochet N, Turchi L, Rezzonico R, Far DF, Auberger P, Rossi B, Ponzio G. Evidence for a p23-caspase-cleaved form of p27(KIP1) Oncogene. 1999;18:3324–3333. doi: 10.1038/sj.onc.1202668. [DOI] [PubMed] [Google Scholar]
- 72.Yan Y, Friesen J, Lee MH, Massague J, Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 73.Hama S, Heike Y, Naruse I, Takahashi M, Yoshioka H, Arita K, Kurisu K, Goldman CK, Curie DT, Saijo N. Adenovirus-mediated p16 gene transfer prevents drug-induced cell death through G1 arrest in human glioma cells. Int J Cancer. 1998;77:47–54. doi: 10.1002/(sici)1097-0215(19980703)77:1<47::aid-ijc9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 74.Sandig V, Brand K, Herwig S, Lukas J, Bartek J, Strauss M. Adenoviral-transferred p16INK4/CDKN2 and p53 genes cooperate to induce apoptotic tumor cell death. Nat Med. 1997;3:313–319. doi: 10.1038/nm0397-313. [DOI] [PubMed] [Google Scholar]
- 75.Schreiber M, Muller WJ, Singh G, Graham FL. Comparison of the effectiveness of adenovirus vectors expressing cyclin kinase inhibitors p16INK4A, p18INK4C, p19INK4D, p21 (WAF1/CIP1) and p27KIP1 in inducing cell cycle arrest, apoptosis and inhibition of tumorigenicity. Oncogene. 1999;18:1663–1676. doi: 10.1038/sj.onc.1202466. [DOI] [PubMed] [Google Scholar]
- 76.Pucci B, Giordano A. Cell cycle and cancer. Clin Ter. 1999;150:135–141. [PubMed] [Google Scholar]