Abstract
Retrovirus budding is greatly stimulated by the presence of Gag sequences known as late or L domains. The L domain of human immunodeficiency virus type 1 (HIV-1) maps to a highly conserved Pro-Thr-Ala-Pro (PTAP) sequence in the p6 domain of Gag. We and others recently observed that the p6 PTAP motif interacts with the cellular endosomal sorting protein TSG101. Consistent with a role for TSG101 in virus release, we demonstrated that overexpressing the N-terminal, Gag-binding domain of TSG101 (TSG-5′) suppresses HIV-1 budding by blocking L domain function. To elucidate the role of TSG101 in HIV-1 budding, we evaluated the significance of the binding between Gag and TSG-5′ on the inhibition of HIV-1 release. We observed that a mutation in TSG-5′ that disrupts the Gag/TSG101 interaction suppresses the ability of TSG-5′ to inhibit HIV-1 release. We also determined the effect of overexpressing a panel of truncated TSG101 derivatives and full-length TSG101 (TSG-F) on virus budding. Overexpressing TSG-F inhibits HIV-1 budding; however, the effect of TSG-F on virus release does not require Gag binding. Furthermore, overexpression of the C-terminal portion of TSG101 (TSG-3′) potently inhibits budding of not only HIV-1 but also murine leukemia virus. Confocal microscopy data indicate that TSG-F and TSG-3′ overexpression induces an aberrant endosome phenotype; this defect is dependent upon the C-terminal, Vps-28-binding domain of TSG101. We propose that TSG-5′ suppresses HIV-1 release by binding PTAP and blocking HIV-1 L domain function, whereas overexpressing TSG-F or TSG-3′ globally inhibits virus release by disrupting the cellular endosomal sorting machinery. These results highlight the importance of TSG101 and the endosomal sorting pathway in virus budding and suggest that inhibitors can be developed that, like TSG-5′, target HIV-1 without disrupting endosomal sorting.
The assembly and release of retroviral particles is driven by the expression of viral Gag precursor proteins (15, 54). Discrete functional domains have been defined within Gag proteins that mediate essential steps in particle formation. Membrane binding (M) domains direct the association of Gag with the lipid bilayer, interaction (I) domains promote Gag-Gag multimerization, and late (L) domains catalyze the pinching off of virus particles from the plasma membrane. In the case of HIV-1, the L domain is encoded by a PTAP motif in the C-terminal, p6 domain of the Gag precursor protein Pr55Gag (21, 25).
L domains, found at a variety of positions in the Gag proteins of a number of retroviruses and in the matrix proteins of the rhabdoviruses and filoviruses, appear to promote virus budding by interacting with cellular host factors (for a review, see reference 17). Three types of sequence motifs have been demonstrated to possess L domain activity: PTAP, Pro-Pro-X-Tyr (PPXY), and Tyr-Pro-Asp-Leu (YPDL). As mentioned above, a PTAP motif in p6 confers HIV L domain activity; the same motif in Ebola VP40 has also been reported to contribute to particle release (36). PPXY motifs, which appear to be the most common sequence associated with L domain function, stimulate budding of Rous sarcoma virus (58, 59), Mason-Pfizer monkey virus (M-PMV) (60), murine leukemia virus (MLV) (63, 64), human T-cell leukemia virus type 1 (32), bovine leukemia virus (57), the rhabdoviruses (10, 24, 26), and the filoviruses (23). Equine infectious anemia virus (EIAV) L domain activity is provided by a YPDL motif (47). A number of retroviruses, and the Ebola filovirus, contain adjacent or overlapping PTAP and PPXY sequences. The presence of both PTAP and PPXY motifs may provide functional redundancy or perhaps enable sequential association of L domains with multiple host factors.
Increasing evidence suggests that L domains interact with cellular ubiquitination and endosomal sorting machinery (17, 56). (i) The L domain-containing proteins of several retroviruses, including HIV-1, HIV-2, MLV, and EIAV are ubiquitinated (41-43). (ii) Proteasome inhibitors disrupt retrovirus and rhabdovirus budding (22, 44, 49, 52). (iii) The L domains of Rous sarcoma virus (29), Mason-Pfizer monkey virus (61), the rhabdoviruses (22), and Ebola virus (23) appear to functionally interact with proteins related to Nedd4, a ubiquitin (Ub) ligase that regulates the cell surface expression of the epithelial sodium channel (51). (iv) The EIAV L domain binds and colocalizes with the AP-50 subunit of the AP-2 complex, which is involved in endocytosis (39). (v) Finally, the host protein TSG101 was identified in Saccharomyces cerevisiae yeast two-hybrid screens as a p6-interacting protein (20, 37, 55). This interaction maps to the N terminus of TSG101, a region that bears sequence and structural similarity with Ub conjugating (E2) enzymes (30, 36, 45, 46).
A functional relevance of the Gag/TSG101 interaction in virus release is supported by the observation of Garrus et al. (20) that depletion of TSG101 using a small interfering RNA approach blocked HIV-1 budding, and our demonstration that overexpression of the N-terminal, E2-like domain of TSG101 (referred to as TSG-5′) impaired particle release (11). Both TSG101 underexpression (20) and TSG-5′ overexpression (11) produced a pinching-off defect highly reminiscent by electron microscopy (EM) of defects observed with p6 L domain mutants (12, 21, 25).
TSG101, and its yeast ortholog Vps23, are members of the so-called class E family of vacuolar protein sorting (Vps) proteins (4, 48). In a wide range of eukaryotic cells, these proteins play an essential role in forming, and sorting cargo into, the multivesicular body (MVB)/late endosome (33, 38). The endosomal sorting pathway controls a variety of cellular processes, including the regulation of cell surface expression of receptors involved in signal transduction, the delivery of lysosomal hydrolases to their appropriate destination (the lysosome in mammalian cells and the vacuole in yeast), and the release of material into the extracellular environment in exosomal vesicles (for reviews, see references 33 and 50). In both yeast and mammalian cells, TSG101/Vps23 associates with a ∼350-kDa complex termed ESCRT-I (for endosomal sorting complex required for transport) (27). tsg101/vps23-deficient cells display a variety of endosomal sorting defects (7, 8, 34). ESCRT-I is the first of three recognized multiprotein complexes (the others being ESCRT-II and -III) that reportedly play a sequential role in the sorting of ubiquitinated cargo proteins into the lumen of the MVB (2, 3, 27). In addition to Vps23, ESCRT-I in yeast contains Vps28 and Vps37. In mammalian cells, a Vps28 ortholog is also expressed (9), whereas a mammalian equivalent of yeast Vps37 has not yet been identified. The activity of the sorting complexes ESCRT-I, -II, and -III also requires the AAA-type (for ATPase associated with a variety of cellular activities) ATPase Vps4 (6, 8, 62); this protein appears to catalyze the dissociation of ESCRT-III at the endosomal membrane (3). vps4 deficiency, or overexpression of a transdominant form of Vps4, induces the formation of aberrant, swollen endosomes that accumulate endosomal cargoes (5, 6, 8, 14, 62). Overexpression of a transdominant form of Vps4 was observed to inhibit both HIV-1 and MLV budding (20).
In this study, we sought to gain mechanistic insights into the ability of TSG-5′ to inhibit HIV-1 budding and to understand in greater detail the role of the endosomal sorting pathway, and TSG101 in particular, in HIV-1 release. To examine the requirement for a direct binding between Gag and TSG-5′ in the inhibition of virus budding, we tested whether a mutant form of TSG-5′ deficient for Gag binding could still interfere with virus release. We also determined the effect of overexpressing several additional C-terminal TSG101 truncation mutants, as well as TSG-F and TSG-3′, on particle release. We demonstrate that both truncated and full-length forms of TSG101 inhibit HIV-1 budding, but they do so by distinct mechanisms. Forms of TSG101 that lack the C-terminal, Vps28-binding domain specifically inhibit HIV-1 budding by interacting with the p6 L domain. In contrast, TSG-F and TSG-3′ overexpression interferes with HIV-1 budding by disrupting the cellular endosomal sorting machinery. The latter effect is not specific for HIV-1, as TSG-3′ also potently inhibits MLV particle production.
MATERIALS AND METHODS
Plasmids, DNA cloning, and tsg101 mutagenesis.
The TSG-5′ expression vector pcGNM2/TSG-5′ (12, 53) was kindly provided by Z. Sun (Stanford University). The full-length TSG101 expression vector (pcGNM2/TSG-F) was constructed by transferring the TSG101 coding region from plasmid pGST2TK/TSG101 (also provided by Z. Sun) into pcGNM2/TSG-5′. C-terminal TSG101 truncation mutants (Fig. 1) were constructed by introducing premature termination codons by PCR using a forward vector primer (TATGACGTGCCTGACTATGCCAGC) and reverse primers (TACGGATCCTCACCCCGTTGCCTGGTA and TACGGATCCTCAGGCTCGGATGGTGTC) to truncate TSG101 at codons 320 and 229, respectively. pcGNM2/TSG-F was used as the PCR template. Fragments were amplified and cloned into the pcGNM2 vector to obtain pcGNM2/TSG-320 and pcGNM2/TSG-229. The TYN(67-69)A mutation (hereafter referred to as TYN−) was introduced into pcGNM2/TSG-F, pcGNM2/TSG-320, pcGNM2/TSG-229, and pcGNM2/TSG-5′ using a two-step PCR strategy. In the first step, two pairs of primers were used; one with the forward vector primer mentioned above and a reverse mutagenic primer (TATTGGAATAGCATTACCTCTATAAGG). In the second step, a forward mutagenic primer (TATAGAGGTAATGCTATTCCAATATGC) and a reverse vector primer (CAACACCCTGAAAACTTTGCCC) were used. The mutagenized tsg101 fragments were cloned into the pcGNM2 vector to obtain TYN− mutant forms of pcGNM2/TSG-F, pcGNM2/TSG-320, pcGNM2/TSG-229, and pcGNM2/TSG-5′. All TSG101-derived expression vectors used in this study encode proteins with N-terminal influenza hemagglutinin (HA) epitope tags. Additional details of vector construction will be made available upon request. pcGNM2/TSG-3′, a kind gift of Z. Sun, encodes the 3′ portion of the TSG101 (Fig. 1). Plasmid eGFP-hVPS4(EQ), which expresses a trans-dominant mutant of Vps4, (8), was kindly provided by P. Woodman (University of Manchester, United Kingdom). For MLV Gag expression, we used plasmid pSV-ψ−MLV-env− (31), obtained through the NIH AIDS Research and Reference Reagent Program from N. Landau (Salk Institute). AIDS patient immunoglobulin (Ig) and HIV neutralizing serum were also obtained from the NIH AIDS Research and Reference Reagent Program.
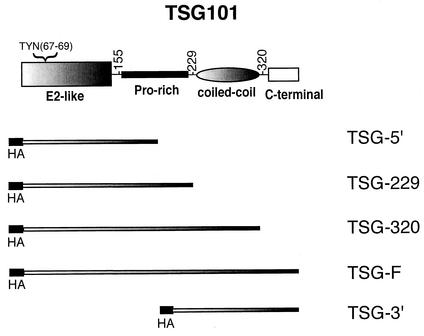
FIG. 1.
Organization of TSG101 and deletion mutant derivatives. At the top is depicted the domain organization of WT TSG101. Numbers denote amino acid positions. The location of the TYN motif eliminated by the TYN− mutation is indicated. The regions encoded by the truncated (TSG-5′, -229, -320, and −3′) and the full-length (TSG-F) expression vectors are shown. HA denotes the N-terminal HA epitope tag.
Cell culture, transfections, radioimmunoprecipitation, Western blotting, and EM analysis.
HeLa cells were maintained in culture and were transfected with the calcium phosphate method as described (18). Transfections were performed in six-well dishes plated at 4 × 105 cells/well. Methods used for metabolic labeling of transfected cells, preparation of cell and viral lysates, immunoprecipitation analysis, and Western blotting have been reported previously (18, 28). HIV-1 proteins were immunoprecipitated with purified Ig derived from AIDS patients (anti-HIV Ig); Western blotting was performed with HIV neutralizing serum (both obtained from the NIH AIDS Research and Reference Reagent Program). Anti-HA was obtained from Sigma (St. Louis, Mo.). MLV Gag was immunoprecipitated with goat anti-Raucher MLV p30 (ViroMed Biosafety Laboratories, Camden, N.J.). Electron microscopy (EM) was performed as previously described (18).
Confocal microscopy.
Confocal microscopy was performed essentially as described previously (40). Briefly, HeLa cells were cultured in chamber slides (Nunc) and transfected by the calcium phosphate precipitation method without glycerol shock. Twenty-four hours posttransfection, cells were washed once with Dulbecco modified Eagle medium supplemented with 5% fetal bovine serum. After another 24 h, cells were rinsed once with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde in 100 mM sodium phosphate buffer (pH 7.2) for 20 min at room temperature. The cells were then permeabilized with methanol at −20°C for 4 min. Subsequently, cells were incubated with 0.1 M glycine in PBS for 10 min and blocked with 3% bovine serum albumin in PBS for 30 min and incubated with primary antibodies, mouse anti-epidermal growth factor receptor (anti-EGF-R) and rabbit anti-HA monoclonal antibody, at 1:100 dilution in 3% bovine serum albumin-PBS for 1 h. Cells were washed with PBS three times and incubated with secondary antibody, Texas Red-conjugated anti-mouse IgG and Alexa-488 conjugated anti rabbit IgG, for 1 h. After being washed with PBS three times, cells were mounted with Fluoromount G (Virotech International, Rockville, Md.) and examined with a Zeiss LSM410 laser scanning microscope. For EGF-Tx uptake studies, cells were incubated with EGF-Tx conjugate (0.4 μg/ml) for 30 min at 37°C prior to fixation. The cells were permeabilized and incubated with anti-HA as described above. Staining using Lysotracker dye was performed as recommended by the manufacturer. Briefly, cells were incubated with Lysotracker Red for 30 min at 37°C prior to fixation, permeabilized, and incubated with anti-HA antibody as described above.
Antibodies and fluorescently labeled reagents were obtained from the following sources: mouse anti-EGF-R antibody, Santa Cruz Biotech (Santa Cruz, Calif.); rabbit polyclonal anti-HA antiserum, Sigma; anti-mouse and Ig antibodies conjugated with horseradish peroxidase, Amersham Pharmacia; and Texas Red-conjugated anti-mouse secondary antibody, Jackson Immunoreagents. Lysotracker Red dye, EGF, Alexa 488-conjugated anti-rabbit secondary antibody, and EGF-Tx conjugate were obtained from Molecular Probes (Eugene, Oreg.).
RESULTS
Inhibition of HIV-1 release by TSG-5′ is promoted by its association with Gag.
We previously reported that overexpression of TSG-5′ potently inhibits HIV-1 budding (11). TSG-5′ is incorporated into WT but not L domain-deficient virions, indicating that it interacts with Gag in an L domain-dependent fashion. To determine whether the inhibition of virus release imposed by TSG-5′ is dependent upon its interaction with Gag, we mutated a region of TSG101 previously shown to be important for Gag/TSG101 interaction (45, 46, 55). The three residues Thr67, Tyr68, and Asn69 (Fig. 1) were substituted for a single Ala to generate the TYN− mutation. To assess the impact of the TYN− mutation on Gag binding, we compared the incorporation of TSG-5′ and TSG-5′/TYN− into virions. HeLa cells were transfected with the full-length molecular clone pNL4-3 (1) or cotransfected with a 1:1 DNA ratio of pNL4-3 and either TSG-5′ or TSG-5′/TYN− expression vectors. Cell and viral lysates were immunoblotted with anti-HIV Ig or anti-HA antiserum. The TYN− mutant form of TSG-5′ was not incorporated into virions (data not shown), consistent with this mutation blocking the Gag/TSG101 interaction. We also introduced single amino acid substitutions at TSG101 residues 110 and 113; these mutations did not block TSG-5′ incorporation (data not shown).
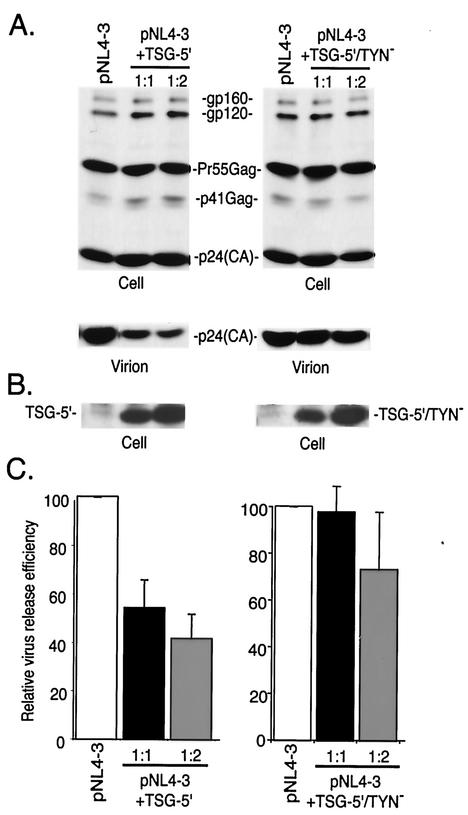
We next tested whether the loss of Gag binding displayed by TSG-5′/TYN− would affect the inhibition of virus release observed with TSG-5′. HeLa cells were transfected with pNL4-3 alone, or were cotransfected with pNL4-3 and either TSG-5′ or TSG-5′/TYN− vectors. Transfected cells were metabolically labeled and cell- and virion-associated lysates were immunoprecipitated with HIV Ig (Fig. 2). As previously reported (11), WT TSG-5′ expression significantly inhibited virus particle production; an approximately 60% reduction was measured at a 1:2 DNA ratio. In contrast, TSG-5′/TYN− reduced virus production only slightly, despite the fact that it was expressed at levels comparable to TSG-5′ (Fig. 2B). EM analysis demonstrated that, as observed previously (11), TSG-5′ caused an accumulation of particles tethered to the plasma membrane and the appearance of numerous doublet particles (Fig. 3B). In contrast, cells transfected with pNL4-3 alone displayed an abundance of released, mature virions (Fig. 3A). Consistent with the biochemical analysis of virus production, the morphology of budding virions produced from cells cotransfected with pNL4-3 and the TSG-5′/TYN− expression vector was essentially identical to that from cells transfected with pNL4-3 alone (Fig. 3C). These results demonstrate that the TSG-5′/TYN− mutant, which disrupts the Gag/TSG-5′ interaction, is impaired relative to WT TSG-5′ in its ability to suppress HIV-1 particle production.
FIG. 2.
Effect of TSG-5′ and TSG-5′/TYN− on HIV-1 release. HeLa cells were transfected with pNL4-3 alone or were cotransfected with pNL4-3 + TSG-5′ or TSG-5′/TYN− at 1:1 and 1:2 DNA ratios. Cells were metabolically labeled, and cell and virion lysates were immunoprecipitated with anti-HIV Ig (A) or cell lysates were immunoblotted with anti-HA antiserum (B). Positions of the Env glycoprotein precursor gp160, the mature surface glycoprotein gp120, the Gag precursor (Pr55Gag), the Gag processing intermediate p41Gag, the mature CA protein p24(CA), and TSG-5′ are indicated. (C) Quantitative analysis of the effect of TSG-5′ and TSG-5′/TYN− on the production of virion-associated Gag. Relative virus release efficiency was calculated as the amount of virion-associated p24(CA) as a fraction of total Gag [(cell-associated Pr55Gag+p41Gag+p24(CA)+virion p24(CA)]. Data are averages from phosphorimager analysis of at least three experiments (error bars, standard errors).
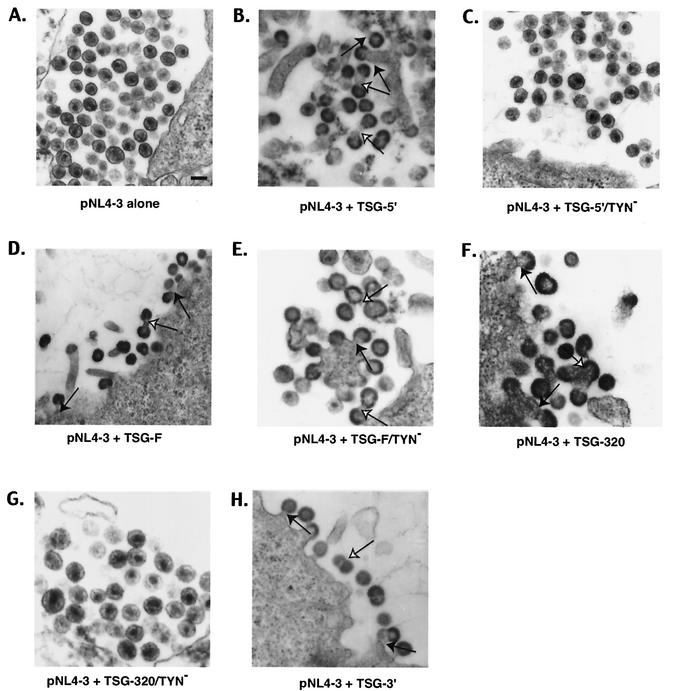
FIG. 3.
EM analysis of HeLa cells transfected with pNL4-3 alone (A) or cotransfected with pNL4-3 + TSG-5′ (B), TSG-5′/TYN− (C), TSG-F (D), TSG-F/TYN− (E), TSG-320 (F), TSG-320/TYN− (G), or TSG-3′ (H). Closed arrows indicate virions tethered to the plasma membrane; open arrows show virion-virion tethers. (A) Bar, ∼100 nm.
Overexpression of full-length TSG101 inhibits virus budding.
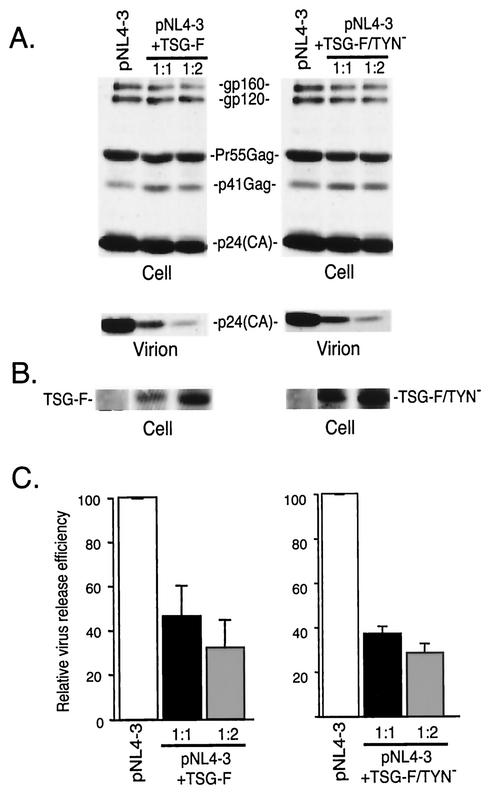
To explore further the role of TSG101 and endosomal sorting in HIV-1 budding, we tested the effect of overexpressing TSG-F on virus release. HeLa cells were transfected with pNL4-3 alone or were cotransfected with pNL4-3 and a TSG-F expression vector (Materials and Methods). As observed with TSG-5′, overexpression of the full-length protein markedly inhibited virus particle production, with an approximately 70% reduction in virus release efficiency observed at a 1:2 DNA ratio (Fig. 4). To determine whether this defect was the result of impaired budding, we examined by EM cells cotransfected with pNL4-3 and the TSG-F expression vector. As seen with TSG-5′, cells overexpressing TSG-F showed a marked pinching-off defect, with particles accumulated at the plasma membrane and frequent doublet virions (Fig. 3D). The results of the biochemical and EM analyses indicate that overexpression of full-length TSG101 inhibits HIV-1 budding.
FIG. 4.
Effect of TSG-F and TSG-F/TYN− on HIV-1 release. HeLa cells were transfected with pNL4-3 alone or cotransfected with pNL4-3 + TSG-F or TSG-F/TYN− at 1:1 and 1:2 DNA ratios. Viral and cell lysates were prepared and analyzed as described in the legend of Fig. 2. (A) Immunoprecipitation of cell and viral lysates with anti-HIV Ig; (B) Western blotting of cell lysates with anti-HA antiserum; (C) quantitation of virus release efficiency. See the legend to Fig. 2 for further details.
Inhibition of budding by TSG-F overexpression does not require Gag/TSG101 binding.
We demonstrated above that a mutation in TSG-5′ that disrupts its binding to Gag also largely eliminates its inhibitory effect on HIV-1 budding. To determine whether inhibition mediated by overexpression of full-length TSG101 also requires Gag binding, we constructed a TYN− mutant version of TSG-F. As observed with TSG-5′, this mutation blocked TSG101 incorporation into virions (data not shown); however, TSG-F/TYN− still inhibited virus release to the same extent as observed upon overexpression of WT TSG-F (Fig. 4). EM data also indicated that a budding defect was imposed by TSG-F/TYN−, with frequent appearance of particles tethered to each other and to the plasma membrane (Fig. 3E). These results demonstrate that overexpression of full-length TSG101 can inhibit HIV-1 budding independently of its interaction with Gag.
Inhibition of HIV-1 budding independently of Gag binding requires the C-terminal domain of TSG101.
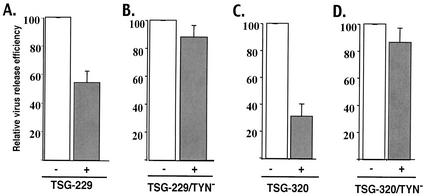
The data presented above indicate that inhibition of budding mediated by TSG-5′ is largely eliminated by the TYN− substitution, whereas inhibition resulting from overexpression of TSG-F is unaffected by this mutation. These results suggest that overexpression of TSG-F can interfere with HIV-1 budding by a mechanism distinct from that imposed by TSG-5′. To identify the domains responsible for this difference, we constructed two additional vectors that express forms of TSG101 intermediate in size between TSG-5′ and TSG-F (Fig. 1). The TSG-229 protein contains the N-terminal E2-like domain as well as the complete Pro-rich region; TSG-320 contains the coiled-coil domain in addition to the E2-like and Pro-rich sequences. These proteins were efficiently expressed, were of the predicted size, and were incorporated into virions (Fig. 5). Immunoprecipitation assays indicated that overexpression of TSG-229 and TSG-320 suppressed virus release (Fig. 6A and C), and EM analysis demonstrated that TSG-320 inhibited particle budding from virus-expressing cells (Fig. 3F). Like TSG-5′, and unlike TSG-F, the TYN− mutation in the context of these truncated forms of TSG101 largely reversed the inhibitory effect (Fig. 6B ad D), a result that was confirmed by examining TSG-320/TYN−-expressing cells by EM (Fig. 3G). The TYN− forms of TSG-229 and TSG-320 were expressed to the same level as were the forms of these truncation mutants containing WT N-terminal domains (data not shown). These data suggest that the ability of TSG101 overexpression to inhibit virus budding in the absence of Gag/TSG101 binding maps to the C-terminal domain of TSG101.
FIG. 5.
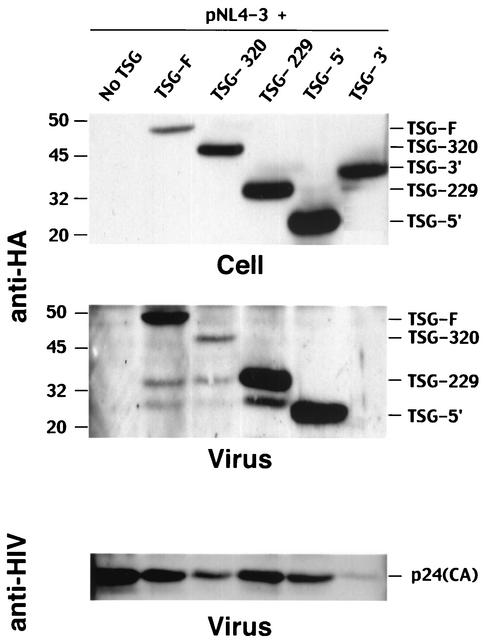
Expression and incorporation of TSG-F and TSG101 truncation mutants. HeLa cells were transfected with pNL4-3 alone (pNL4-3 + no TSG) or were cotransfected with pNL4-3 plus TSG-F, TSG-320, TSG-229, TSG-5′, or TSG-3′ expression vectors. Cell and viral lysates were prepared and subjected to Western blotting with anti-HA antisera (top and middle panels) or anti-HIV Ig (bottom panel). Levels of cell- and virion-associated TSG and virion p24(CA) are shown in top, middle, and bottom panels, respectively. Positions of TSG101 derivatives are indicated on the right; TSG-3′ is not detected in virion lysates. Molecular mass markers, in kilodaltons, are shown on the left.
FIG. 6.
Effect of TSG-229, TSG-229/TYN−, TSG-320, and TSG-320/TYN− on HIV-1 release. HeLa cells were transfected with pNL4-3 alone or were cotransfected with pNL4-3 + TSG-229 (A), TSG-229/TYN− (B), TSG-320 (C), or TSG-320/TYN− (D) at a 1:1 DNA ratio. Viral and cell lysates were prepared and analyzed as described in the legend of Fig. 2. The efficiency of virus release was determined by phosphorimager analysis of at least three experiments (error bars, standard errors).
Overexpression of the C-terminal portion of TSG101 disrupts retrovirus budding.
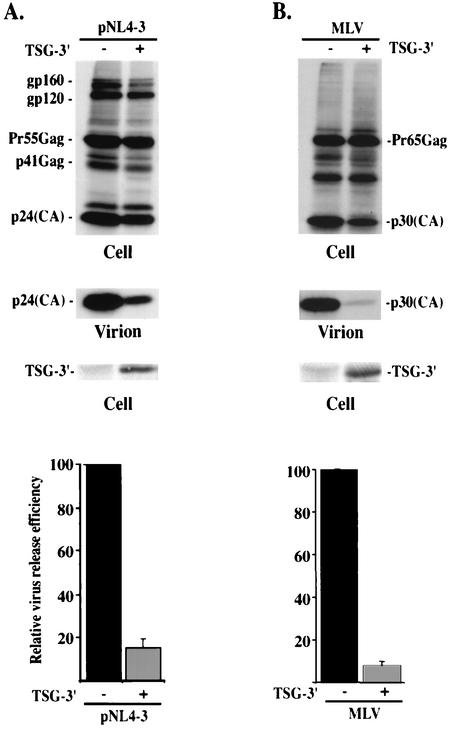
The finding that TSG-F overexpression inhibits HIV-1 budding in a manner independent of Gag binding, whereas inhibition by C-terminally truncated forms of TSG101 is largely reversed by the TYN− mutation, raises the possibility that overexpressing the C-terminal portion of TSG101 might disrupt virus release. To examine this possibility, we cotransfected pNL4-3 with the TSG-3′ vector, which expresses the C-terminal portion of TSG101 (Fig. 1). Even at a 1:1 DNA ratio, virus production was severely inhibited (Fig. 7A); the efficiency of virus release was reduced by approximately 10-fold. To determine whether the defect in virus production is due to a block in the budding step of the assembly-release pathway, we examined cells cotransfected with pNL4-3 and TSG-3′ by EM. Again, numerous virus particles were observed tethered to the plasma membrane, and doublet particles were common (Fig. 3H). These results indicate that TSG-3′ inhibits HIV-1 budding.
FIG. 7.
TSG-3′ inhibits HIV-1 and MLV particle release. (A) HeLa cells were transfected with pNL4-3 alone or a 1:1 ratio of pNL4-3 + TSG-3′. Cells were metabolically labeled, cell and viral lysates were immunoprecipitated with anti-HIV Ig (top and middle) or immunoblotted with anti-HA antiserum (lower). (B) HeLa cells were transfected with an MLV molecular clone alone or a 1:1 ratio of molecular clone plus TSG-3′. Cells were metabolically labeled, cell and viral lysates were immunoprecipitated with anti-MLV p30 antiserum (top and middle) or immunoblotted with anti-HA antiserum (lower). In both panels, relative virus release efficiency was calculated as described in the legend for Fig. 2.
The observation that the TSG-3′ mutant inhibits HIV-1 budding, despite the fact that it completely lacks the E2-like domain responsible for TSG101 interaction with HIV-1 p6, suggests the possibility that this N-terminally truncated protein might disrupt the budding of retroviruses other than HIV-1. To test this possibility, we cotransfected the TSG-3′ vector with an MLV Gag expression construct (Materials and Methods). Cell- and virion-associated proteins were radioimmunoprecipitated with an anti-MLV CA antibody. As seen with HIV-1, virus production was potently suppressed, in this case by more than 10-fold (Fig. 7B). These data indicate that, unlike TSG-5′ (11), TSG-3′ broadly disrupts retrovirus particle release.
Effects of overexpressing full-length and truncated forms of TSG101 on endosomal sorting.
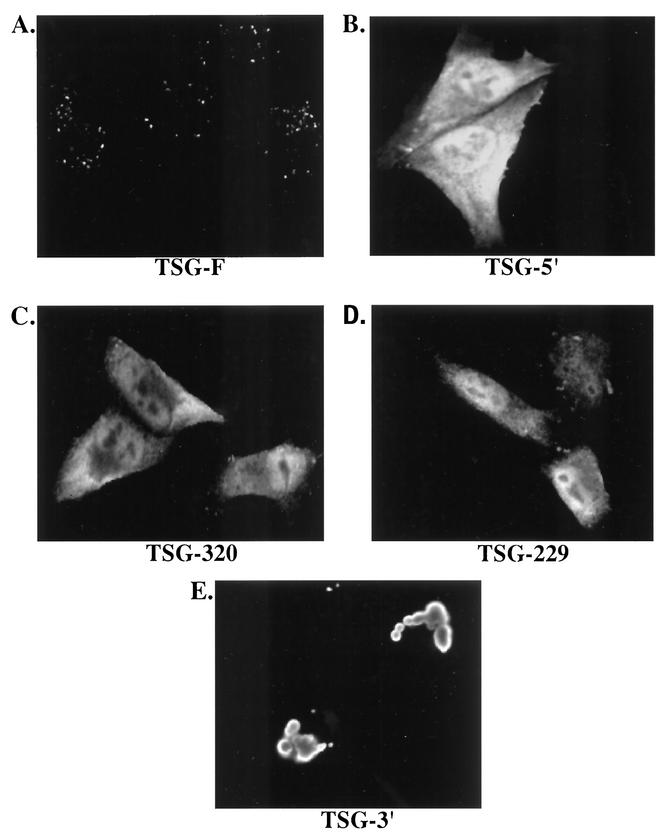
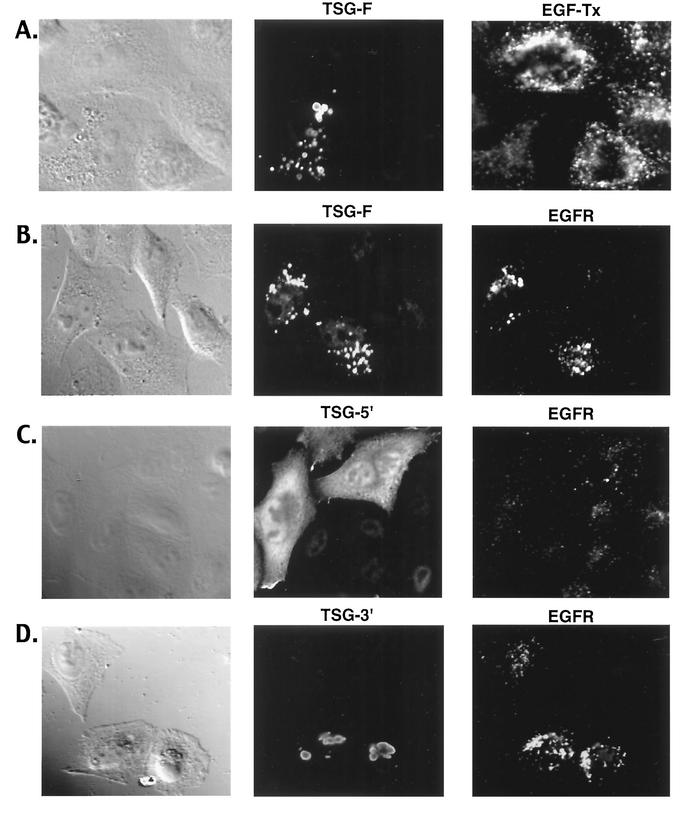
To examine further the mechanism by which overexpression of full-length and truncated forms of TSG101 inhibits virus release, we analyzed their subcellular localization and their effect on the endosomal sorting pathway by confocal microscopy. TSG-F displayed a highly punctate, putatively endosomal expression pattern (Fig. 8A). In contrast, the C-terminally truncated TSG101 mutants (TSG-5′, TSG-229, and TSG-320) were distributed diffusely throughout the cytoplasm (Fig. 8B-D). Transfection of cells with TSG-3′ resulted in the formation of large vacuolar structures strikingly different from the compartment in which TSG-F was localized (Fig. 8E).
FIG. 8.
Subcellular localization of TSG-F and truncated forms. HeLa cells were transfected with plasmids encoding TSG-F, TSG-5′, TSG-320, TSG-229 or TSG-3′ (A to E). The cells were fixed, permeabilized and stained using anti-HA antiserum and visualized by confocal microscopy as described in Materials and Methods.
Following the binding of EGF to the EGF-R, the latter undergoes ubiquitination, internalization, and sorting into the MVB for eventual degradation in the lysosome (19, 35). The internalization and trafficking of EGF and EGF-R thus provide a probe for monitoring the endosomal sorting pathway. To determine the effects of TSG-F and TSG-5′ overexpression on endosomal sorting, we analyzed the binding and uptake of fluorescently labeled EGF. Cells that overexpressed TSG-F exhibited a reduced binding and uptake of EGF compared with neighboring untransfected cells (Fig. 9A). In contrast, TSG-5′ and the other C-terminally truncated forms of TSG101 did not detectably affect EGF trafficking (data not shown). To corroborate the results obtained with EGF, we also examined the localization of EGF-R following EGF stimulation. A striking increase in the intracellular levels of EGF-R was observed in cells overexpressing TSG-F (Fig. 9B); the receptors accumulated in a compartment that partially colocalized with TSG-F. Interestingly, the intense intracellular localization of EGF-R was evident in TSG-F-expressing cells even in the absence of added ligand (data not shown). Unlike the pattern observed upon TSG-F overexpression, cells transfected with TSG-5′ (Fig. 9C), TSG-229, or TSG-320 (data not shown) showed a distribution of EGF-R essentially indistinguishable from that in untransfected cells. TSG-3′ overexpression led to an increase in intracellular levels of EGF-R (Fig. 9D), though this effect was less pronounced than observed upon TSG-F overexpression. TSG-3′ did not significantly colocalize with EGF-R.
FIG. 9.
Effect of overexpressing TSG101 and derivatives on uptake of EGF and localization of EGF-R. (A) HeLa cells were transfected with the TSG-F expression vector. Cells were incubated with 0.4 μg/ml EGF-Tx for 30 min at 37°C, fixed, permeabilized with methanol, and stained for TSG using anti-HA antiserum. (B to D) The cells transfected with TSG-F, TSG-5′ and TSG-3′ expression vectors were stimulated using EGF (0.4 μg/ml) for 30 min at 37°C and analyzed for intracellular EGF-R using EGF-R antibody and TSG using anti-HA.
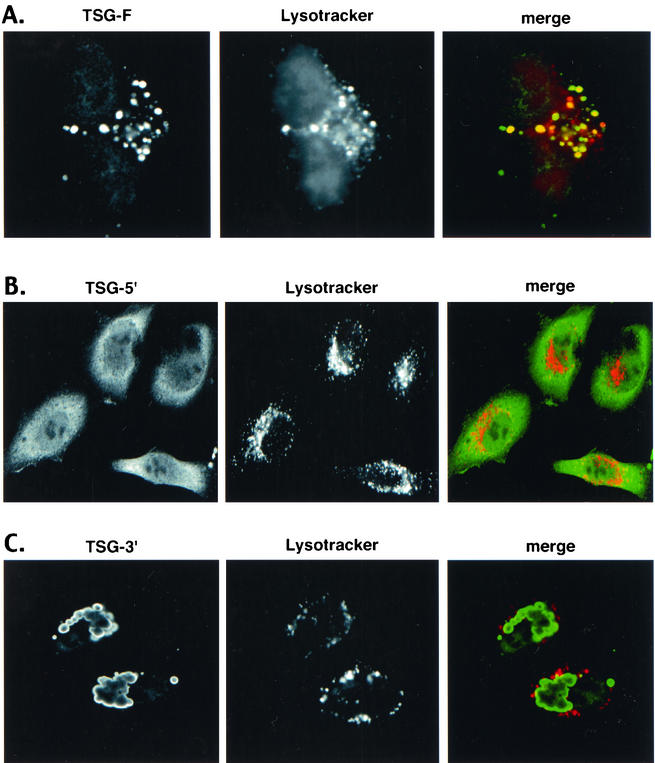
To identify the subcellular compartment in which TSG-F and TSG-3′ are localized, we probed transfected cultures with Lysotracker, a fluorescent acidotropic labeling agent. TSG-F was found to be largely sequestered in an acidic compartment, as evidenced by significant colocalization between TSG-F and Lysotracker Red (Fig. 10A). TSG-5′, on the other hand, did not colocalize with Lysotracker Red (Fig. 10B). The aberrant enlarged vacuolar structures formed by TSG-3′ were not acidic in nature, as no colocalization between TSG-3′ and Lysotracker Red was observed (Fig. 10C). These results suggest that TSG-F is localized in a swollen endosomal compartment distinct from that induced by TSG-3′.
FIG. 10.
TSG-F localizes to an acidic compartment. HeLa cells were transfected with plasmids expressing TSG-F (A), TSG-5′ (B) , or TSG-3′ (C). Transfected cells were incubated with Lysotracker Red dye for 30 min at 37°C prior to fixation. The cells were then permeabilized and stained for TSG using anti-HA antibody and visualized by confocal microscopy as described in Materials and Methods.
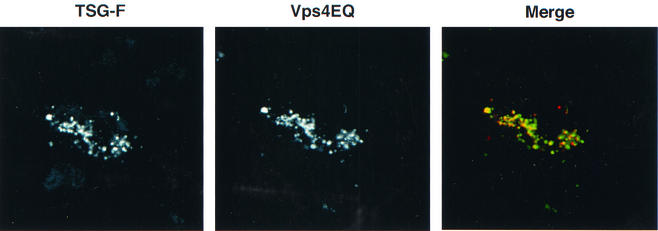
Previous studies indicated that overexpression of an ATPase-defective form of human Vps4, Vps4EQ, induced the formation of an aberrant endosomal compartment (4). To test whether the compartment induced by Vps4EQ resembled that formed upon TSG-F overexpression, we cotransfected cells with Vps4EQ and TSG-F expression vectors and examined the localization of these two proteins. As indicated in Fig. 11, a high degree of colocalization was observed between TSG-F and Vps4EQ. In contrast, no colocalization was observed between TSG-3′ and Vps4EQ (data not shown). These results suggest that overexpression of TSG-F leads to the formation of an enlarged endosomal compartment similar to that induced upon Vps4EQ expression, or that coexpression of TSG-F and Vps4EQ results in the retention of both proteins in the same aberrant endosomal compartment. These data highlight the distinct nature of the compartments in which TSG-F and TSG-3′ are localized.
FIG. 11.
TSG-F colocalizes with Vps4EQ. HeLa cells were cotransfected with plasmids expressing TSG-F and Vps4EQ-GFP. The cells were fixed, permeabilized and stained for TSG using anti-HA antibody.
DISCUSSION
Efficient budding of HIV-1 is promoted by the PTAP motif in p6, apparently via a direct interaction with the host endosomal sorting protein TSG101. We previously reported that overexpression of TSG-5′ markedly inhibits HIV-1 budding in an L-domain dependent manner (11). In this study, we observe that potent inhibition of virus release by TSG-5′ is dependent upon its interaction with Gag. A mutation that abolishes the Gag/TSG101 interaction largely eliminates its inhibitory activity. Two additional C-terminal TSG101 truncation mutants, TSG-229 and TSG-320, also inhibit HIV-1 budding in a manner that is greatly reduced or eliminated by disrupting the Gag/TSG101 interaction. We also demonstrate that overexpression of full-length TSG101 inhibits HIV-1 budding. However, in contrast to results obtained with the C-terminal truncation mutants (i.e., TSG-5′, TSG-229, and TSG-320), disruption of the Gag/TSG101 interaction has no effect on the ability of TSG-F to inhibit virus release. This finding indicates that overexpression of TSG-F interferes with HIV-1 budding by a mechanism distinct from that imposed by the C-terminally truncated TSG101 mutants. Strong inhibition by the C-terminal truncation mutants requires an interaction with Gag, whereas inhibition by TSG-F does not. Overexpression of TSG-3′, which lacks the Gag binding domain of TSG101, was observed to severely inhibit both HIV-1 and MLV budding.
As mentioned in the introduction, defects in class E Vps proteins give rise to the formation of an aberrant, exaggerated endosomal compartment in both yeast and mammalian cells (8, 48). Previous studies by Babst et al. (4) demonstrated a defect in endosomal sorting in cells deficient for TSG101 expression; this defect was reflected by a delayed down-regulation of cell surface EGF-R in tsg101 mutant cells. The results presented in this study indicate that overexpression of full-length TSG101 induces a similar aberrant endosome phenotype, perhaps by disrupting the stoichiometry of ESCRT-1 components. The exaggerated endosomal phenotype observed in cells overexpressing TSG-F is reminiscent of the defect induced by the ATPase-defective mutant of mammalian Vps4 (Vps4EQ) (8). In addition, we have observed a specific colocalization of TSG-F and Vps4EQ. Together, these results suggest that overexpression of TSG-F and Vps4EQ results in the formation of a similar exaggerated endosomal compartment. Alternatively, expression of Vps4EQ may result in the retention of TSG-F in an aberrant endosomal compartment, or TSG-F overexpression may induce the retention of Vps4EQ in this compartment. It has been noted previously (9) that endogenous TSG101 is retained in swollen endosomes induced by Vps4EQ. In contrast to what we observe with TSG-F, TSG101 truncation mutants that lack the C-terminal, Vps28 binding domain are diffusely localized in the cytoplasm and do not appear to adversely affect endosome formation or sorting. TSG-3′ retains the Vsp28 binding domain and could disrupt the ESCRT-1 complex by sequestering Vps28 and preventing its interaction with endogenous TSG101. Alternatively, the presence in TSG-3′ of the so-called steadiness box, which regulates the levels of intracellular TSG101 (13) could result in a TSG-3′-mediated downregulation of endogenous TSG101. However, the phenotype observed in TSG-3′-expressing cells appears to be differ substantially from that observed upon TSG101 depletion.
It is important to note that the defects induced by overexpression of full-length TSG101 and TSG-3′ differ in several significant respects. (i) TSG-3′ induces the formation of very large vacuolar structures that fail to stain with Lysotracker. (ii) Despite the extreme nature of the morphological alterations induced by TSG-3′, its expression appears to have a less adverse effect on EGF-R trafficking than does overexpression of TSG-F. (iii) While overexpression of TSG-F potently inhibits HIV-1 budding, its effect on the release of MLV virions is relatively mild (Goila-Gaur and Freed, unpublished). In contrast, TSG-3′ severely inhibits both HIV-1 (Fig. 7A) and MLV (Fig. 7B) budding. (iv) Finally, TSG-F shows a high degree of colocalization with Vps4EQ (Fig. 11) whereas TSG-3′ does not. We postulate that TSG-3′ induces the formation of novel endosomal-like structures that fail to undergo acidification. Further studies will be required to define more precisely the impact of TSG-3′ and TSG-F overexpression on both the endosomal sorting pathway and virus budding.
Taken together, our findings demonstrate that TSG-5′ and other C-terminal TSG101 truncation mutants inhibit HIV-1 budding primarily by directly binding and inactivating the p6 L domain, whereas TSG-F and TSG-3′ overexpression inhibits virus release by disrupting the cellular endosomal sorting pathway. The data reported here highlight the importance of TSG101 in HIV-1 budding, and shed new light on the interplay between the endosomal sorting pathway and retrovirus budding. The ability of TSG-5′ to inhibit virus budding without any apparent adverse effect on cellular sorting machinery demonstrates that antiviral agents can be developed that interfere with virus replication by specifically targeting the Gag/TSG101 interaction (16).
Acknowledgments
We thank S. Ablan for expert technical assistance, Alicia Buckler-White for DNA sequencing, and P. Woodman and Z. Sun for plasmids.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 5.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, N., A. Horman, and P. Woodman. 2002. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, G. H., C. J. Lih, and S. N. Cohen. 2000. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 60:1736-1741. [PubMed] [Google Scholar]
- 14.Finken-Eigen, M., R. A. Rohricht, and K. Kohrer. 1997. The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Curr. Genet. 31:469-480. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 2003. The HIV-TSG101 interface: Recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 17.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futter, C. E., A. Pearse, L. J. Hewlett, and C. R. Hopkins. 1996. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132:1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for hiv-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 21.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-98127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin, E. V., and R. A. Abagyan. 1997. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat. Genet. 16:330-331. [DOI] [PubMed] [Google Scholar]
- 31.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 66:5110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemmon, S. K., and L. M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457-466. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., T. Kane, C. Tipper, P. Spatrick, and D. D. Jenness. 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 19:3588-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 37.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase e2, interacts specifically with human immunodeficiency virus type 2 gag polyprotein and results in increased levels of ubiquitinated gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odorizzi, G., M. Babst, and S. D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847-858. [DOI] [PubMed] [Google Scholar]
- 39.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269:1872-1875. [DOI] [PubMed] [Google Scholar]
- 40.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 42.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, 2nd, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, R. C. Sowder, 2nd, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 46.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 50.Stahl, P. D., and M. A. Barbieri. 2002. Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sci STKE 2002:PE32. [DOI] [PubMed]
- 51.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 52.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, Z., J. Pan, W. X. Hope, S. N. Cohen, and S. P. Balk. 1999. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer 86:689-696. [DOI] [PubMed] [Google Scholar]
- 54.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 55.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimori, T., F. Yamagata, A. Yamamoto, N. Mizushima, Y. Kabeya, A. Nara, I. Miwako, M. Ohashi, M. Ohsumi, and Y. Ohsumi. 2000. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11:747-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]