Abstract
Three-finger toxins are a family of low-molecular-mass toxins (<10 kDa) having very similar three-dimensional structures. In the present study, 19 novel cDNAs coding three-finger toxins were cloned from the venom gland of Ophiophagus hannah (king cobra). Alignment analysis showed that the putative peptides could be divided into six kinds of three-finger toxins: LNTXs (long-chain neurotoxins), short-chain neurotoxins, cardiotoxins (CTXs), weak neurotoxins, muscarinic toxins and a toxin with a free SH group. Furthermore, a phylogenetic tree was established on the basis of the toxin cDNAs and the previously reported similar nucleotide sequences from the same source venom. It indicated that three-finger-toxin genes in O. hannah diverged early in the course of evolution by long- and short-type pathways. Two LNTXs, namely rLNTX1 (recombinant LNTX1) and rLNTX3, were expressed and showed cytolytic activity in addition to their neurotoxic function. By comparing the functional residues, we offer some possible explanations for the differences in their neurotoxic function. Moreover, a plausible elucidation of the additonal cytolytic activity was achieved by hydropathy-profile analysis. This, to our knowledge, is the first observation that recombinant long chain α-neurotoxins have a CTX-like cytolytic activity.
Keywords: cardiotoxins, cytotoxicity, α-neurotoxins, nicotinic acetylcholine receptors (nAChRs), Ophiophagus hannah (king cobra), phylogenetic tree
Abbreviations: α-Cbtx, α-cobratoxin; CTXs, cardiotoxins; GST, glutathione S-transferase; HUVEC, human umbilical-vein endothelial cells; IPTG, isopropyl β-D-thiogalactoside; (r)LNTXs, (recombinant) long-chain neurotoxins; mAChRs, muscarinic acetylcholine receptors; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; MTXs, muscarinic toxins; nAChRs, nicotinic acetylcholine receptors; SEC, size-exclusion chromatography; SNTXs, short-chain neurotoxins; WNTXs, weak neurotoxins
INTRODUCTION
Three-finger toxins in snake venoms consist of a multitude of pharmacologically active peptides having a very similar three-dimensional structure, with three loops extending from a very conserved core rich in disulphide bridges [1]. According to the diversity of their functions, three-finger toxins can be classified into two major groups: neurotoxins and CTXs (cardiotoxins). LNTXs (long-chain neurotoxins), SNTXs (short-chain neurotoxins), WNTXs (weak neurotoxins) and MTXs (muscarinic toxins) belong to the neurotoxin group, members of which interfere in the transmission of nervous impulses by selectively binding to particular receptors in the nerve or muscular membrane. LNTXs and SNTXs are also called α-neurotoxins. They have a high affinity for nAChRs (nicotinic acetylcholine receptors), whereas MTXs bind to mAChRs (muscarinic acetylcholine receptors) [2,3]. WNTXs are characterized by their being able to act on nAChRs with a lower affinity than LNTXs [4]. The major members of the CTX group have no known effect on nAChRs or mAChRs, but exhibit quite different pharmacological properties, such as cytolysis, haemolysis and heart failure [5].
Up until now the functional residues of some LNTXs and SNTXs have been elucidated on the basis of the crystal structures of binding complexes and mutation analysis, but further structure–function validations in more α-neurotoxins are needed [8,9]. In the interim, rather fewer findings have been obtained in research on CTXs, WNTXs and MTXs. This is partly because the wide distribution of hydrophobic regions renders most of the CTXs difficult to crystallize, and few WNTXs and MTXs have been purified [7,10]. In the present study we cloned 19 novel cDNAs coding for the abovementioned three-finger toxins from Ophiophagus hannah (king cobra). Two LNTXs were expressed and their neurotoxic effects on muscular nAChRs were tested by means of electrophysiological experiments. By comparing the functional residues we are able to offer some plausible explanation for the differences in their neurotoxic functions. The work partly supports the location of functional residues related to neurotoxic activity. Moreover, these cDNAs will be helpful in the structure–function elucidation of three-finger toxins by sequence alignment and recombinant-mutation technology.
The evolutionary relationship of three-finger toxins is another ‘hot’ topic dating back to the 1990s. Most of the phylogenetic trees constructed previously were based on the amino acid sequences of snake toxins [11]. However, Strydom [12] concluded that no particular choice of snake toxins could be used to build a phylogenetic tree on the basis of their amino acid sequences alone. Since the first three-finger toxin was sequenced in 1985, phylogenetic analysis has been focused on the construction of phylogenetic trees of nucleotide sequences and the analysis of gene structures [13,14]. However, little work has been done on the evolutionary relationship of the six kinds of three-finger toxins, since toxins from different snakes may exhibit different evolutionary tendencies, and it is relatively difficult to get multi-sequences of three-finger toxins in a specific snake [13].
Now, for the first time, many cDNAs of three-finger toxins have been cloned from O. hannah. The phylogenetic tree of these cDNAs, and previously published sequences from the same source venom, indicated that three-finger toxin genes in O. hannah evolved from a common ancestor and diverged early in the course of evolution. Do these evolutionary related toxins have similar functions? Recent reports showed that some of these three-finger toxins had ‘crossed’ functions. It was reported that a SNTX with the cytotoxic function of CTXs was isolated from Daboia russelli russelli (Sri Lankan Russell's viper) [15]. Zhou et al. [16] provided evidence that WNTXs, in the presence of CTXs, may exhibit a synergic cytolytic effect. In our study, two recombinant LNTXs showed a cytolytic activity in addition to their neurotoxic function. A possible elucidation has been carried out by means of hydropathy-profile analysis.
MATERIALS AND METHODS
All experiments were carried out in accordance with Chinese Animal Welfare legislation and were approved by the Committee on Ethics in the Care and Use of Laboratory Animals in the National Institute for the Control of Pharmaceutical and Biological Products.
Isolation of mRNA, reverse-transcription PCR and identification of cDNA
On the basis of the seven amino acids in the N-terminal sequences of the signal peptides of neurotoxins in the elapid-snake venoms, the sense primer 1 was designed as 5′-AGATGGCAAGATGAAAACTCTGCTGCTGACC-3′. The double-stranded cDNAs were amplified using primer1 and the universal adaptor primer according to the 3′-RACE (rapid amplification of cDNA ends) core set procedures. The PCR products were cloned into pMD18-T vector. Randomly selected transformants, 145 in all, were screened, and 137 positive clones were sequenced on both strands by TaKaRa Biotechnology Co. Ltd, Dalian, China. Nucleic acid sequences were analysed using DNASTAR 4.0. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (http://www.megasoftware.net/).
Construction of fusion expression plasmids
Two cDNAs encoding LNTX were named as lntx1 and lnxt3 respectively. Primer2 (5′-CGTGGATCCACAAAATGCTACAAAACCGGTG-3′) and primer3 (5′-CGACTCGAGTCAACGTTGTTTCATTTTTGGATG-3′) were used to amplify lntx1; primer4 (5′-CGTGGATCCTTGATATGCTTCATATCTTCTCATG-3′) and primer5 (5′-CGACTCGAGTCAAGGTCTCAATTTCGGATGTGGG-3′) were used to amplify lntx3. Underlined nucleotides include the restriction sites of BamHI and XhoI respectively. PCR products were subcloned into vector pGEX4T-1. Plasmids with the correct sequences were expressed in the Escherichia coli host cell Origami B.
Expression and purification of rLNTXs (recombinant LNTXs)
The bacteria harbouring vector-lntx1/lntx3 were induced with 0.1mM IPTG (isopropyl β-D-thiogalactoside) and the culture further incubated at 16 °C for 20 h. The cell pellets were lysed using 0.5 mg/ml lysozyme for 15 min, followed by mild sonication. After centrifugation, the supernatant was mixed with glutathione–Sepharose 4B and incubated at 4 °C overnight. The mixture was then loaded on to an empty column and the fusion proteins were eluted with buffer containing 50 mM Tris/HCl and 10 mM GSH. Elution fractions were loaded on to a Superdex75 column (1.6 cm internal diameter×100 cm long) at 1 ml/min and peaks were desalted on a Sephadex G25 HiPrep 26/10 column. Fusion proteins were freeze-dried, then dissolved in the cleavage buffer prior to the addition of 10 units of thrombin for each 1 mg of fusion protein. Fusion protein LNTX1 was cleaved at room temperature for 8 h, whereas LNTX3 was cleaved at 37 °C for 12 h. After cleavage, proteins were loaded on a benzamidine column placed in series below the GSTrap column at a rate of 0.5 ml/min.
Characterization of rLNTX1 and rLNTX3
Purity tests on rLNTX1 and rLNTX3 were carried out using reversed-phase HPLC (Alltech-C8 column; 4.6 mm internal diameter×150 mm long) and SEC (size-exclusion chromatography) HPLC (TSK G2000 SWXL; 7.8 mm×300 mm). The tip of the goal peak eluted from reversed-phase HPLC was collected for MS and amino acid sequencing. The molecular mass was determined by MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS (AXIMA-CFR™) under the mode of linear 2 GHz positive ionization. Ten amino acids at the N-terminus of the proteins were sequenced by the Department of Biochemistry, School of Life Science, Peking University (Beijing, China) on an ABI Procise 491 protein sequencer.
CD spectra were recorded on a Jasco J715 spectropolarimeter and averaged from three recordings. Both samples were dissolved in PBS at 0.3 mg/ml and the measurements taken at 22 °C using 1-mm-light-path cell. The molecular ellipticity [17] was used as the y co-ordinate.
Lethality assay (LD50)
A total of 15 Kunming mice were injected intravenously with 200 μl of recombinant toxins dissolved in 0.9% NaCl. An LD50 test was performed using the up-and-down method [18] and according to the AOT425StatPgm1.0 procedure (http://www.epa.gov/oppfead1/harmonization/). All mice were observed for up to 48 h.
Electrophysiological experiments
Skeletal myocytes were obtained by the method of Yaffe [19], with some modifications. Nicotine-induced currents were recorded with a single microelectrode voltage clamp at −70 mV, using electrodes with approx. 2 MΩ resistances when filled with internal solution (2.36% CaCl2/0.24% Hepes/0.38% EGTA/0.11% ATP, pH 7.2). The external bath composition was 0.82% NaCl/0.04% KCl/0.02% MgCl2/0.24% Hepes/0.2% D-glucose/0.03% CaCl2, pH 7.2. All drugs were applied on to the surface of experimental cells by low-pressure injection with a micro-injector (IM-5B; Narishige). At 5 min after impalement with the electrodes, the myoball was exposed to various concentrations (40–120 μM) of nicotine to select a suitable concentration of nicotine for control current, and peak current was recorded on Clampex 7.0 workstation.
Before toxins were tested, the cell was exposed to 80 μM nicotine for 5 s and the current was recorded. The same cell was then exposed to various concentrations of toxins dissolved in external solution containing 80 μM nicotine. For each concentration of toxin this procedure was repeated five times on five different myoballs. The currents recorded either for nicotine or for the toxin were averaged as control current and test current respectively. The curves of concentration-dependent inhibition were fitted according to the median effect equation [20].
Cytotoxity assay
HUVEC (human umbilical-vein endothelial cells) and L929 cells were maintained respectively in glucose-supplemented Dulbecco's modified and in RPMI 1640 medium containing 10% (v/v) Cosmic calf serum (HyClone) under monolayer conditions. The loss of ability of the cells to oxidize MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] was used as a measure of the toxin's effect on cell viability [21]. The IC50 (50% inhibitory concentration of toxins; μg/ml) is defined as the amount of toxins needed to inhibit 50% viability of cells compared with the negative control. The median effect equation previously described was used to calculate the IC50 value.
RESULTS
cDNA sequence analysis
The complete sequences of 19 cDNAs with GenBank® accession numbers from DQ273567 to DQ273585 were obtained from 137 clones and named on the basis of the NCBI (National Center for Biotechnology Information) Blastx results. These novel genes encoded for six kinds of three-finger toxins: LNTXs (five isoforms), SNTXs (four isoforms), WNTXs (two isoforms), MTXs (two isoforms), CTXs (five isoforms) and a neurotoxin with a free SH group.
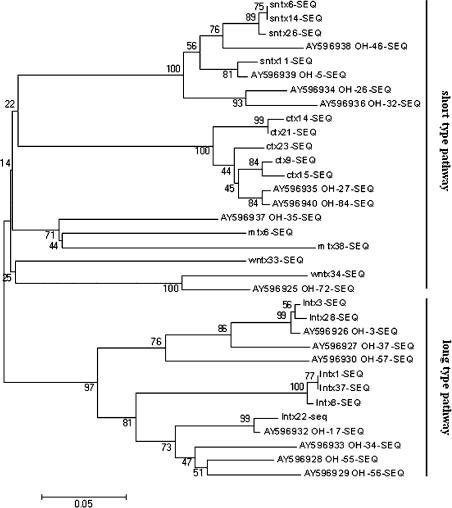
Phylogenetic analysis of the cDNAs obtained, and of published cDNAs also amplified from O. hannah, suggested that three-finger toxin genes in O. hannah diverged early in the course of evolution by long-type (70–73 residues) and short-type (57–65 residues) pathways (Figure 1). SNTXs, CTXs, MTXs and WNTXs belonging to the short-type-pathway group evolved with LNTXs belonging to the long-type-pathway from a common ancestor. This was suggested by Fujimi et al. [22], who explained the molecular evolution of snake-toxin genes by the analysis of intron sequences. Among genes in the short-type pathway toxins, SNTXs and CTXs showed a greater divergence at a later stage of evolution than did MTXs and WNTXs, and so did LNTXs among the long-type-pathway toxins. It appears that SNTXs, CTXs and LNTXs genes were relatively active in the evolution of three-finger toxins in O. hannah. Although WNTXs have a similar peptide length, and the same numbers of disulphide bridges, as LNTXs, sequences coding for WNTXs were phylogenetically closer to the SNTXs group than to the LNTXs group.
Figure 1. Phylogenetic analysis of O. hannah cDNAs obtained in the present study and of already registered cDNAs.
Multiple sequences were aligned using CLUSTALW (version 1.83). The tree was reconstructed using distance methods of Neighbour-joining on the basis of the Poisson-corrected amino acid distance. The reliability of clusters was estimated using a Bootstrap test. The GenBank® accession numbers of cDNAs whose sequences have already been published are indicated. The scale bar means 0.05 amino acid substitution per site.
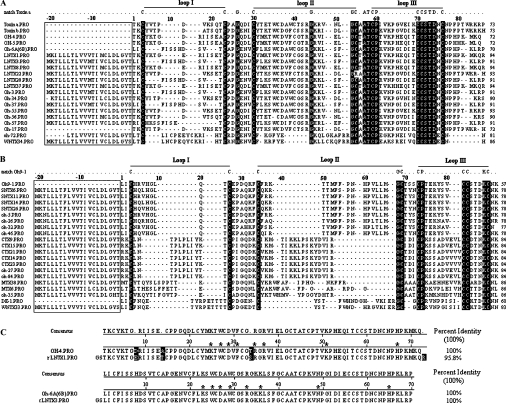
The hypothetical proteins corresponding to the cDNAs obtained were compared with sequence-published toxins from O. hannah and classified into two groups on the basis of numbers of disulphide bridges. Six LNTXs and WNTX34 formed group A and all retained ten conserved cysteine residues (Figure 2A). Four SNTXs, five CTXs, two MTXs and WNTX33 formed group B and all had eight conserved cysteine residues (Figure 2B). Both groups have a highly conserved 21-residue hydrophobic signal peptide. The putative mature peptides of six LNTXs comprise 70–73 amino acids, whereas WNTX34 comprises 65. The fifth disulphide bridge of WNTX34 is located in loop I, whereas that of LNTXs is located in loop II. This is the structural basis with which to classify WNTX34 as a weak neurotoxin. Although WNTX33 consists of 62 residues and four disulphide bridges, just like SNTXs, and lacks the identifying disulphide of WNTX in loop I, it has 85% identity (53/62) and 90% positive homology (56/62) with the weak toxin DE-1 (Swiss-Prot. number P01412). On the basis of these results, we term the protein ‘WNTX33’ rather than ‘SNTX33’.
Figure 2. Multiple sequence alignments of deduced amino acids with those of toxins from O. hannah.
Natural peptides for which sequences have already been published were Toxin a (Swiss-Prot. number P01387), Toxin b (P01386), OH-4 (P80516), OH-5 (P80965), Oh-6A/6B (P82662), Oh9-1 (P83302) and DE-1 (P01412). The sequences of Oh-3, -5, -17, -26, -27, -32, -34, -35, -37, -46, -55, -56, -57 and -84 were deduced from their corresponding cDNAs with GenBank® accession numbers from AY596925 to AY596940 [23]. Signal peptides are boxed. Amino acid residues that matched the template residues in the first line of each group reversed-out (i.e. white-on-black) in (A) and (B), whereas residues reversed-out in (C) are unmatched ones. The last number in each row indicates the numbers of residues in each toxin. (A) Group A toxins having ten conserved cysteine residues (five disulphide bridges). (B) Group B toxins having eight conserved cysteine residues (four disulphide bridges). (C) Alignments of rLNTX1 and rLNTX3 to their corresponding natural proteins. Functional residues at homologous sites are indicated with asterisks (*).
LNTX22 is an interesting toxin, having 83% identity with toxin b (P01386), one of the first two LNTXs purified from O. hannah. Arg42 replaces Cys42 in LNTX22, which is conserved in other LNTXs and participates in forming the disulphide bridge of loop II [7]. To date, only schistosa 4 and schistosa 5, two principle neurotoxins purified from Enhydrina schistosa (beaked sea snake), have the same free thiol group as LNTX22 [24]. These toxins are the most unusual members of the neurotoxin group and, because of their unique structural characteristics, may constitute a new subfamily of three-finger proteins.
Because LNTX3 and LNTX1 show high similarity to other known proteins (Figure 2C), and lntx1 was the most abundant gene among the 19 cDNAs, we expressed LNTX3 and LNTX1 for further research into their function.
Expression and purification of rLNTX3 and rLNTX1
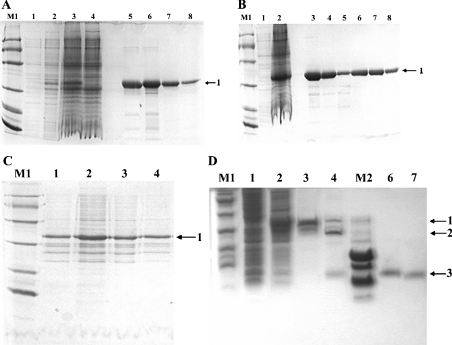
Tris/glycine/SDS/16%-(w/v)-PAGE analysis of total proteins extracted from uninduced and induced culture, together with a two-step purification of fusion proteins, are shown in Figure 3. Bands of about 34 kDa corresponding to fusion proteins (labelled ‘1’) appeared in the induced cultures. No significant difference in expression of the lntx3 gene was observed on increasing the incubation temperature to 37 °C (Figure 3A). Proteins obtained from the culture had more soluble forms in the lysate supernatant at 16 °C and with 0.1 mM IPTG (Figure 3C). Thrombin was used to cleave the GST (glutathione S-transferase) tag from the fusion protein (Figure 3D). The released recombinant peptides were further purified on a benzamidine FF column placed in series below the GSTrap FF column (Figure 4).
Figure 3. SDS/PAGE analysis of rLNTX3 and rLNTX1.
(A)–(C) employed Tris/glycine/SDS/16%-(w/v)-PAGE and (D) employed Tris/tricine/SDS/16%-PAGE. Lane M1, protein markers of low molecular mass (kDa): 97, 66, 43, 31, 20 and 14; lane M2, peptide markers (kDa): 16, 14, 10, 0.8, 0.6 and 0.2; bands labelled 1, 2 and 3 are fusion protein, GST-tag and recombinant peptides respectively. (A) Total proteins of uninduced and induced cultures together with GST–LNTX3 obtained from each of the purification steps. Lanes 1 and 2, total proteins of culture at 37 °C after 0 and 4 h induction respectively; lanes 3 and 4, total proteins of culture at 16 °C after 0 and 20 h induction respectively; lanes 5 and 6, elution fractions from the affinity column; lanes 7 and 8, purified GST–LNTX3 from the Superdex75 column. (B) Total proteins of uninduced and induced culture together with GST–LNTX1 obtained from each of the purification steps. Lanes 1 and 2, total proteins of culture at 16 °C after 0 and 20 h induction respectively; lanes 3–5, elution fractions from the affinity column; lanes 7 and 8, purified GST–LNTX1 from the Superdex75 column. (C) Proteins from supernatant and pellet of lysate after the induction of IPTG for 20 h at 16 °C. Lanes 1 and 2, the pellet and supernatant of LNTX1 respectively; lanes 3 and 4, the pellet and supernatant of LNTX3 respectively. (D) Cleavage of fusion protein and purified recombinant peptides. Lanes 1 and 2, total proteins of uninduced and induced culture of LNTX1; lanes 3 and 4, GST–LNTX1 after being mixed with thrombin for 0 and 8 h; lanes 6 and 7, the purified rLNTX1 and rLNTX3 respectively.
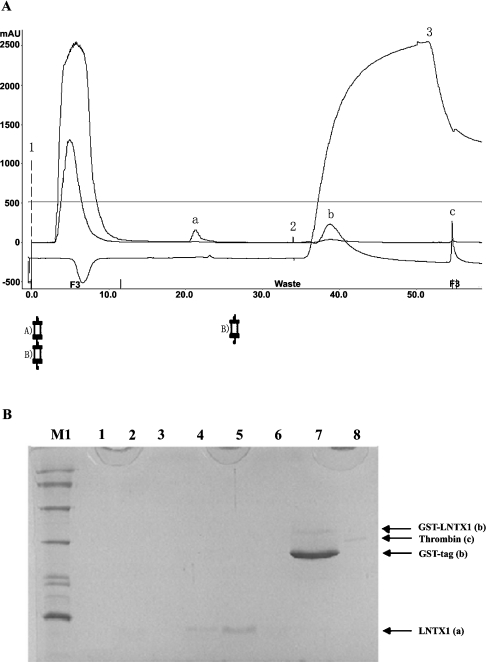
Figure 4. Purification of recombinant peptides on a benzamidine FF column placed in series below the GSTrap FF column.
(A) Chromatography of rLNTX1. Position 1 indicates the sample application. Positions 2 and 3 show the beginning of the high-salt elution and low-pH-buffer elution respectively. The high-salt-wash buffer contained 20 mM sodium phosphate and 1.0 M NaCl, pH 7.0; the low-pH-wash buffer contained 10 mM HCl and 0.5 M NaCl, pH2.0. Column A was a GSTrap FF 1-ml-volume column; column B was a HiTrap Benzamidine FF 1 ml column; ‘a’ is the released rLNTX1 peptide; ‘b’ is a mixture of GST-tag and some non-cleaved fusion proteins; ‘c’ is bovine thrombin. Abbreviation: mAU, milli-absorbance unit. (B) Tris/glycine/SDS/16%-PAGE analysis of each peak in the above chromatography. Lane M1, protein markers of low molecular mass; lanes 1–3, flow-through during sample loading; lanes 4–6, peak ‘a’ in (A); lane 7, peak ‘b’ in (A); lane 8, A chain (31 kDa) of bovine thrombin (‘c’ in A).
Characterization of rLNTX1 and rLNTX3
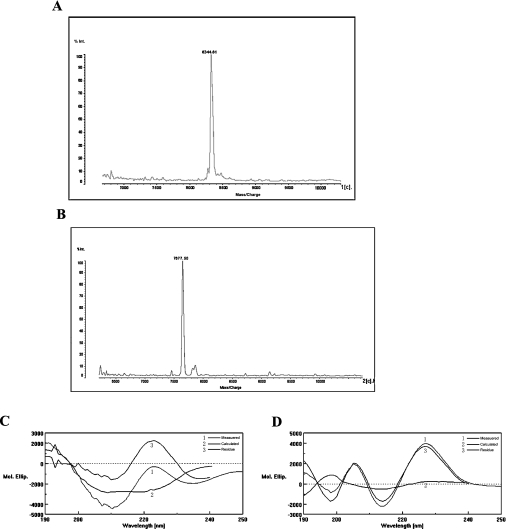
Both proteins existed as monomers according to SEC–HPLC chromatography, with molecular masses of 8343.68 Da and 7676.87 Da respectively as assessed by MALDI–TOF-MS (Figures 5A and 5B). These values corresponded to those calculated on the basis of amino acid sequence. Ten amino acids at the N-terminus of rLNTX1 and rLNTX3 corresponded to the expected amino acids in those positions, with the first two amino acid residues being Gly-Ser and the remainder in the cleavage site of thrombin.
Figure 5. Characterizations of rLNTX1 and rLNTX3.
(A) and (B) MALDI–TOF-MS of rLNTX1 and rLNTX3, respectively. (C) and (D) CD spectra showing the molecular ellipticity (Mol. Ellip.) of rLNTX1 and rLNTX3 respectively.
In the secondary structures of rLNTX1 and rLNTX3 determined by CD spectroscopy (Figures 5C and 5D), the negative ellipticity extrema near 200 and 215 nm indicated a major β-sheet conformation with little random-coil structure and little α-helix, which corresponded to the conformation of known α-neurotoxins [25].
Bioassays
LD50 values for rLNTX1 and rLNTX3 are listed in Table 1. During the in vivo toxicity test (LD50), we found that, when rLNTX1 was injected at the high dose of 0.7 mg/kg, mice became quiet, sluggish and showed deep and rapid breathing. Most of the mice died within 10 min, and some excreted bloody urine. Dissection of the mice excreting bloody urine showed no necrosis, but some haemorrhage points in kidneys and lungs as compared with other mice. The phenomenon of bloody urine was not observed in mice injected with rLNTX3, but, during the dissection of dead mice injected with rLNTX3, the same hemorrhage points at the surface of lungs were observed in several mice. This was an unusual observation in the α-neurotoxin toxicity test.
Table 1. Comparison of some pharmaceutical functions of the toxins.
| Cytotoxicity (IC50, μg/ml) | ||||
|---|---|---|---|---|
| Toxin | LD50 (mg/kg) | Patch clamp (IC50, μM) | HUVEC | L929 cells |
| rLNTX1 | 0.5114 | 0.763 | 1.58 | 0.31 |
| Oh-4 | 0.25* | − | − | − |
| rLNTX3 | 0.4150 | 0.385 | 0.84 | 0.30 |
| Oh-6A/6B | 0.17/0.14* | − | − | − |
| Crude venom | − | − | 4.2 | 1.8 |
| Cobrotoxin | − | − | >58 | >58 |
* Data from [26].
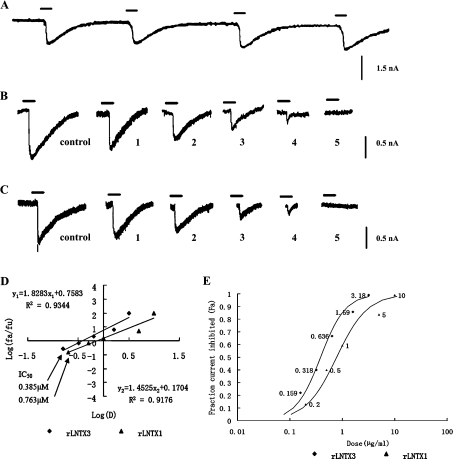
A whole-cell patch-clamp technique was used to test the neurotoxic activity of the recombinant peptides. Nicotine at 80 μM was chosen to excite the repeatable control current (Figure 6A). As shown in Figure 6B, trace 5, the presence of 3.18 μM rLNTX3 caused an almost complete block in the nicotine-evoked current. After an interval of 5 min, 80 μM nicotine was applied again, but the current could not be restored. The same phenomenon was observed when rLNTX1 was added to the cell at a concentration of 10 μM (Figure 6C, trace 5). The IC50 values for rLNTX3 and rLNTX1 were 0.385 and 0.763 μM (Figure 6D) respectively, which was similar to that of two long-chain neurotoxins: P9-CTX from O. hannah and α-bungarotoxin from Bungarus multicinctus (many-banded krait) [27,28]. The value for the latter was about twice as high as that of the former. The results of their in vivo lethality determination corresponded with the LD50 value for rLNTX1 being higher than that for rLNTX3. The results of whole-cell patch-clamp experiment showed that these recombinant peptides could lead to a functional block of the muscle-type AChRs and that they could be classified as α-neurotoxins.
Figure 6. Inhibitory effects of rLNTX1 and rLNTX3 on nAChR-enriched skeletal myocytes.
Cells were held throughout the experiment at −70 mV. (A) Reproducible control currents excited by 80 μM nicotine. (B) Currents inhibited by rLNTX3 at a concentration of: 1, 0.159 μM; 2, 0.318 μM; 3, 0.636 μM; 4,1.59 μM; and 5, 3.18 μM. (C) Currents inhibited by rLNTX1 at a concentration of: 1, 0.20 μM; 2, 0.50 μM; 3, 1.00 μM; 4, 5.00 μM; and 5, 10.00 μM. (D) Analysis of the data by the median-effect plot. The median effect concentrations (IC50) of each peptide are marked. (E) Fraction of inhibited current to control current (fa) as functions of recombinant peptides at a series of concentrations (labelled beside the data points).
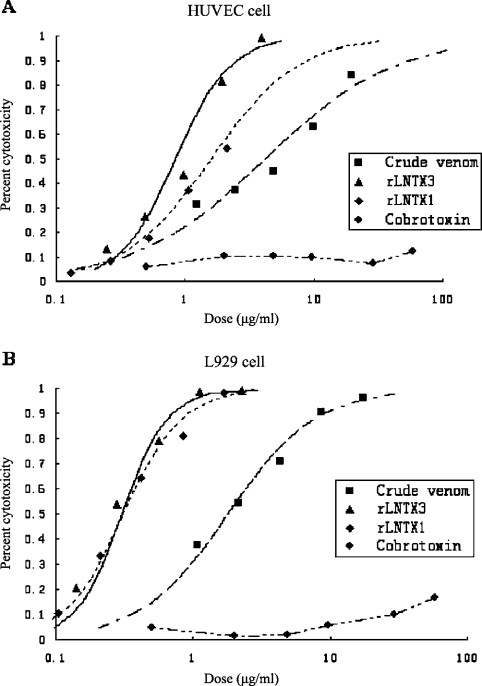
The MTT assay was used to measure the effect of two peptides on cell viability. Crude venom and cobrotoxin, an SNTX from Naja naja atra (Taiwan cobra), were compared with rLNTX1 and rLNTX3. To date there are no published reports that cobrotoxin has cytolytic activity.
In general, both cell types were susceptible to crude venom, rLNTX1 and rLNTX3. However, the recombinant peptides had more cytolytic effect than did the crude venom (Figure 7). Furthermore, each showed more potent cytotoxicity towards L929 than to HUVEC. rLNTX3 was about twice as cytotoxic to HUVEC than was rLNTX1. However, they showed almost equal cytotoxicity towards L929 cells. The IC50 values for rLNTX1, rLNTX3, crude venom and cobrotoxin are listed in Table 1. When observed under the microscope (×400) at 24 h after administration of toxins, the cells treated with crude venom at a middle/high dose shrunk to a darker dot, while the membranes of cells were intact. A portion of the cells treated with rLNTX1 and rLNTX3 at middle/high dose were unable to retain an intact membrane, and much cell debris was observed. This indicated that the mechanism of lethality to cells might be different. The last curves in Figures 7(A) and 7(B) showed the cytolytic effect of cobrotoxin. At a comparable concentration, cobrotoxin was not able to kill either cell type (cytotoxicity <15%).
Figure 7. Lethal effect of crude venom, rLNTX1, rLNTX3 and Cbtx on HUVEC and L929 cells.
DISCUSSION
It is known that LNTXs are dual-function peptides that are able to bind to both muscular and neuronal nAChRs [29]. It has been suggested that eight functionally conserved residues are necessary for binding to nAChRs. Trp25, Asp27, Phe29, Arg33, Arg36 and Phe65 are involved in binding to both types of nAChRs, whereas Lys23 and Lys49 are required only for muscular nAChRs [8,30]. rLNTX1 has all of the eight residues at homologous positions, with the exception of Phe65, which is replaced by His67, whereas, rLNTX3 has intriguing substitutions at four of the above eight homologous positions: a Trp29 substitution for Phe29, Lys36 for Arg36, Asn49 for Lys49 and His65 for Phe65 (Figure 2C).
Phe29 is a very important residue protruding from loop II (middle finger) of the toxin and filling the binding pocket through aromatic interactions with Tyr185 of nAChRs [9]. Remarkably, Trp29 occupies the position of Phe29 in rLNTX3. Although both tryptophan and phenylalanine are aromatic amino acids, the indole ring of tryptophan provides more potent electron density than does the sole phenyl ring of phenylalanine, which may enhance the aromatic interaction with Tyr185 of the receptor. It suggests that the affinity of rLNTX3 to nAChRs is likely to be higher than that of rLNTX1. Our electrophysiological experiments support this supposition (Figures 6D and 6E).
Another obvious difference between the functional residues of rLNTX1 and rLNTX3 is that Asn49 replaces Lys49 at the tip of loop III in rLNTX3. Lys49 is an important residue required for LNTX binding to muscular nAChRs. In α-Cbtx (α-cobratoxin), an LNTX from Naja kaouthia (monocled cobra), Antil et al. [31] mutated Lys49 to Glu49. The mutation caused a 53-fold decrease in the binding affinity of α-Cbtx to muscular nAChRs. The authors concluded that a positive charge at this position is critical for binding. Nevertheless, Asn49 occupies the position of Lys49 in rLNTX3. Our patch-clamp experiments showed that the ability of rLNTX3 to inhibit the current evoked by nicotine is almost twice as high as that of rLNTX1 (Figure 6D). It is known that asparagine plays a special role in forming the two-dimensional structure of proteins, for Oγ in its side chain always forms an H-bond with NHn+3 in the main chain, which could change the orientation of the main chain (the subscript n+3 means the third residue behind asparagine in the backbone peptide). Looking at the structure of LNTXs, we can see that Asn49 occupies the position next to the tip of loop III [31]. One can conjecture that Asn49 would be helpful in maintaining the tip of loop III by forming an H-bond with the NHn+3 of Ile52, and the positive charge at this site needed for binding is provided by the nearby Lys47. Anyway, site-directed-mutagenesis experiments are required to elucidate the precise function of Asn49 and Phe29.
Our experiments showed that rLNTX1 and rLNTX3 were able to functionally block the muscle-type nAChRs in the same way as SNTXs. Shelke et al. [15] isolated a postsynaptic SNTX with a cytotoxic function from D. r. russelli venom. It is an example of an SNTX having a lytic activity. Do these recombinant neurotoxins also have this ability?
In expressing stages, E. coli harbouring vector pGEX4T-1-lntx3 grew much more slowly than E. coli having blank vector pGEX4T-1. In the in vivo LD50 test, bloody urine and hemorrhage points were observed in some mice. It indicated that rLNTX1 and rLNTX3 might have cytotoxic abilities. The results of testing on HUVEC and L929 cells showed that both of them have a more potent cytotoxic activity than has crude venom and cobrotoxin (an SNTX with no known lytic activity). Further analysis of cytolytic-function-related residues, comparing sequences of rLNTX1 and rLNTX3 with those of CTXs, is difficult. The major reason is that the functional sites in CTXs are still a matter of controversy [7,32]. However, the presence of both a cationic and a hydrophobic site would seem to be essential for lytic activity [7].
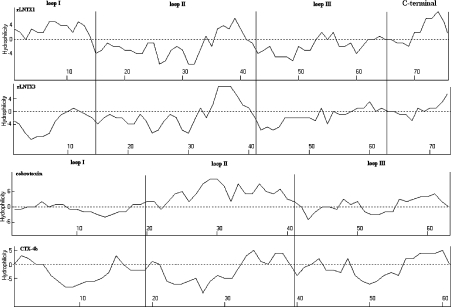
Figure 8 shows a hydrophilicity analysis of rLNTX1, rLNTX3, cobrotoxin and CTX-4b. CTX-4b is a CTX from Naja Naja sputatrix (Southern Indonesian spitting cobra) having the greatest cytolytic activity in its isoforms [33]. The hydrophobic residues distribute in the three loops of CTX-4b as clusters. rLNTX3 has a hydropathy profile very similar to that of CTX-4b, except that the hydrophobic residues located in loop II show less of a tendency, compared with CTX-4b, to get clustered. Nevertheless, rLNTX1 has different hydropathic features, for the loop I of rLNTX1 is full of hydrophilic residues, and its hydrophobic residues in other loops are also fewer in number than those in rLNTX3. These observations correspond to the experimental finding that the cytolytic ability of rLNTX1 is less than that of rLNTX3. An obvious fact is that a predominant distribution of hydrophilic residues throughout three loops of cobrotoxin, especially in loop II. This is quite different from rLNTX1, rLNTX3 and CTX-4b. an NMR solution study and a graphic modelling study of CTX indicated that loop II plays an important role in its toxic action [34]. Thus the broad distribution of hydrophobic residues on the three loops of recombinant LNTXs might be one of reasons for their extra cytolytic activity.
Figure 8. Hydropathy profiles for rLNTX1, rLNTX3, Cbtx and CTX-4b.
Hydropathy profiles were analysed according to the Kyte–Doolittle method [35]. Values above the axis denote hydrophilic regions, whereas those below the axis indicate hydrophobic regions. Analyses were carried out using ANTHEPROT V5.0 (Institut de Biologie et Chimie des Protéines, Lyon, France).
Acknowledgments
We thank Dr Chenggang Liang for his great help in collecting the venom glands and insightful opinions on the whole experiment, Dr Fawei He and Mrs Xuechun Luo for performing the CD spectroscopy, and Dr Sheng Yang for completing the whole-cell patch-clamp experiments.
References
- 1.Ricciardi A., le Du M.-H., Khayati M., Dajas F., Jean-Claude B., Menez A., Ducancel F. Do structural deviations between toxins adopting the same fold reflect functional differences? J. Biol. Chem. 2000;275:18302–18310. doi: 10.1074/jbc.275.24.18302. [DOI] [PubMed] [Google Scholar]
- 2.Hall Z. W. α-Neurotoxins and their relatives: foes and friends? Neuron. 1999;23:4–5. doi: 10.1016/s0896-6273(00)80745-x. [DOI] [PubMed] [Google Scholar]
- 3.Jerusalinsky D., Harvey A. L. Toxins from mamba venoms: small proteins with selectivities for different subtypes of muscarinic acetylcholine receptors. Trends Pharmacol. Sci. 1994;15:424–430. doi: 10.1016/0165-6147(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 4.Utkin Y. N., Kukhtina V. V., Kryukva E. V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V. I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of α7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001;276:15810–15815. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 5.Harvey A. L. Cardiotoxins from cobra venoms: possible mechanism of action. J. Toxicol. Toxin Rev. 1985;4:41–69. [Google Scholar]
- 6.Qin G. P. 2nd edn. Guangxi: Guangxi Scientific and Technical Publishers; 1998. Chinese poisonous snake research; pp. 44–121. [Google Scholar]
- 7.Rees B., Bilwes A. Three-dimensional structures of neurotoxins and cardiotoxins. Chem. Res. Toxicol. 1993;6:385–406. doi: 10.1021/tx00034a001. [DOI] [PubMed] [Google Scholar]
- 8.Antl S., Servent D., Menez A. Variability among the sites by which curaremimetic toxins bind to Torpedo acetylcholine receptor, as revealed by identification of the functional residues of α-cobratoxin. J. Biol. Chem. 1999;274:34851–34858. doi: 10.1074/jbc.274.49.34851. [DOI] [PubMed] [Google Scholar]
- 9.Bourne Y., Talley T. T., Hansen S. B., Taylor P., Marchot P. Crystal structure of a Cbtx–AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nirthanan S., Gopalakrishnakone P., Gwee M. C., Khoo H. E., Kini R. M. Non-conventional toxins from elapid venoms. Toxin. 2003;41:397–407. doi: 10.1016/s0041-0101(02)00388-4. [DOI] [PubMed] [Google Scholar]
- 11.Yang C. C. Chemistry and evolution of toxins in snake venoms. Toxcion. 1974;12:1–43. doi: 10.1016/0041-0101(74)90096-8. [DOI] [PubMed] [Google Scholar]
- 12.Strydom D. J. The evolution of toxins found in snake venoms. In: Lee C. Y., editor. Snake Venom: Handbook of Experimental Pharmacology. Berlin: Springer-Verlag; 1979. pp. 258–275. [Google Scholar]
- 13.Yee Phui J. S., Gong N., Afifiyan F., Ma D., Lay P. S., Armugam A., Jeyaseelan K. Snake postsynaptic neurotoxins: gene structure, phylogeny and applications in research and therapy. Biochimie. 2004;86:137–149. doi: 10.1016/j.biochi.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Gong N., Armugam A., Jeyaseelan K. Molecular cloning, characterization and evolution of the gene encoding a new group of short-chain α-neurotoxins in an Australian elapid, Pseudonaja textiles. FEBS Lett. 2000;473:303–310. doi: 10.1016/s0014-5793(00)01549-0. [DOI] [PubMed] [Google Scholar]
- 15.Shelke R. R. J., Sathish S., Gowda T. V. Isolation and characterization of a novel postsynaptic/cytotoxic neurotoxin from Daboia russelli russelli venom. J. Peptide Res. 2002;59:257–263. doi: 10.1034/j.1399-3011.2002.02969.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H., Meng Q. X., Xie L. P., Zhang R. Q. A weak neurotoxin exhibits synergic effect with cardiotoxins when co-applied to two non-neural cell lines. Biol. Pharm. Bull. 2004;27:1241–1244. doi: 10.1248/bpb.27.1241. [DOI] [PubMed] [Google Scholar]
- 17.Sivaraman T., Kumar T. K., Jayaraman G., Han C. C., Yu C. Characterization of a partially structured state in an all-β-sheet protein. Biochem. J. 1997;321:457–464. doi: 10.1042/bj3210457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Organisation for Economic Co-operation and Development (OECD) Paris, France: OECD; 2001. Guideline 425: Acute Oral Toxicity – Up-and-Down Procedure OECD guidelines for testing of chemicals. [Google Scholar]
- 19.Yaffe D. Rat skeletal muscle cells. In: Kruse P. F., Patterson M. K., editors. Tissue Culture: Methods and Applications. London: Academic press; 1974. pp. 106–114. [Google Scholar]
- 20.Chou T. C., Hayball M. Cambridge, U.K.: Biosoft; 1996. CalcuSyn for Windows 3.1 and 95: Multiple Dose Effect Analyzer and Manual for IBM-PC. [Google Scholar]
- 21.Carmichael J., DeGraff W. G., Gazdar A. F., Minna J. D., Mitchell J. B. Evaluation of a tetrazolium-based semi automated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 22.Fujimi T. J., Nakajyo T., Nishimura E., Ogura E., Tsuchiya T., Tamiya T. Molecular evolution and diversification of snake toxin genes, revealed by analysis of intron sequences. Gene. 2003;313:111–118. doi: 10.1016/s0378-1119(03)00637-1. [DOI] [PubMed] [Google Scholar]
- 23.He Y. Y., Lee W. H., Zhang Y. Cloning and purification of α-neurotoxins from king cobra (Ophiophagus Hannah) Toxicon. 2004;44:295–303. doi: 10.1016/j.toxicon.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Fryklund L., Eaker D., Karlsson E. Amino acid sequences of the two principal neurotoxins of Enhydrina Schistosa venom. Biochemistry. 1972;11(24):4633–4640. doi: 10.1021/bi00774a034. [DOI] [PubMed] [Google Scholar]
- 25.Lin S. R., Leu L. F., Chang L. S., Chang C. C. Amino acid sequence and chemical modification of a novel α-neurotoxins (Oh-5) from King Cobra (Ophiophagus hannah) Venom. J. Biochem. (Tokyo) 1997;121:690–695. doi: 10.1093/oxfordjournals.jbchem.a021641. [DOI] [PubMed] [Google Scholar]
- 26.Lin S. R., Chang L. S., Chang C. C. Disulfide isomers of α-neurotoxins from king cobra (Ophiophagus hannah) venom. Biochem. Biophys. Res. Commun. 1999;254:104–108. doi: 10.1006/bbrc.1998.9888. [DOI] [PubMed] [Google Scholar]
- 27.Dong Y., Tang Y. G., Jiang W. L., Sun Z. F., Huang Y., Xie Z. P. The inhibition effect of purified P9-CTX on nACh receptor. Chin. J. Appl. Physiol. 1995;11:113–116. [Google Scholar]
- 28.Xie Z. P., Wang D., Zhu Y. Z. Research of whole-cell patch clamp on nicotinic neuron of Xenopus embryo. Sci. Sin. B. 1988;11:1169–1173. [Google Scholar]
- 29.Antil-Delbeke S., Gaillard C., Tamiya T., Corringer P. J., Changeux J. P., Servent D., Menez A. Molecular determinants by which a long chain toxin from snake venom interacts with the neuronal α7 nicotinic acetylcholine receptor. J. Biol. Chem. 2000;275:9594–9601. doi: 10.1074/jbc.M909746199. [DOI] [PubMed] [Google Scholar]
- 30.Malany S., Osaka H., Sine S. M., Taylor P. Orientation of a-neurotoxin at the subunit interfaces of the nicotinic acetylcholine receptor. Biochemistry. 2000;39:15388–15398. doi: 10.1021/bi001825o. [DOI] [PubMed] [Google Scholar]
- 31.Antil S., Servent D., Ménez A. Variability among the sites by which curaremimetic toxins bind to Torpedo acetylcholine receptor, as revealed by identification of the functional residues of α-cobratoxin. J. Biol. Chem. 1999;274:34851–34858. doi: 10.1074/jbc.274.49.34851. [DOI] [PubMed] [Google Scholar]
- 32.Kini R. M. Invited paper: Animal toxins of Asia and Australia. Clin. Exp. Pharmacol. Physiol. 2002;29:815–823. [Google Scholar]
- 33.Jeyaseelan K., Armugan A., Lachumanan R., Tan C. H., Tan N. H. Six isoforms of cardiotoxin in Malayan spitting cobra (Naja naja sputatrix) venom: cloning and characterization of cDNAs. Biochim. Biophs. Acta. 1998;1380:209–222. doi: 10.1016/s0304-4165(97)00143-8. [DOI] [PubMed] [Google Scholar]
- 34.Chiou S. H., Hung C. C., Huang H. C., Chen S. T., Wang K. T., Yang C. C. Sequence comparison and computer modeling of cardiotoxins and cobrotoxin isolated from Taiwan cobra. Biochem. Biophys. Res. Commun. 1995;206:22–32. doi: 10.1006/bbrc.1995.1004. [DOI] [PubMed] [Google Scholar]
- 35.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]