Abstract
Budded virions (BV) of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) contain a major envelope glycoprotein known as GP64, which was previously shown to be palmitoylated. In the present study, we used truncation and amino acid substitution mutations to map the palmitoylation site to cysteine residue 503. Palmitoylation of GP64 was not detected when Cys503 was replaced with alanine or serine. Palmitoylation-minus forms of GP64 were used to replace wild-type GP64 in AcMNPV, and these viruses were used to examine potential functions of GP64 palmitoylation in the context of the infection cycle. Analysis by immunoprecipitation and cell surface studies revealed that palmitoylation of GP64 did not affect GP64 synthesis or its transport to the cell surface in Sf9 cells. GP64 proteins lacking palmitoylation also mediated low-pH-triggered membrane fusion in a manner indistinguishable from that of wild-type GP64. Cells infected with viruses expressing palmitoylation-minus forms of GP64 produced infectious virions at levels similar to those from cells infected with wild-type AcMNPV. In combination, these data suggest that virus entry and exit in Sf9 cells were not significantly affected by GP64 palmitoylation. To determine whether GP64 palmitoylation affected the association of GP64 with membrane microdomains, the potential association of GP64 with lipid raft microdomains was examined. These experiments showed that: (i) AcMNPV-infected Sf9 cell membranes contain lipid raft microdomains, (ii) GP64 association with lipid rafts was not detected in infected Sf9 cells, and (iii) GP64 palmitoylation did not affect the apparent exclusion of GP64 from lipid raft microdomains.
The Baculoviridae are a family of large, enveloped, double-stranded DNA viruses with rod-shaped nucleocapsids. Historically, these viruses have been used as agents for biological control of certain insect pest species and as important protein expression vectors. More recently, certain properties of baculoviruses have stimulated an interest in their use as potential agents for gene therapy. The best-studied baculoviruses (Autographa californica multicapsid nucleopolyhedrovirus [AcMNPV], Orgyia pseudotsugata MNPV [OpMNPV], and Bombyx mori NPV [BmNPV]) are closely related and are classified within the so-called “group I NPVs” of this virus family. In the baculovirus life cycle, two types of infectious virus particles (virion phenotypes) are generated. During the very late phase of infection, nucleocapsids are enveloped within the nucleus, and these virions are subsequently embedded within a viral occlusion body protein. These occluded virions are referred to as “occlusion-derived virions” (ODV). Occlusion bodies are highly stable in the environment, but when consumed by a susceptible insect host, they disassemble in the midgut and ODV are released. ODV are highly infectious to midgut epithelial cells but are not highly infectious to other tissues. The second virion phenotype is formed during the late phase when progeny nucleocapsids are transported to the plasma membrane, where they bud from the cell surface to generate budded virions (BV). BV of AcMNPV appear to be highly promiscuous and may enter many cell types in the infected animal. BV thus serve to disseminate infection from cell to cell within the infected animal. Laboratory studies have shown that AcMNPV BV can enter many types of cultured cells, including many mammalian cells (18, 39). Thus, in recent years, there has been considerable interest in cellular entry by BV and the role of the major envelope protein.
Budded virions of group I NPVs such as AcMNPV contain a highly abundant envelope protein known as GP64, which is required for virion entry and exit. GP64 is initially involved in virion attachment at the cell surface (9). After uptake of BV by receptor-mediated endocytosis, GP64 mediates a pH-triggered membrane fusion event that releases the nucleocapsid into the cytoplasm (4, 7, 17, 19, 23, 26, 43). The GP64 protein is also necessary for egress of the virus from infected cells. Deletion of the gp64 gene from the AcMNPV genome demonstrated that the GP64 protein is essential (27). GP64-null viruses exhibited an approximately 98% reduction in virion budding (30), indicating that in addition to its essential role in virus entry, GP64 is necessary for efficient budding and production of infectious virions. GP64 is a type I integral membrane protein that is present on the infected cell surface and in the virion as a trimer (31). Homotrimers of GP64 are associated by both intra- and intermolecular disulfide bonds, and these trimers appear to comprise the spike structures observed at the ends of BV and at the sites of BV budding (43). Studies of the synthesis of the OpMNPV GP64 protein show that GP64 is trimerized rapidly (within approximately 15 min after synthesis), and monomeric GP64 appears to be degraded within the cell (31). GP64 is also extensively modified posttranslationally. In OpMNPV-infected cells, GP64 is rapidly glycosylated, but subsequent processing of carbohydrates occurs over an extended period with halftimes of 45 to 75 min (31). However carbohydrate processing does not appear to be required for GP64 transport or function (15). The AcMNPV GP64 protein is glycosylated at four of the five predicted N-linked glycosylation sites (16), and no single N-linked glycosylation site is necessary for GP64 synthesis, transport, or association with BV. However, glycosylation at certain sites contributes to virion attachment to host cells (16). GP64 is also posttranslationally modified by phosphorylation (24, 41) and acylation (34). While little is known regarding GP64 phosphorylation, palmitate was identified as a covalently linked acyl moiety, and several sites near the transmembrane domain were suggested as possible sites for GP64 palmitoylation (34). In many cases, acylation of viral proteins affects protein function or association with specific membrane domains. In the present study, we used C-terminal truncations and amino acid substitutions of GP64 to identify the GP64 palmitoylation site. Genes encoding palmitoylation-minus forms of GP64 were inserted into a gp64-null AcMNPV virus genome to generate recombinant viruses. Using these viruses, we compared the activities of wild-type and palmitoylation-minus forms of GP64 proteins in the context of a viral infection. We found that GP64 palmitoylation is not necessary for GP64 synthesis or transport, BV production or infectivity, or membrane fusion activity. We also examined Sf9 cells for the presence of membrane microdomains known as lipid rafts and examined the localization of GP64 in cell membranes. Lipid rafts are regions of cell plasma membranes that are rich in sphingolipid and cholesterol and are believed to exist in a state similar to a liquid-ordered (lo) phase described in artificial membranes (6). The high concentrations of cholesterol and long-chain saturated fatty acids in lipid raft regions are believed to result in increased bilayer thickness and lower fluidity of proteins and phospholipids. Many, although not all, palmitoylated proteins are found in lipid rafts. A number of important viral envelope proteins are palmitoylated and are associated with lipid raft domains, including the influenza virus hemagglutinin (HA) protein and the human immunodeficiency virus type 1 (HIV-1) gp120/41 envelope protein. In those cases, lipid rafts appear to be important for viral budding (29, 37, 38). In this study, we identified lipid raft microdomains in AcMNPV-infected Sf9 cells. However, GP64 was not detected in lipid rafts, and GP64 palmitoylation did not affect the apparent exclusion of GP64 from lipid rafts.
MATERIALS AND METHODS
Cells, transfections, and infections.
Spodoptera frugiperda Sf9 cells were maintained in complete TNMFH medium (TNMFH medium plus 10% fetal bovine serum) at 27°C (11). For transient transfections, Sf9 cells (9 × 105) were plated in each well of 35-mm-diameter six-well plates, and 10 μg of plasmid or bacmid was used to transfect cells for 4 to 5 h as described previously (2-4). For viral infection, virus was incubated on cells for 1 h, and afterwards cells were washed three times in TNMFH. Times postinfection (p.i.) were calculated from the end of the 1-h viral absorption period.
Drosophila melanogaster S2 cells were maintained in complete Schneider's medium (Invitrogen) plus 10% fetal bovine serum (FBS) at 27°C. For transient transfections, S2 cells (3 × 106) were plated, allowed to grow for 6 to 16 h, and then transfected with plasmid pRmHa3-FasI as described previously for Sf9 cells (2-4). At 24 h posttransfection, copper sulfate was added to a final concentration of 500 μM to induce protein expression, and cells were harvested 24 h after induction.
Site-specific mutagenesis and construction of bacmids.
Modified gp64 genes were generated from plasmid p166B+1AcSpe/Bgl (26) by site-directed mutagenesis with the QuikChange site-directed mutagenesis kit (Stratagene). The following oligonucleotides were used to generate substitutions at cysteine residue 503: C503A+ (5′-GTGATTTTATTTTTGTACgcTATGATTcGAAACCGTAATAGAC-3′), C503A− (5′-GTCTATTACGGTTTCgAATCATAgcGTACAAAAATAAAATCAC-3′), C503S+ (5′-GTGATTTTATTTTTGTACTcTATGATTcGAAACCGTAATAGAC-3′), and C503S− (5′-GTCTATTACGGTTTCgAATCATAgAGTACAAAAATAAAATCAC-3′). Modified codons are underlined, and nucleotide changes are indicated in lowercase. A TfiI site (double underlined) was also introduced by adding a single nonsense nucleotide change. The C-terminal region of the mutated gp64 open reading frame (ORF) was excised as a 540-bp restriction fragment by digestion with AatII and SpeI. The C-terminal region of the wild-type gp64 ORF in the Fastbac vector pdFB/gus(R)Acgp64 (21) was subsequently removed by AatII and AvrII and then replaced with each of the modified C-terminal regions to generate mutant gp64 ORFs containing either an alanine substitution at cysteine 503 (pFB-gp64C503A) or a serine substitution at cysteine 503 (pFB-gp64C503S). The mutant gp64 genes were confirmed by restriction enzyme digestion and DNA sequencing.
To introduce the modified gp64 genes into the AcMNPV genome, each modified gp64 gene was introduced into a gp64-null bacmid. The gp64-null bacmid was originally generated by deleting the gp64 gene from the AcMNPV genome in bacmid bMON14272 (20) by homologous recombination in Escherichia coli (21). The modified gp64 genes were inserted into the polyhedrin locus of the gp64-null bacmid (AcMNPV gp64-null bacmid) by Tn7-mediated transposition as described previously (20). Viral DNAs from mutant viruses were purified, and the modified regions were sequenced to confirm the modified codon.
To construct a recombinant baculovirus expressing the D. melanogaster Fasciclin I (FasI) protein, the D. melanogaster FasI gene (47) was excised from plasmid pRmHa3-FasI (14) as an EcoRI fragment and used to replace the gp64 gene in vector pdFB/gus(R)Acgp64 (21). The construct was confirmed by restriction enzyme digestion and DNA sequencing across the cloning sites. The resulting construct contained the Drosophila FasI coding region under the transcriptional control of the AcMNPV gp64 promoter. The FasI gene was inserted into the polyhedrin locus of bacmid bMON14272 by Tn7-mediated transposition as described earlier.
Metabolic labeling and immunoprecipitation.
For [35S]methionine labeling of viral proteins, Sf9 cells (2 × 106) were infected at a multiplicity of infection (MOI) of 30, and at 34 h p.i., cells were starved by incubation in Grace's medium without methionine (Grace's − met) for 1.5 h. Cells were then incubated in 800 μl of the same medium containing 100 μCi of [35S]methionine (EasyTag EXPRE 35S protein labeling mix; 1,175 Ci/mmol; Perkin-Elmer) per well for 8 h. For [3H]palmitate labeling of viral proteins, cells were similarly infected and then starved for 1.5 h by incubation in Grace's medium without FBS. Viral proteins were then labeled by incubation of infected cells in 800 μl of Grace's medium containing 0.5% FBS and 500 μCi of [3H]palmitate per well. For immunoprecipitations, labeled Sf9 cells were lysed in 700 μl of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 8]) containing leupeptin (10 μg/ml) by agitation for 1 h at 4°C. Insoluble material was cleared by centrifugation (16,000 × g, 10 min), 50 μl of an anti-GP64 monoclonal antibody (MAb; a supernatant from hybridoma line AcV1) was added to the cleared supernatant, and the supernatant was then incubated for 1 h at 4°C, followed by addition of 50 μl of ImmunoPure Plus (G) immobilized protein G (Pierce) and incubation for 1 h at 4°C. Immunocomplexes were pelleted (16,000 × g, 10 min), washed twice with RIPA buffer, and then resuspended in 40 μl of 1× disruption buffer (125 mM Tris-HCl, 1% SDS, 2.5% mercaptoethanol, 10% glycerol, 0.2% bromophenol blue) and incubated at 100°C for 10 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) in 1.5-mm 12% polyacrylamide gels and then fixed in a 25% isopropanol-10% acetic acid solution for 30 min. Gels containing [3H]palmitate-labeled proteins were impregnated with Amplify (Amersham) for 30 min according to the manufacturer's instructions and then dried. After drying, gels containing [3H]palmitate-labeled proteins were used to expose Kodak BIOMAX film at −70°C for 60 to 180 days. Proteins labeled with [35S]methionine were detected on storage phosphor screens (Molecular Dynamics).
Western blot analysis, CELISA, and syncytium formation assays.
To detect GP64 or FasI by Western blot analysis, infected or transfected Sf9 cells (2 × 106) and D. melanogaster S2 cells (3 × 106) were washed with phosphate-buffered saline (PBS) before lysis in 1× disruption buffer and prepared as described previously (4, 22). Blots were incubated with either an anti-GP64 MAb (AcV5; 1:1,000 dilution) or an anti-FasI MAb (h5F7; 1:1,000 dilution) (13), and proteins were detected with an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. For analysis of cell surface localization of GP64 proteins by cell surface enzyme-linked immunosorbent assays (CELISA), transfected or infected cells were fixed in glutaraldehyde, and cell surface-localized GP64 was detected with MAb AcV5 as described previously (26). Briefly, infected cells were rinsed twice in PBS (pH 7.4) and fixed in 0.5% glutaraldehyde for 10 min at room temperature so that cells were not permeabilized. Fixed cells were washed once with PBS and blocked by incubation for 2 h in PBS containing 1% gelatin at 27°C. Cells were then incubated in MAb AcV5 (hybridoma culture supernatant diluted 1:25 in PBS containing 0.5% gelatin) for 45 min at 27°C. Cells were washed three times in PBS and then incubated in a secondary goat anti-mouse antibody conjugated to β-galactosidase (GAM-βgal; diluted 1:750 in PBS containing 0.5% gelatin) for 45 min at 27°C. Cells were washed four times in PBS and then incubated in 1 mM o-nitrophenyl-β-d-galactopyranoside (ONPG) in ONPG substrate buffer (10 mM Tris base, 10 mM NaCl, 10 mM MgCl2, 10 mM β-mercaptoethanol) at 37°C. Aliquots were removed, and absorbance at an optical density 405 nm (OD405) was determined at multiple time points. For comparisons of relative GP64 cell surface expression levels, averaged readings from triplicate wells containing cells expressing wild-type GP64 were assigned a value of 1.0.
For syncytium formation assays of membrane fusion activity, Sf9 cells were infected with each virus at an MOI of 10. At 36 h postinfection, TNMFH medium was removed, and cells were washed once with PBS at pH 7.4. The PBS at pH 7.4 was then replaced with PBS at various pHs from 5.0 to 6.0. After a 20-min incubation, cells were washed again with PBS at pH 7.4 and then returned to TNMFH. After a 4-h incubation at 27°C, cells were fixed with 2% paraformaldehyde in PBS for 10 min and scored for the presence of syncytia. The criterion for identification of syncytia was the presence of at least five nuclei.
Virus growth curves.
Sf9 cells were infected in triplicate with AcMNPV, vAcGP64wt, vAcGP64C503A, or vAcGP64C503S virus at an MOI of 5. After an initial 1-h infection period, cells were washed three times with TNMFH and then incubated in TNMFH at 27°C. Supernatants were collected from infected cells at the indicated time points. Data from each time point represent accumulated infectivity from infection through the indicated time. The titers of all supernatants were determined by 50% tissue culture infective dose (TCID50) assay on Sf9 cells.
Detergent extraction and membrane flotation assays.
For analysis of GP64 association with membrane microdomains, Sf9 (2 × 106) or S2 (3 × 106) cells were infected or transfected in 35-mm-diameter plates. At 72 h p.i. or posttransfection, cells were washed three times with PBS and then incubated in 1 ml of extraction buffer (0.5% Triton X-100, 10 mM PIPES, 0.1 M KCl, 3 mM MgCl2, 10 mM EDTA, 0.3 M sucrose [pH 7.5]) for 3 min at either 4, 27, or 37°C. The soluble and insoluble fractions were then separated by centrifugation at 10,000 × g for 10 min. Insoluble fractions (pellets) were dissolved in 40 μl of 1× disruption buffer at 100°C for 10 min and used for Western blot analysis. Proteins in the soluble fraction (supernatant) were precipitated by adding 0.5 ml of ice-cold 20% trichloroacetic acid (TCA) and incubation on ice for 30 min, followed by centrifugation (2,000 × g) for 20 min. To the precipitated material, 1 ml of ice-cold acetone was added. After brief mixing and then centrifugation (2,000 × g, 10 min), the resulting pellet was dissolved in 40 μl of 1× disruption buffer, incubated at 100°C for 10 min, and then used for Western blot analysis.
For membrane flotation assays, 5.3 × 106 Sf9 or S2 cells were transfected or infected (MOI of 10) in a T25 flask. At 72 h p.i. or posttransfection, cells were washed once in PBS and resuspended in 0.25 ml of PBS and 0.25 ml of ice-cold 2× lysis buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 2 mM sodium orthovandate, 60 mM sodium pyrophosphate · 10H2O, 20 mM sodium glycerophosphate, 0.04 U of aprotinin per ml, 0.02% sodium azide) containing 0.1% Triton X-100 and 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF). After a brief mixing, cells were lysed on ice for 10 min. An equal volume (0.5 ml) of 80% sucrose (wt/vol) was added to the lysate and mixed well, and then this mixture was overlaid with 2 ml of 30% sucrose and 1 ml of 5% sucrose. The step gradient was then centrifuged (250,000 × g) for 16 h. Fractions of 0.5 ml were collected from the top of the gradient, and proteins from each fraction were precipitated with TCA and acetone as described earlier. Proteins were denatured in 40 μl of 1× disruption buffer and examined by SDS-PAGE and Western blot analysis.
RESULTS
Identification of the GP64 palmitoylation site.
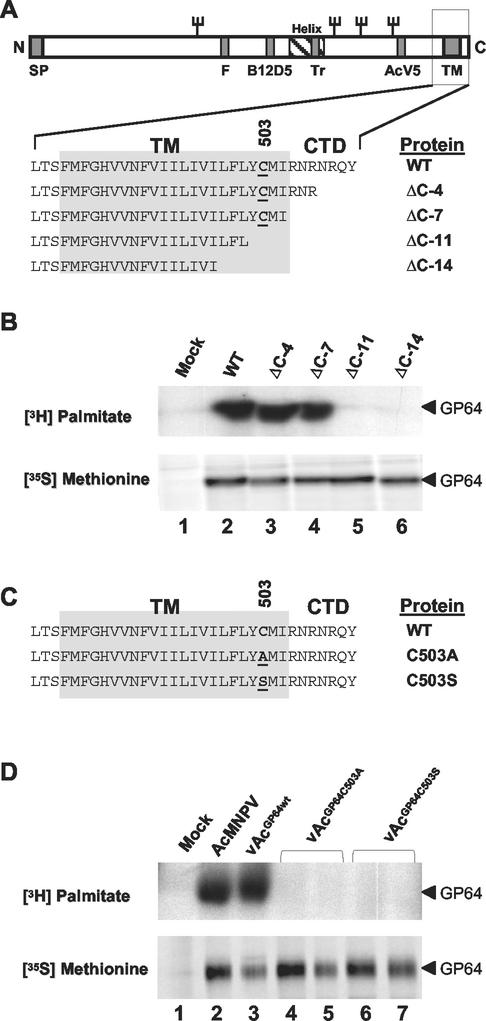
Many viral envelope proteins are palmitoylated at cysteine residues within or near their membrane-spanning domains (32). To map the GP64 palmitoylation site, we first used a series of recombinant viruses in which the wild-type gp64 gene was replaced with genes encoding C-terminally truncated forms of GP64 (30). To determine if the palmitoylation site was located near the C terminus, wild-type or truncated GP64 proteins were metabolically labeled with [3H]palmitate and immunoprecipitated from infected cell lysates with anti-GP64 MAb AcV1 (12) (Fig. 1A and B). To determine whether the truncations of GP64 affected synthesis or accumulation of the GP64 protein, infected cells were also labeled in parallel with [35S]methionine and similarly immunoprecipitated. All forms of GP64 were equivalently labeled with [35S]methionine, indicating that the truncations did not affect GP64 synthesis or accumulation (Fig. 1B, lower panel). In [3H]palmitate labeling experiments, we found that wild-type GP64 or constructs containing C-terminal deletions of 4 and 7 amino acids were labeled with [3H]palmitate (Fig. 1B, WT, ΔC-4, and ΔC-7), indicating that the palmitoylation site was not removed by deletion of seven C-terminal amino acid residues. However, when 11 or 14 amino acids were deleted from the C terminus (Fig. 1B, constructs ΔC-11 and ΔC-14), incorporation of [3H]palmitate into the GP64 protein was not detected. Thus, the AcMNPV GP64 palmitoylation site was mapped between amino acids 501 and 506 of the AcMNPV GP64 protein. The predicted transmembrane (TM) and cytoplasmic tail domain (CTD) of AcMNPV GP64 contains a single cysteine residue located at position 503 (Fig. 1A), and this cysteine residue (Cys503) is highly conserved among GP64 proteins. Therefore, we hypothesized that Cys503 was the likely palmitoylation site.
FIG. 1.
Palmitoylation of truncated or substituted GP64 proteins. (A) A diagrammatic representation of the GP64 protein shows the locations of the signal peptide (SP), fusion domain (F), epitopes (B12D5 and AcV5), predicted amphipathic alpha helix (Helix), trimerization domain (TR), transmembrane domain (TM), and cytoplasmic tail domain (CTD). The names of GP64 constructs containing deletions in the CTD and TM domain are indicated on the right, and Cys503 is underlined. (B) GP64 labeling with [3H]palmitate or [35S]methionine. Cells infected with wild-type AcMNPV (WT) or recombinant viruses expressing truncated forms of GP64 (ΔC-4, ΔC-7, ΔC-11, and ΔC-14) were pulse-labeled with [3H]palmitate or [35S]methionine, and then GP64 was immunoprecipitated with MAb AcV1. Results from [3H]palmitate labeling are shown in the upper panel, and those from [35S]methionine labeling are shown in the lower panel. (C) Sequences of the transmembrane and cytoplasmic tail domain of AcMNPV GP64 proteins. The wild-type AcMNPV (WT) sequence is shown above sequences of two mutant GP64 proteins containing substitutions at Cys503 (C503A and C503S). Substitutions are indicated by underlined amino acids. (D) GP64 labeling with [3H]palmitate or [35S]methionine and immunoprecipitation were performed as described for panel B. Cells were infected with viruses expressing wild-type GP64 (AcMNPV and vAcGP64wt) or GP64 proteins with substitutions at Cys503 (vAcGP64C503A and vAcGP64C503S). The lower panel shows GP64 labeling with [35S]methionine, and the upper panel shows labeling with [3H]palmitate. For immunoprecipitations from cells infected with vAcGP64C503A and vAcGP64C503S, duplicate infection and immunoprecipitation experiments are shown (lanes 4 and 5 and 6 and 7, respectively).
To test this hypothesis, the Cys503 codon of the gp64 gene was modified by site-directed mutagenesis to encode either alanine or serine (Fig. 1C, C503A and C503S). Each modified gp64 gene (C503A or C503S) was inserted into the polyhedrin locus of a gp64-null AcMNPV genome (21), resulting in viruses vAcGP64C503A and vAcGP64C503S, respectively. As an additional control, a virus containing the wild-type gp64 gene reinserted into the polyhedrin locus of a gp64-null AcMNPV genome was also used, and this virus is referred to as a “repair” virus (vAcGP64wt) (21). In each case, the gp64 coding region was under the transcriptional control of the wild-type gp64 promoter and was inserted into the polyhedrin locus by Tn7-mediated transposition as described in Materials and Methods. To examine the GP64 proteins expressed from the mutant and control viruses, Sf9 cells were infected with either wild-type AcMNPV, the repair virus expressing wild-type GP64 (vAcGP64wt), or viruses expressing GP64 with substitutions at Cys503 (vAcGP64C503A and vAcGP64C503S). As described above, infected cells were pulse-labeled with either [35S]methionine or [3H]palmitate, and GP64 was immunoprecipitated and examined by SDS-PAGE. Examination of proteins labeled with [35S]methionine shows that wild-type and modified GP64 proteins were expressed and accumulated at similar levels (Fig. 1D, lower panel, lanes 2 to 7). We also examined GP64 expression by Western blot analysis of lysates from infected cells and observed similar results (data not shown). Thus, the alanine and serine substitutions at Cys503 do not appear to influence GP64 expression or accumulation. In [3H]palmitate labeling experiments, [3H]palmitate labeling was not detected in GP64 proteins containing either the C503A or C503S substitution (Fig. 1D, upper panel, lanes 4 to 7), although wild-type GP64 was labeled with [3H]palmitate (Fig. 1D, upper panel, lanes 2 and 3). These data indicate that the palmitoylation site was eliminated in GP64 proteins containing substitutions at Cys503. Thus, the AcMNPV GP64 palmitoylation site was mapped to Cys503.
GP64 palmitoylation and cell surface localization.
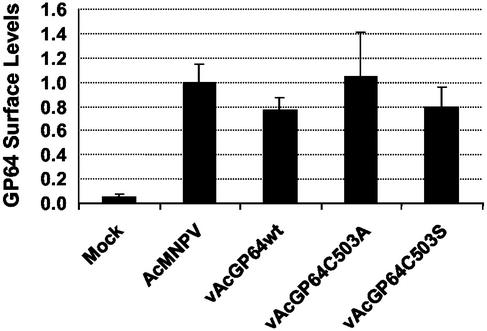
To examine possible effects of GP64 palmitoylation on GP64 transit through the secretory pathway and localization at the cell surface, we used a semiquantitative CELISA (26) to compare the relative levels of wild-type and palmitoylation-minus GP64 at the cell surface. Sf9 cells were infected with wild type AcMNPV, a repair virus expressing wild-type GP64 (vAcGP64wt), or two viruses expressing palmitoylation-minus GP64 proteins (vAcGP64C503A and vAcGP64C503S). At 48 h p.i., cells were fixed by using a protocol that does not permeabilize cells (see Materials and Methods), and GP64 on the cell surface was detected with an anti-GP64 MAb (AcV5) by CELISA. No significant differences in GP64 surface localization were detected when the wild-type GP64 construct was compared with palmitoylation-minus GP64 constructs (Fig. 2). Thus, palmitoylation does not appear to influence GP64 surface localization or accumulation in cultured Sf9 cells.
FIG. 2.
Relative levels of GP64 at the cell surface. Relative levels of cell surface GP64 were determined on infected Sf9 cells expressing either wild-type AcMNPV GP64 or GP64 proteins containing substitutions at Cys503. Sf9 cells were infected with either wild-type AcMNPV, a GP64 repair virus (vAcGP64wt), or viruses expressing modified GP64 proteins (vAcGP64C503A and vAcGP64C503S). Relative levels of surface-localized GP64 were determined by CELISA analysis with MAb AcV5, as described previously (26). Data points represent results from triplicate infections, and error bars represent the standard deviation from the mean. Relative GP64 expression levels are indicated on the y axis, with all constructs normalized to wild-type AcMNPV-infected cells (assigned a value of 1).
GP64 palmitoylation and pH-triggered membrane fusion.
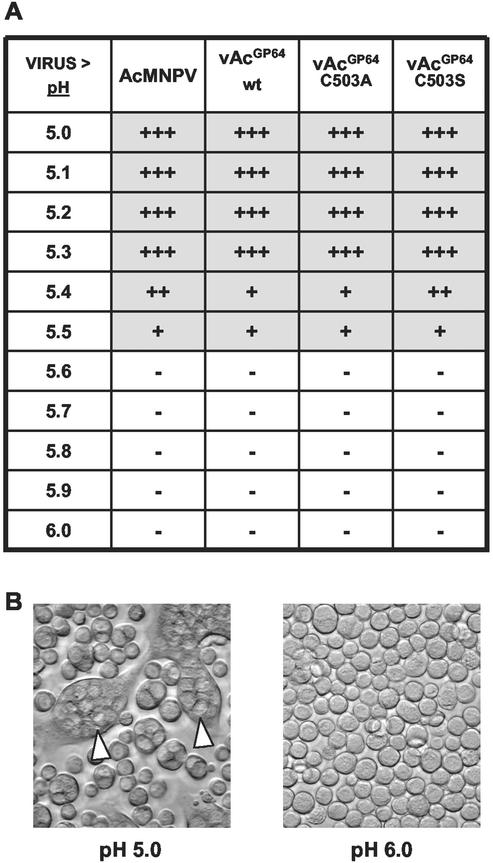
Previous studies established that GP64 is necessary and sufficient for pH-triggered membrane fusion activity (4, 42). Cells expressing the GP64 protein mediated membrane fusion and syncytium formation in a pH-dependent manner. To determine whether palmitoylation influences the membrane fusion function of GP64, we compared membrane fusion in cells expressing either wild-type or palmitoylation-minus GP64 proteins. For these studies, we compared Sf9 cells infected with AcMNPV viruses expressing various forms of GP64 by using a standard syncytium formation assay (30). Viruses used for these studies included wild-type AcMNPV and a control GP64 repair virus (both expressing the wild-type GP64 protein) and viruses expressing the two palmitoylation-minus GP64 mutants (vAcGP64C503A and vAcGP64C503S). Results from syncytium formation assays showed that the two mutant GP64 proteins (C503S and C503A) mediated pH-triggered membrane fusion activity in a manner similar to wild-type GP64 (Fig. 3). In all cases, syncytia were formed after exposure to a pH of 5.5 or lower. In these studies, syncytia were not observed when GP64-expressing cells were exposed to a pH of 5.6 or higher. Thus, GP64 palmitoylation did not alter the threshold pH required for membrane fusion in syncytium formation assays in Sf9 cells.
FIG. 3.
Analysis of GP64-mediated membrane fusion. (A) Summary of results from syncytium formation assays of cells infected with viruses expressing wild-type GP64 (AcMNPV and vAcGP64wt) or viruses expressing palmitoylation-minus GP64 proteins (vAcGP64C503A and vAcGP64C503S). Sf9 cells were infected with the indicated viruses at an MOI of 10. At 36 h p.i., cells were incubated in PBS buffer that was adjusted to the indicated pH value (from pH 5.0 to 6.0) for 20 min. Cells were scored as positive for syncytium formation when at least five nuclei were detected in a syncytial mass (+++, >20 syncytia per well; ++, 11 to 20 syncytia per well; +, 1 to 10 syncytia per well; −, no syncytia observed). (B) Examples of syncytium formation in Sf9 cells expressing wild-type GP64 and exposed to PBS at pH 5.0 or 6.0. Syncytial masses are indicated by open arrowheads.
AcMNPV budded virus production.
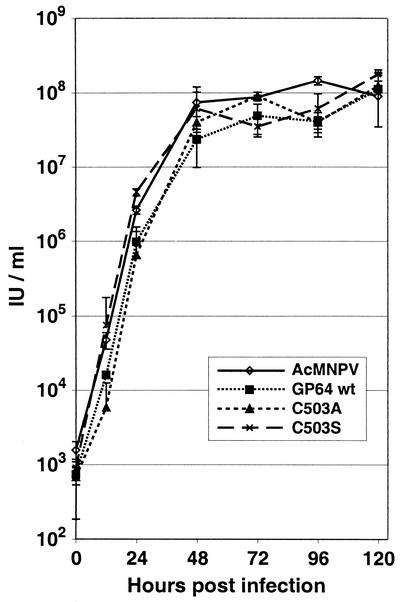
The GP64 protein is necessary for both viral entry and exit from the host cell. In the absence of GP64, virion budding from AcMNPV-infected Sf9 cells is reduced by approximately 98% (30). In addition, virions that bud in the absence of GP64 are completely noninfectious. To determine if GP64 palmitoylation affected production of infectious AcMNPV in Sf9 cells, we used one-step growth curve experiments to compare viruses expressing wild-type GP64 proteins to those expressing palmitoylation-minus GP64 proteins. For those experiments, two control viruses (wild-type AcMNPV and a GP64 repair virus, vAcGP64wt) and two viruses expressing palmitate-minus forms of GP64 (vAcGP64C503A and vAcGP64C503S) were used to infect Sf9 cells, and titers of infectious progeny budded virus were determined at various times p.i. (Fig. 4). BV production from cells infected with viruses expressing palmitate-minus GP64 was similar to that from cells infected with the control viruses expressing wild-type GP64. Therefore, we concluded that palmitoylation of GP64 was not required for efficient production of infectious BV in cultured Sf9 cells.
FIG. 4.
Budded virus production. Viral one-step growth curves for AcMNPV viruses expressing wild-type GP64 (AcMNPV and vAcGP64wt) and viruses expressing palmitoylation-minus GP64 proteins (vAcGP64C503A and vAcGP64C503S). Data points represent titers derived from triplicate infections, and error bars represent the standard deviation from the mean. IU, infectious units.
Analysis of GP64 and cellular membrane microdomains.
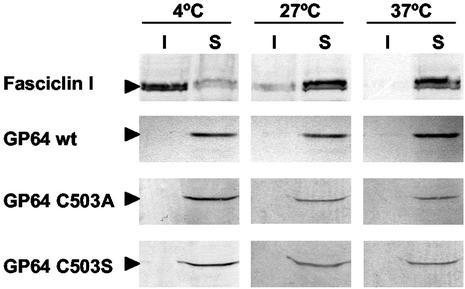
A number of well-studied viral envelope proteins associate with cellular membrane microdomains commonly referred to as “lipid rafts” or “detergent-resistant membranes” (DRMs). Examples of viral envelope proteins identified in lipid rafts include HIV GP120/41 (36) and the influenza virus HA protein (38). Proteins associated with the dense lipid raft microdomains of membranes are resistant to extraction with detergents (such as Triton X-100) at low temperatures, and this property is believed to result from insolubility of these membrane domains due to the presence of highly ordered acyl chains. Many proteins associated with lipid rafts are palmitoylated, and in some cases, palmitoylation is required for raft association (25). To determine if such membrane microdomains are found in membranes of AcMNPV-infected Sf9 cells, we constructed a recombinant baculovirus expressing FasI, a protein that is associated with lipid rafts in D. melanogaster cells (13, 33). FasI was expressed under the transcriptional control of the AcMNPV gp64 early-late promoter (see Materials and Methods). We then performed cold detergent membrane solubility assays with Sf9 cells infected with the FasI-expressing baculovirus. Cell membranes were treated with Triton X-100 at 4, 27, or 37°C. Insoluble and soluble membrane fractions were separated by centrifugation and examined for the presence of FasI by using FasI-specific antibodies. FasI was found primarily in detergent-soluble fractions at 37 and 27°C, but was found primarily in the detergent-insoluble fraction when extractions were performed at 4°C (Fig. 5, top panel). Experiments examining FasI that was transiently expressed in Drosophila S2 cells produced results identical to those illustrated in Fig. 5 (data not shown). Because FasI expressed in Sf9 cells behaved as expected for a typical lipid raft-associated protein, these data suggest that AcMNPV-infected Sf9 cells contain DRMs or lipid rafts and that the FasI lipid raft protein was associated with those raft domains. To determine whether the AcMNPV GP64 protein was associated with lipid rafts in AcMNPV-infected Sf9 cells, we examined the same detergent extracts for the presence of GP64 by using an anti-GP64 MAb. In contrast to the control lipid raft protein (FasI), GP64 was solubilized in Triton X-100 at all temperatures examined (Fig. 5, GP64 wt). This suggested that GP64 is not a lipid raft-associated membrane protein.
FIG. 5.
Analysis of cold detergent extraction of FasI and GP64 proteins in Sf9 cells and the effect of GP64 palmitoylation. (Top two panels) Infected Sf9 cells expressing both FasI and wild-type GP64 were lysed in 0.5% Triton X-100 at 4, 27, or 37°C. The soluble (S) and insoluble (I) fractions were separated by centrifugation as described in Materials and Methods and subjected to Western blot analysis with either an anti-FasI MAb (top panel) or anti-GP64 MAb AcV5 (second panel). Sf9 cells infected with viruses expressing palmitoylation-minus forms of GP64 (C503A or C503S) were treated as described above, and GP64 was detected with MAb AcV5 (bottom two panels).
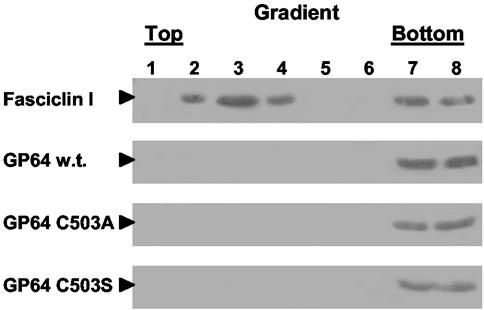
To confirm the results presented above, membrane fractions containing GP64 and the FasI raft-associated protein were also examined with membrane flotation gradients. For these studies, infected cells were lysed, and membranes were solubilized in Triton X-100 at 4°C, adjusted to 40% sucrose and placed at the bottom of a 5/30/40% sucrose step gradient, and then centrifuged at 250,000 × g for 16 h. The resulting gradients were fractionated, and each fraction was examined for the presence of GP64 or FasI with the appropriate antibodies. Typically, fractions in the upper portion of the gradient are enriched for lipid rafts because the lower density (buoyancy) of the insoluble lipid rafts causes them to “float.” In contrast, proteins that are solubilized in the detergent extraction remain in the lower portion of the gradient. FasI, the lipid raft marker, was detected in fractions 2 to 4 (which correspond to the lipid raft fraction) and fractions 7 and 8 (the soluble protein fraction) (Fig. 6, top panel). This type of distribution is typical for lipid raft proteins (25, 46). Similar results were observed when the FasI protein was transiently expressed in Drosophila S2 cells and analyzed in the same manner (data not shown). In contrast to the results with FasI, GP64 was detected only in fractions 7 and 8 at the bottom of the gradient, which represents proteins solubilized in the cold detergent extraction. GP64 was not detected in the upper portion of the gradient. Thus, in both membrane solubility studies and analysis by flotation gradients, AcMNPV GP64 was not detected in fractions associated with lipid raft proteins, suggesting that GP64 is not found in lipid raft microdomains in infected Sf9 cells.
FIG. 6.
Flotation gradient analysis of GP64 and FasI association with lipid raft microdomains. Sf9 cells infected with a virus expressing wild-type (w.t.) GP64 and the FasI protein (top two panels) were subjected to Triton X-100 extraction at 4°C. Insoluble membranes were isolated by flotation gradient fractionation. Proteins from each sucrose gradient fraction were examined by Western blot analysis with either an anti-FasI MAb (top panel) or an anti-GP64 MAb (lower three panels). Fractions 2 to 4 represent the more buoyant insoluble membranes associated with lipid rafts, and fractions 7 and 8 represent soluble non-raft portions of the membrane. Sf9 cells infected with viruses expressing palmitoylation-minus forms of GP64 (C503A and C503S) were treated as described above and probed with anti-GP64 MAb AcV5 (lower two panels).
Because protein palmitoylation is known to affect the association of proteins with lipids in membranes, we next asked whether GP64 palmitoylation might result in exclusion of GP64 from lipid rafts. To address this question, detergent solubility experiments were carried out as described above, but with cells infected with viruses expressing either the wild-type GP64 protein, or palmitoylation-minus GP64 proteins. The localization of GP64 in raft or nonraft domains was then determined for each GP64 construct. Results from initial experiments showed that both of the palmitate-minus GP64 proteins were soluble in Triton X-100 at lower (4°C) and higher (27 and 37°C) temperatures (Fig. 5, lower panels). In addition, data from flotation gradients showed that the palmitate-minus GP64 proteins were detected only in the lower (non-raft) fractions (Fig. 6, lower panels) and did not “float” with the control raft protein, FasI. Therefore, the membrane localization of these palmitate-minus GP64 proteins was indistinguishable from that of wild-type GP64, suggesting that palmitoylation does not cause the apparent exclusion of GP64 from lipid rafts in infected cells.
DISCUSSION
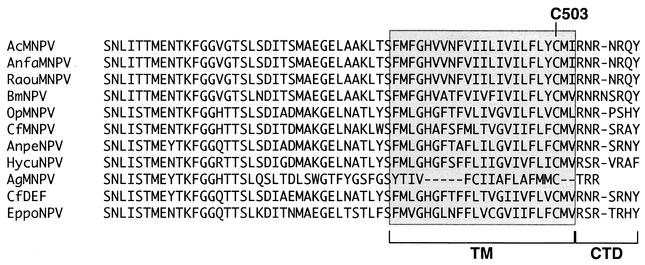
GP64 is an essential AcMNPV BV structural protein that is important for viral entry into and exit from host cells. Previous studies demonstrated that GP64 was posttranslationally modified by the addition of palmitate (34). However, the palmitoylation site of GP64 was not determined, and the function of palmitoylation in GP64 was not previously examined. In the studies described here, we used a combination of truncation and site-specific mutagenesis to map the AcMNPV GP64 palmitoylation site to a single cysteine residue at amino acid 503. GP64 proteins are found in a subgroup (group I NPVs) of the Baculoviridae (1, 10), and within this subgroup, GP64 proteins are very highly conserved. Among the GP64 proteins, the highest degree of conservation is found among the predicted GP64 ectodomains, and a lower degree of conservation is found among the predicted signal peptides and transmembrane domains. The predicted GP64 cytoplasmic tail domains are very short (3 to 8 amino acids), not highly conserved, and arginine rich (Fig. 7). The AcMNPV Cys503 is highly conserved, since each GP64 protein contains a single cysteine residue at the same location near the cytoplasmic border of the predicted transmembrane domain. While palmitoylation of other GP64 proteins has not been examined, the conservation of this residue suggests that other GP64 proteins may be similarly palmitoylated at this single site.
FIG. 7.
Alignments of the cytoplasmic tail domains (CTD) and transmembrane (TM) domains of baculovirus GP64 proteins. The predicted transmembrane (shaded) and cytoplasmic tail domains of 11 baculovirus GP64 proteins are aligned. Cysteines corresponding to Cys503 in the AcMNPV transmembrane domain are indicated (C503). Predicted GP64 amino acid sequences are from viruses AcMNPV, Anagrapha falcifera MNPV (AnfaMNPV), Rachiplusia ou MNPV (RaouMNPV), BmNPV, OpMNPV, Choristoneura fumiferana MNPV (CfMNPV), Antheraea pernyi NPV (AnpeNPV), Hyphantria cunea NPV (HycuNPV), Anticarsia gemmatalis MNPV (AgMNPV), C. fumiferana DEF NPV (CfDEF), and Epiphyas postvittana NPV (EppoNPV).
By inserting genes encoding palmitoylation-minus forms of GP64 into a recombinant baculovirus in which the wild-type gp64 gene was deleted, we examined several potential functions of GP64 palmitoylation in the context of the AcMNPV infection cycle. Palmitoylation of GP64 did not substantially affect GP64 synthesis or the transport of GP64 to the cell surface. In addition, we detected no effects of GP64 palmitoylation on yields or infectivity of BV generated from infected Sf9 cells. GP64 proteins lacking palmitoylation were capable of mediating pH-triggered membrane fusion in syncytium formation assays, and the pH required for triggering was unaffected by the absence of palmitoylation. These data suggest that virus entry and exit in Sf9 cells were not dependent on or significantly affected by GP64 palmitoylation. However, we cannot rule out the possibility that more subtle effects of GP64 palmitoylation on viral entry or exit were present but not detected. Because protein palmitoylation may affect the interaction of proteins with cellular membranes, we also examined infected Sf9 cells to determine (i) if lipid raft microdomains are present in AcMNPV-infected Sf9 cells, (ii) if GP64 is localized to lipid raft microdomains, and (iii) if GP64 association with or exclusion from such domains was affected by palmitoylation. We show that a previously characterized raft protein from D. melanogaster (FasI) was identified in cold DRM fractions in AcMNPV-infected Sf9 cells, suggesting the presence of lipid raft domains. However, in the same experiments, GP64 partitioned into detergent-soluble membrane fractions, suggesting that GP64 is not associated with DRMs or lipid rafts. When palmitoylation-minus forms of GP64 were examined in a similar manner, those GP64 proteins were also found in soluble membrane fractions, suggesting that GP64 palmitoylation plays no substantial role in excluding GP64 from lipid rafts in the Sf9 cell membrane.
Lipid raft domains have been shown to play roles in the infection cycles of viruses such as HIV-1 and influenza virus. The influenza virus HA protein is palmitoylated at three conserved cysteine residues located at the C-terminal end of the transmembrane domain and in the cytoplasmic domain (40). Palmitoylation at all three cysteine residues is necessary for lipid raft localization of HA (25). Influenza virus budding occurs at the apical surface of polarized epithelial cells, with viruses appearing to utilize lipid raft domains as budding sites. However, in viruses containing cysteine substitutions that disrupt palmitoylation and lipid raft association of HA, virion budding continues to be associated with apical surfaces. Therefore, localization of HA to lipid rafts does not appear to determine the polarity of budding (28, 46), and the full significance of the HA association with lipid raft domains is not yet clear. The HIV-1 envelope protein gp160 is palmitoylated at two cysteine residues in the cytoplasmic tail domain, and palmitoylation at those residues is important for its association with lipid raft domains (36, 45). Budding of HIV-1 is thought to occur selectively from lipid rafts (29). Since gp160 is not necessary for virion budding, the association of gp160 with lipid rafts is believed to be important for its inclusion in budding virions. Palmitoylation-minus forms (mutants) of gp160 are not efficiently targeted to lipid rafts and are found in virions at much reduced levels compared to wild-type gp160 (36). This also results in decreased infectivity of the resulting virions. Thus, palmitoylation of gp160 appears to be important for recruitment of the envelope protein into virions and thus for virion infectivity. The vesicular stomatitis virus (VSV) G protein is also palmitoylated through a cysteine residue in the cytoplasmic tail domain (35), but in contrast to influenza virus HA and HIV-1 gp160, palmitoylation at this site does not appear to result in localization of G to lipid raft domains (37). In addition, palmitoylation of VSV G protein is not necessary for either membrane fusion activity or assembly of G protein into VSV virions (44). Thus, palmitoylation of some viral envelope proteins appears to play an important role in lipid raft association, which in turn may affect either virion assembly or infectivity. In other cases, envelope protein palmitoylation appears to have no detectable role in cultured cells. A major difference between the above examples is the addition of palmitate at multiple sites within each protein. Proteins that localize to lipid rafts may require two or more palmitoylation sites. In this regard, palmitoylation of the AcMNPV GP64 protein appears to be similar to that of the VSV G protein, with both proteins having a single palmitoylation site and with both proteins absent from lipid raft domains. We detected no effect of GP64 palmitoylation on protein trafficking, membrane fusion activity, or BV production or infectivity, and this was also similar to the results from studies of VSV G protein (35, 44).
Our observation that GP64 was not associated with DRM fractions is also consistent with prior studies of the membrane composition of AcMNPV BV. Lipid rafts are characterized by high concentrations of cholesterol, sphingolipid, and long-chain saturated fatty acids. Previous studies (5) indicated that BV envelopes contained low ratios of saturated to unsaturated fatty acids and that cholesterol levels were relatively low. Although it has been reported that the levels of cholesterol in the plasma membranes of Sf9 cells are lower than those from most mammalian cells (8), we were able to isolate cold DRMs containing a known lipid raft protein (the FasI protein) in Sf9 cell membranes, indicating that Sf9 cells contain the membrane microdomains described as lipid rafts. Thus, the present study in combination with prior studies suggests that while lipid rafts are present in Sf9 membranes, they are not likely to be a major component of AcMNPV BV envelopes. In addition, we did not detect the lipid raft protein FasI in isolated AcMNPV BV (data not shown), whereas cellular lipid raft proteins have been identified in the envelopes of viruses such as HIV-1, which bud from lipid raft domains (29). Thus, current data are consistent with a model in which the AcMNPV BV envelope is comprised largely or entirely of non-raft domain membrane.
In this study, the single palmitoylation site in GP64 was mapped, and potential roles of GP64 palmitoylation were examined. GP64 palmitoylation had no detectable effect on GP64 synthesis, transport, or membrane fusion activity and was not required for BV production or infectivity in Sf9 cells. GP64 was not detected in lipid raft domains, and GP64 palmitoylation was found to have no detectable effect on its membrane microdomain localization. That GP64 was not identified in lipid rafts suggests GP64 may be located in more fluid portions of the membrane. Therefore, it is possible that GP64 palmitoylation may serve to modulate or regulate the mobility of GP64 in the membrane. Because all studies described here were performed with cultured Sf9 cells, an intriguing additional possible role for GP64 palmitoylation is the targeting of GP64 in polarized epithelial cells of the insect midgut, where GP64 is targeted basolaterally for budding of virions into the hemocoel. Future studies will examine these and other possible functions of GP64 palmitoylation.
Acknowledgments
We thank Oliver Lung for providing the gp64-null bacmid and assistance, Michael Hortsch for providing the Drosophila FasI gene and anti-FasI antibodies, and Basil Arif and Hilary Lauzon for providing the CfDEF gp64 nucleotide sequence. We also thank Guangyun Lin, Gretchen Hoffmann, and Irina Lukina for technical assistance and Barbara Baird and Ryan Young for advice on membrane solubility studies. We thank Tom Oomens, Erik Burnett, and Oliver Lung for comments on the manuscript.
This work was supported by NIH grant AI33657.
REFERENCES
- 1.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2000. Baculoviridae: taxonomic structure and properties of the family, p. 195-202. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Seventh Report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 2.Blissard, G. W., P. H. Kogan, R. Wei, and G. F. Rohrmann. 1992. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology 190:783-793. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 65:5820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315-328. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and E. London. 1998. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164:103-114. [DOI] [PubMed] [Google Scholar]
- 7.Chernomordik, L., E. Leikina, M.-S. Cho, and J. Zimmerberg. 1995. Control of baculovirus gp64-induced syncytium formation by membrane lipid composition. J. Virol. 69:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimpl, G., U. Klein, H. Reilander, and F. Fahrenholz. 1995. Expression of the human oxytocin receptor in baculovirus-infected insect cells: high-affinity binding is induced by a cholesterol-cyclodextrin complex. Biochemistry 34:13794-13801. [DOI] [PubMed] [Google Scholar]
- 9.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455-468. [DOI] [PubMed] [Google Scholar]
- 10.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hink, W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466-467. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 13.Hortsch, M., and C. S. Goodman. 1990. Drosophila fasciclin I, a neural cell adhesion molecule, has a phosphatidylinositol lipid membrane anchor that is developmentally regulated. J. Biol. Chem. 265:15104-15109. [PubMed] [Google Scholar]
- 14.Hortsch, M., Y. M. Wang, Y. Marikar, and A. J. Bieber. 1995. The cytoplasmic domain of the Drosophila cell adhesion molecule neuroglian is not essential for its homophilic adhesive properties in S2 cells. J. Biol. Chem. 270:18809-18817. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis, D. L., and A. Garcia. 1994. Biosynthesis and processing of the Autographa caIifornica nuclear polyhedrosis virus GP64 protein. Virology 205:300-313. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis, D. L., L. Wills, G. Burow, and D. A. Bohlmeyer. 1998. Mutational analysis of the N-linked glycans on Autographa californica nucleopolyhedrovirus gp64. J. Virol. 72:9459-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingsley, D. H., A. Behbahani, A. Rashtian, G. W. Blissard, and J. Zimmerberg. 1999. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol. Biol. Cell 10:4191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kost, T. A., and J. P. Condreay. 2002. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol. 20:173-180. [DOI] [PubMed] [Google Scholar]
- 19.Leikina, E., H. O. Onaran, and J. Zimmerberg. 1992. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 304:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangor, J. T., S. A. Monsma, M. C. Johnson, and G. W. Blissard. 2001. A GP64-null baculovirus pseudotyped with vesicular stomatitis virus G protein. J. Virol. 75:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markovic, I., H. Pulyaeva, A. Sokoloff, and L. V. Chernomordik. 1998. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 143:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruniak, J. W., and M. D. Summers. 1981. Autographa californica nuclear polyhedrosis virus phosphoproteins and synthesis of intracellular proteins after virus infection. Virology 109:25-34. [DOI] [PubMed] [Google Scholar]
- 25.Melkonian, K. A., A. G. Ostermeyer, J. Z. Chen, M. G. Roth, and D. A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274:3910-3917. [DOI] [PubMed] [Google Scholar]
- 26.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 69:2583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mora, R., E. Rodriguez-Boulan, P. Palese, and A. Garcia-Sastre. 2002. Apical budding of a recombinant influenza A virus expressing a hemagglutinin protein with a basolateral localization signal. J. Virol. 76:3544-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 31.Oomens, A. G. P., S. A. Monsma, and G. W. Blissard. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209:592-603. [DOI] [PubMed] [Google Scholar]
- 32.Ponimaskin, E., and M. F. Schmidt. 1998. Domain-structure of cytoplasmic border region is main determinant for palmitoylation of influenza virus hemagglutinin (H7). Virology 249:325-335. [DOI] [PubMed] [Google Scholar]
- 33.Rietveld, A., S. Neutz, K. Simons, and S. Eaton. 1999. Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J. Biol. Chem. 274:12049-12054. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, T. E., and P. Faulkner. 1989. Fatty acid acylation of the 67K envelope glycoprotein of a baculovirus Autographa californica nuclear polyhedrosis virus. Virology 172:377-381. [DOI] [PubMed] [Google Scholar]
- 35.Rose, J. K., G. A. Adams, and C. J. Gallione. 1984. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc. Natl. Acad. Sci. USA 81:2050-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 38.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoji, I., H. Aizaki, H. Tani, K. Ishii, T. Chiba, I. Saito, T. Miyamura, and Y. Matsuura. 1997. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 78:2657-2664. [DOI] [PubMed] [Google Scholar]
- 40.Veit, M., E. Kretzschmar, K. Kuroda, W. Garten, M. F. G. Schmidt, H. D. Klenk, and R. Rott. 1991. Site-specific mutagenesis identifies three cysteine residues in the cytoplasmic tail as acylation sites of influenza virus hemagglutinin. J. Virol. 65:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkman, L. E., and P. A. Goldsmith. 1984. Budded Autographa californica NPV 64K protein: further biochemical analysis and effects of postimmunoprecipitation sample preparation conditions. Virology 139:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143:185-195. [DOI] [PubMed] [Google Scholar]
- 43.Volkman, L. E., P. A. Goldsmith, R. T. Hess, and P. Faulkner. 1984. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology 133:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitt, M. A., and J. K. Rose. 1991. Fatty acid acylation is not required for membrane fusion activity or glycoprotein assembly into VSV virions. Virology 185:875-878. [DOI] [PubMed] [Google Scholar]
- 45.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinn, K., L. McAllister, and C. S. Goodman. 1988. Sequence analysis and neuronal expression of fasciclin I in grasshopper and Drosophila. Cell 53:577-587. [DOI] [PubMed] [Google Scholar]