Abstract
The Atlas human cDNA expression array was used to evaluate gene expression profile changes in the genesis of human lung adenocarcinomas and squamous cell carcinomas. Gene expression changes between adenocarcinomas and squamous cell carcinomas were also analyzed. Of the 588 gene targets, 262 genes were expressed in these tissues and, of these, 45 genes were differentially expressed by at least two-fold in tumor tissues compared to corresponding normal tissues. Semiquantitative reverse-transcriptase polymerase chain reaction was used to confirm gene expression changes. Only those genes that reflected changes in >50% of the analyzed tissues were included in the final analysis. Ultimately, 26 genes were evaluated with 14 genes overexpressed and 12 genes underexpressed compared to matching normal lung tissues. Although similar expression changes were detected in adenocarcinomas and squamous cell carcinomas for most of the genes analyzed, some subtype-specific differences were also found. Genes encoding cell cycle regulators, intracellular signal transducers, cell receptor and adhesion molecules, growth factors, oncogenes, and apoptotic effectors were differentially expressed in this study. These gene expression changes may directly contribute to the initiation or progression of human lung cancer or may be secondary effects of the tumorigenesis process. Regardless, many of these differences may be useful in the diagnosis and/or treatment of this deadly disease.
Keywords: non-small cell lung cancer, cDNA microarray, expression profile, differential changes, cancer genes
Introduction
An estimated 553,400 Americans will lose their lives to cancer in the year 2001. Lung cancer is the most common type of cancer death among men and women in the United States, with an estimated 169,500 new cases and 157,400 deaths predicted for the year 2001 [1]. The molecular pathogenesis of human cancer involves the accumulation of genetic and epigenetic alterations in cancer-related genes [2,3]. Progression to lung cancer is believed to involve 10 to 20 alterations, including the activation of proto-oncogenes and the inactivation of tumor suppressor genes [4]. In addition to overriding growth control mechanisms, transformed cells must also avoid programmed cell death that can ensue when cell cycle checkpoint control is lost. Although most tumors have high rates of apoptotic death, cancer cells frequently inactivate components of apoptotic pathways and induce cell survival genes [5]. Additional alterations may include disruption of DNA repair genes and chromosomal fragile sites as well as activation of invasive, metastatic, and/or angiogenic factors. Genetic susceptibility may also contribute to the development of human lung cancer [4,6].
Lung cancers are broadly categorized into two histologic groups. Small cell lung cancer (SCLC) is diagnosed in approximately 20% to 25% of all lung cancer cases and non-small cell lung cancer (NSCLC) is diagnosed in the remaining 75% to 80% of cases [7,8]. NSCLC is further subdivided into three groups: 1) squamous cell carcinoma is characterized by rapidly growing epidermoid cells that produce keratin (∼25%); 2) adenocarcinoma arises peripherally and is composed of cuboidal or columnar cells that can form glandular patterns and produce mucin (∼30%); 3) large cell carcinoma consists of a heterogeneous mix of poorly differentiated cells that do not resemble cells from the other two categories (∼10%) [7,8].
Cytogenetic and molecular studies have revealed different genetic alterations in these two histologic subgroups. For example, the expression frequency of Bcl-2 is much higher in SCLC (75% to 95%) than in NSCLC (10% to 35%) [6,9–11]. Additionally, within NSCLC, squamous cell carcinomas express higher levels of Bcl-2 more frequently than adenocarcinomas (25% to 35% vs. ∼10%, respectively) [6,10,12,13]. Abnormal Rb protein expression is detected in over 90% of SCLC but only 15% to 30% of NSCLC [14–16]. p16INK4a inactivation is more frequently detected in NSCLC (∼10% to 40%) and very rarely in SCLC (<1%) [6,17,18]. Activating point mutations in the ras gene family occur in 20% to 30% of adenocarcinomas and 15% to 20% of all NSCLC but in <1% of SCLC [6,19]. In contrast, the frequency of abnormal p53 expression detected by immunohistochemistry is ∼40% to 70% for both SCLC and NSCLC [20–22]. Most studies have reported a higher frequency of abnormal expression of the p53 gene in squamous cell carcinomas compared to adenocarcinomas [6]. In SCLC, losses from chromosomes 1p, 3p, 5q, 17p, and 10q predominate [23,24]. Common alterations in NSCLC include 3p, 6q, 8p, 9p, 9q, 13q, 17p, 18q, 19p, 21q, 22q, as well as +7, i(5)(p10) and i(8)(q10) [23,25]. Genetic alterations appear to differ between SCLC and NSCLC and possibly even between NSCLC subtypes.
Using a human cDNA expression array, we determined gene expression profile changes between human lung NSCLCs and normal human lung tissues. We also analyzed the gene expression profile changes between human lung squamous cell carcinomas and adenocarcinomas with respect to their matching normal lung tissues. The cDNA expression arrays allow for rapid, high-throughput analyses of gene expression patterns in tissues. Analysis of differentially expressed genes in human lung tumors may reveal additional, unsuspected genes involved in lung tumorigenesis, provide novel diagnostic markers or chemopreventive targets, and/or implicate the involvement of other molecular pathways in cancer development.
Materials and Methods
Tissue Specimens
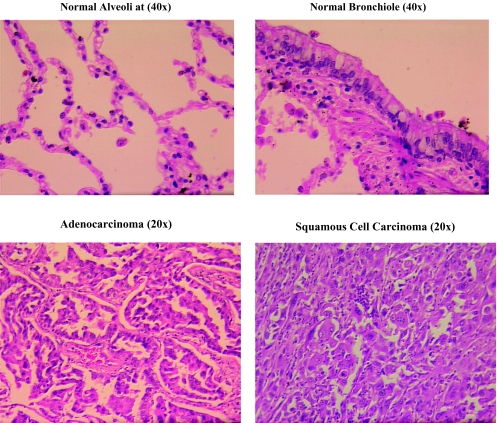
Frozen lung cancer specimens and matching normal tissues were obtained from Cooperative Human Tissue Network through The Ohio State University Department of Pathology. Fourteen pairs of clinical samples (seven squamous cell carcinomas with their normal controls and seven adenocarcinomas with their normal controls) were used in this study (Table 1). Frozen tumor tissues were microdissected to determine the tumor versus normal cell ratio for each specimen. Tissues were embedded in Tissue Tek OCT compound (VWR Scientific Products, West Chester, PA), cryostat sectioned, and stained with hemotoxin and eosin for microscopy. Tumor tissue sections corresponding to the microscopic sections containing ≥70% tumor cells were isolated and stored at -80°C for subsequent RNA isolation. Matching normal tissues were also microdissected to ensure that specimens consisted of purely normal lung tissue. Figure 1 shows the typical morphology of normal alveoli and an adenocarcinoma as well as a normal bronchiole and a squamous cell carcinoma used in this study.
Table 1.
Description of Adenocarcinomas and Squamous Cell Carcinomas Used in This Study.
| Patient No. | Description | Location | Diagnosis | Other Relevant Facts |
| 8322 | 48Y, W, F | RLL | Invasive, moderately differentiated SCC | Smoker: 1 ppdx25 years; prior SCC of the lung treated with radiation; mother died of breast cancer |
| 8326 | 72Y, W, F | RLL | Moderately differentiated SSC; vascular invasion; lymph node metastasis |
Smoker: 1/2 ppdx30 years; prior SCC of the lung treated with chemotherapyx5 |
| 8599 | 62Y, F | RA Lung | Moderately to poorly differentiated SCC; pleural and intrapulmonary lymphatic invasion |
Prior right lung cancer |
| 8611 | 67Y, M | LUL | Moderately differentiated bronchogenic SSC with desmoplastic reaction |
Atelectasis and inflammation with cavitation and hemorrhage |
| 8706 | 74Y, M | LUL | Poorly differentiated SSC with extensive necrosis | Emphysematous change with focal chronic interstitial pneumonitis and fibrosis; atelectasis |
| 94-10-A039 | 68Y, W, M | RUL | Moderately differentiated SCC with no evidence of keratin formation; adenosquamous in some areas |
Moderate emphysema in normal lung tissue |
| 94-11-A120 | 58Y, W, M | LLL | Poorly differentiated endobronchial SSC with lymph node metastasis |
Prior asbestos exposure |
| 5796 | 69Y, F | RUL | Moderately to poorly differentiated alveolar AdC with extensive central necrosis |
Moderate lymphocytic host response |
| 8252 | 70Y, W, F | RML | Poorly differentiated AdC with bronchioalveolar pattern at its periphery; lymph node metastasis |
Prior cervical carcinoma but compatible with primary lung carcinoma |
| 8606 | 52Y, W, F | LLL | Poorly differentiated AdC with lymph node metastasis |
Smoker: 1.5 ppdx32 years; prior RU lobe tumor; post-obstructive chronic pneumonia |
| 8607 | 45Y, W, F | LUL; LLL |
Adenocarcinoma; mass present at hilum and junction of LU lobe and LL lobe |
Brain metastasis |
| 8641 | 68Y, B, F | RML | Moderately differentiated AdC invading into bronchus and vasculature |
Tumor destroys and fills one bronchus; chronic bronchitis and centrilobular emphysema |
| 8712 | 76Y, W, F | RUL | Moderately differentiated AdC with lymph node metastasis |
History of skin cancer and primary RU lobe lung cancer |
| 94-11-C066 | 51Y, M | RLL | Poorly differentiated AdC | Smoker: 1.5 ppdx30 years |
Abbreviations: AdC, adenocarcinoma. SCC, squamous cell carcinoma. W, white. B, black. F, female. M, male. Y, years old. ppd, packs per day.
Location: first position: R, right; L, left; second position: U, upper; M, middle; L, lower; third position: L, lobe.
Figure 1.
Histology of normal and tumor tissues used in this study.
PolyA+ RNA Isolation
Total RNA was extracted using TRI-Reagent according to standard protocol (Molecular Research Center, Cincinnati, OH). Briefly, after homogenization in 5.0 ml TRI-Reagent, 1.0 ml chloroform was added per sample, shaken vigorously, and centrifuged for 15 minutes at 4°C, 12,000g. The clear, upper phase containing total RNA was collected for isolation of the polyA+ RNA fraction. One volume of aqueous phase was mixed with 0.1 volume of 1 M Tris (free base) and 0.8 volume isopropanol and stored at room temperature for approximately 5 minutes. Meanwhile, oligo(dT)-cellulose columns (Molecular Research Center) were washed with 1.0 ml binding buffer (0.5 M LiCl, 50 mM sodium citrate, 0.1% SDS). The RNA suspension was applied to the cellulose column to which the polyA+ RNA adheres. The columns were then washed with 0.5 ml 75% ethanol and 1.0 ml binding buffer. Finally, the polyA+ RNA fraction was eluted from the column with 0.6 ml elution buffer (1 mM sodium citrate, 0.1% SDS), quantitated by spectrophotometry, and precipitated with a final concentration of 0.3 M NaCl and 1.5 volumes isopropanol. Pellets were dissolved in nucleasefree water to a final concentration of 1 µg/µL.
cDNA Expression Array Hybridization
A total of 5 µg of polyA+ RNA was reverse transcribed using 0.2xCDS primer mix (Clontech Laboratories, Palo Alto, CA); 10 mM DTT; 1 x reverse transcription buffer (50 mM Tris-HCl, pH 8.3; 75 mM KCl; 3 mM MgCl2); 1 mM each of dATP, dTTP, and dGTP; 80 U RNAsin (Promega, Madison, WI); 50 µCi [α-32P]dCTP and 800 units MMLV reverse transcriptase (Gibco BRL Life Technologies, Carlsbad, CA). The mixture was incubated at 37°C for 1 hour and then purified by centrifugation through G50 Sephadex columns, denatured at 95°C for 10 minutes, and hybridized to Atlas cDNA expression arrays (Clontech Laboratories, Palo Alto, CA) for 16 to 20 hours at 42°C. Membranes were then washed twice, each for 10 minutes in 2xSSC, 0.1% SDS at room temperature, and twice, each for 30 minutes in 0.1xSSC, 0.1% SDS at 55°C, followed by exposure to Molecular Dynamics Phosphorimager screens (Sunnyvale, CA) overnight. Densitometry was performed using ImageQuant software.
Reverse Transcription PCR
Primers for each differentially expressed gene were designed and ordered from Gibco BRL Life Technologies. Each primer was diluted to 1 OD/100 µL and forward primers were then end-labeled with [γ-32P]dATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). A total of 4 µg normal/tumor tissue total RNA and 2.5 µg oligo-dT primer were denatured at 65°C for 10 minutes and placed on ice. To this mixture the following components were added: 1 x reverse transcription buffer (50 mM Tris-HCl, pH 8.3; 75 mM KCl; 3 mM MgCl2); 10 mM DTT; 1.0 mM of each dATP, dTTP, and dGTP; 100 µCi [α-32P]dCTP or 1.0 mM dCTP; 60 U RNAsin (Promega); and 800 U MMLV reverse transcriptase (Gibco BRL Life Technologies). Reaction mixtures were incubated at 37°C for 1 hour. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control in each PCR amplification. The coamplification of the control cDNA and the target gene cDNA in tumor and normal tissues provided a means to control for PCR amplification and enabled the relative level of the target gene expression to be quantified. PCR reactions using 1 µL cDNA, 1xreaction buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.75 units of Taq DNA Polymerase (Promega), labeled forward primers, and unlabeled reverse primers were subjected to 18 to 24 cycles of amplification at 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute per cycle. To determine the linear range of each set of PCR reactions, a series of 3 to 4 PCR reactions using 18, 20, 22, or 24 cycles were performed for each target gene, and one of the reactions that fitted into the linear range was used for further quantitation. PCR products were then electrophoresed on 8% polyacrylamide gels with urea and exposed to Molecular Dynamics Phosphorimager screens for analysis. Densitometry was performed by ImageQuant software.
Results
The 588 human cDNAs immobilized on each Atlas human cDNA expression array consisted of 200 to 600 base pair fragments amplified from mRNA regions without polyA tails, highly homologous sequences, or base sequence repeats. This ensured that nonspecific background and cross hybridization of closely related gene family members was minimized. The low background levels acquired from use of primers specific for the genes immobilized on the nylon membrane allowed for the detection of mRNAs as rare as 10 to 20 copies per cell [26]. The cDNA spot signal intensity was compared to three housekeeping gene spot signal intensities (GAPDH, β-actin, and 60S ribosomal RNA gene) to determine the relative abundance of target cDNA in the hybridized sample.
Six different sets of cDNAs were immobilized on cDNA expression arrays at approximately 10 ng per spot including: 1) oncogenes, tumor suppressor genes, and cell cycle regulators; 2) ion channel and transport, stress response, and modulators, effectors, and intracellular transducer genes; 3) genes involved in apoptosis, DNA synthesis, repair and recombination; 4) transcription factors and DNA binding proteins; 5) receptors, cell surface antigens, and cell adhesion molecules; 6) growth factors, cytokines, chemokines, hormones, interleukins, and interferons [26].
Using the Altas human cDNA expression array, four sets of hybridizations were performed with reverse-transcribed polyA+ RNA of two squamous cell carcinomas and their matching normal tissues (patients 8611 and 8706) and two adenocarcinomas and their matching normal tissues (patients 8641 and 94-11-C066/65R). Representative examples of the pathologic features of an adenocarcinoma and its paired normal alveoli as well as a squamous cell carcinoma and its paired normal bronchiole are shown in Figure 1.
Of the 588 cDNAs present on the cDNA expression array, an average of 262 spots (45%) were detected after hybridization. After normalization to housekeeping genes GAPDH, β-actin, and 60S ribosomal protein L13A, sequences that were differentially expressed by at least two-fold between tumor and normal tissues were selected for further confirmation. Because each housekeeping gene standardized the cDNA spot intensities to different relative levels, only those cDNAs that were differentially expressed by at least two-fold when normalized to each of these housekeeping genes were examined further. The two-fold expression differences also needed to be detected in at least two of the four individual hybridizations that were performed, with two expression differences occurring in the same lung cancer subtype. Forty-five genes (17%) were differentially expressed in the cDNA expression array screening and selected for reverse-transcriptase polymerase chain reaction (RT-PCR) confirmation. Of these, 22 were overexpressed and 23 were underexpressed in the tumor tissues compared to their normal counterparts. The initial RT-PCR analysis was performed using seven pairs of matching normal/tumor tissues (three squamous cell carcinomas and four adenocarcinomas). Only the genes that displayed differential expression in >50% of the squamous cell carcinomas and/or adenocarcinomas were reanalyzed using the second set of seven matching normal/tumor tissues (four squamous cell carcinomas and three adenocarcinomas). Final analysis included 26 of the initial 45 genes (58%) that were differentially expressed by cDNA expression array analysis.
Of the 26 genes, 14 genes were overexpressed and 12 genes were underexpressed in the tumor tissues compared to their corresponding normal tissues (Table 2). Most of the differentially expressed genes were detected in both squamous cell carcinomas and adenocarcinomas. Eight genes were overexpressed or underexpressed in at least 70% of adenocarcinomas and squamous cell carcinomas. Of these, two were overexpressed (CD27L antigen receptor and ephrin A3) whereas six were underexpressed (c-kit, protein tyrosine kinase receptor tyro3, c-fgr proto-oncogene, TR3 orphan receptor, amphiregulin (AR), and macrophage inflammatory protein 2 alpha). However, some expression differences were more common in adenocarcinomas (c-jun, ephrin type A receptor 1, and interleukin 2 receptor alpha [IL2Rα]). Differential expression of intercellular adhesion molecule 1 (ICAM1) was more frequently detected in squamous cell carcinomas. The rasrelated gene, rab2, was only differentially expressed in adenocarcinomas. Some genes were overexpressed in one NSCLC subtype but were variable or underexpressed in the other subtype. Cdc25A was overexpressed by at least two-fold in some squamous cell carcinomas but underexpressed in some adenocarcinomas compared to their respective normal tissues. Caspase 8 and insulin receptor were overexpressed in most adenocarcinomas but underexpressed in squamous cell carcinomas. Because sample sizes were small, more NSCLC subtypes need to be analyzed to substantiate these tendencies. Figures 2–4 show the representative results of differential expression of c-jun, AR, and IL2Rα. Further verification of differential gene expression in either squamous cell carcinomas or adenocarcinomas by Northern blot analysis could not be performed due to the limited amount of microdissected tumor tissues available for the present study.
Table 2.
Differentially Expressed Genes in Adenocarcinomas and Squamous Cell Carcinomas.
| No. | Gene | GenBank No. | RT | RT Frequency |
| Oncogenes, tumor supressor genes, and cell cycle regulators | ||||
| 1 | protein tyrosine kinase c-kit | X06182 | - | 5/7S, 6/7A, 1S&1A(+) |
| 2 | c-jun | J04111 | - | 2/7S, 5/7A |
| 3 | protein tyrosine kinase receptor tyro3 | D17517 | - | 5/7S, 5/7A |
| 4 | c-src | K03214 | - | 5/7S, 3/7A, 4S(+) |
| 5 | c-fgr proto-oncogene | M19722 | - | 5/7S, 4/7A, 1A(+) |
| 6 | TR3 orphan receptor | L13740 | - | 6/7S, 6/7A, 1S(+) |
| 7 | cdc25A | M81933 | + | 5/7S, 3/7A, 2A(-) |
| Stress response and modulators, effectors, and intracellular transducers | ||||
| 8 | ephrin type-A receptor 1 | M18391 | + | 3/7S, 6/7A, 1S&2A(-) |
| 9 | LIM domain kinase 1 (LIMK-1) | D26309 | + | 3/7S, 4/7A, 1S(-) |
| 10 | ras-related protein RAB6 | M28212 | + | 3/7S, 3/7A, 1S(-) |
| 11 | ras-related protein RAB2 | M28213 | + | 0S, 5/7A |
| 12 | janus kinase 3 (JAK3) | U09607 | + | 4/7S, 3/7A, 1S&2A(-) |
| 13 | c-jun N-terminal kinase1 (JNK1) | L26318 | + | 4/7S, 3/7A, 1S(-) |
| 14 | ephrin A3 (EFNA3) | U14187 | + | 7/7S, 6/7A |
| 15 | transmembrane receptor PTK7 | U40271 | + | 3/7S, 5/7A, 1S(-) |
| Apoptosis and DNA synthesis, repair, and recombination | ||||
| 16 | adenosine A1 receptor (ADORA1) | S56143 | - | 5/7S, 3/7A, 2S(+) |
| 17 | caspase 8 | U60520 | - | 3/7S, 4/7A, 3A(+) |
| Receptors, cell surface antigens, and cell adhesion | ||||
| 18 | insulin receptor (INSR) | M10051 | + | 3/7S, 4/7A, 2S(-) |
| 19 | IL2Rα | X01057 | + | 4/7S, 7/7A |
| 20 | CD27L antigen receptor | M63928 | + | 5/7S, 5/7A |
| 21 | ICAM1 | J03132 | - | 7/7S, 2/7A, 1S&1A(+) |
| Growth factors, cytokines, chemokines, and interferons | ||||
| 22 | HGF activator | D14012 | + | 4/7S, 6/7A, 2S&1A(-) |
| 23 | AR | M30704 | 6/7S, 5/7A, 2A(+) | |
| 24 | vascular endothelial growth factor C | U43142 | - | 4/7S, 2/7A, 1S&1A(+) |
| 25 | platelet-derived growth factor A (PDGFA) | X06374 | + | 4/7S, 4/7A, 2S&1A(-) |
| 26 | macrophage inflammatory protein 2 alpha | X53799 | - | 5/7S, 6/7A |
A, adenocarcinoma. S, squamous cell carcinoma. -, underexpressed; +, overexpressed compared to normal lung tissue from the same patient. RT, reverse transcription-PCR analysis.
RT Frequency, ratio of the number of tumors with differential expression over the total number of normal/tumor tissue pairs analyzed; S/A(+/-), differential expression detected in tumors that did not reflect what was found in the majority of normal/tumor tissue pairs.
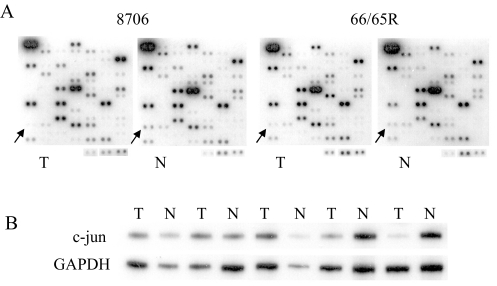
Figure 2.
Differential expression of c-jun. (A) cDNA expression array hybridization with the squamous cell carcinoma 8706 and its matching normal tissue and the adenocarcinoma 94-11-C066R (66R) and its matching normal tissue 94-11-C065R (65R). (B) RT-PCR confirmation. The first two pairs of tumor/normal tissues from the left are squamous cell carcinomas of the lung. The remaining three pairs of tumor/normal tissues are lung adenocarcinomas.
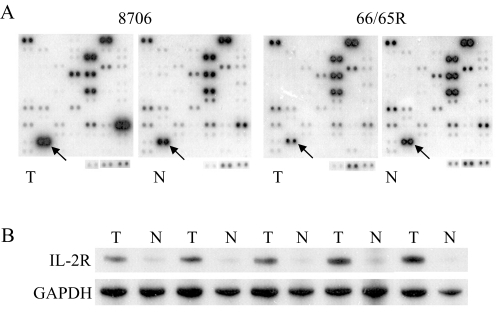
Figure 4.
Differential expression of IL-2Rα. (A) cDNA expression array hybridization with the squamous cell carcinoma 8706 and its matching normal tissue and the adenocarcinoma 94-11-C066R (66R) and its matching normal tissue 94-11-C065R (65R). (B) RT-PCR confirmation. The first two pairs of tumor/normal tissues from the left are squamous cell carcinomas of the lung. The remaining three pairs of tumor/normal tissues are lung adenocarcinomas.
Discussion
In this study, we compared the gene expression patterns of two subtypes of human NSCLC, adenocarcinomas and squamous cell carcinomas, to their matching normal lung tissues using Atlas human cDNA expression arrays. The use of this limited cDNA microarray with known genes, which were potentially related to cancer, certainly enriched for genes that might appear to be related directly to the cancer process. Approximately half of the 588 spotted cDNAs were detected in these normal and tumor tissues. Forty-five of the 262 cDNAs were differentially expressed according to the parameters set at the beginning of the study and were selected for further RT-PCR confirmation. Twenty-six genes including oncogenes, cell cycle regulators, intracellular signal transducers, apoptotic genes, cell receptors and adhesion molecules, and growth factors were differentially expressed in adenocarcinomas and/or squamous cell carcinomas compared to normal lung tissues obtained from the same patient. We found eight genes that were reproducibly (>70% of samples) altered in both adenocarcinomas and squamous cell carcinomas when compared with their normal controls. In general, the expression of genes identified in both adenocarcinomas and squamous cell carcinomas were routinely increased or decreased by more than two-fold.
Proto-oncogenes
Several members of this group were routinely altered in expression in both adenocarcinomas and squamous cell carcinomas. Interestingly, most of the genes with aberrant expression had decreased expression relative to their own controls including c-kit, c-fgr, c-src, and c-jun. c-Kit encodes a transmembrane tyrosine kinase receptor. It functions in cell survival, proliferation, migration, and differentiation [27]. The c-kit and stem cell factor (SCF) autocrine loop in SCLC may mediate chemotaxis and/or provide a growth advantage in these tumors [28,29]. Chemotactic and chemokinetic mobility was detected in lung cancer cell lines expressing c-kit and SCF [30]. Tonary et al. reported that c-kit expression was decreased in advanced stages of ovarian cancer and was associated with decreased survival [31].
Non-receptor tyrosine kinase, c-src, has been implicated in the tumorigenesis of multiple human cancers. Reported alterations involved both increases and decreases in enzymatic activity, gene copy number, and protein levels [32]. In lung cancer, elevated c-src expression was found in 60% of tumors of different histologic types, including adenocarcinomas and squamous cell carcinomas, compared to normal bronchial tissue [33,34]. Budde et al. reported high c-src activity for NSCLC cell lines but low activity in SCLC cell lines [35]. In our study, c-src levels were elevated in the majority of squamous cell carcinomas but were decreased in three of seven adenocarcinomas. This may suggest different functional roles for this proto-oncogene in these NSCLC subtypes.
c-Jun is a tightly regulated component of the activator protein-1 transcription factor complex. It is involved in cell proliferation and survival but can also induce apoptosis, growth arrest, and differentiation [36]. Expression analysis of c-jun in lung cancer has yielded conflicting results. Koomagi et al. reported elevated levels of c-jun in tumor tissues of lung cancer patients compared to adjacent normal tissues [37]. However, in an additional study comparing 101 NSCLC specimens to adjacent normal lung tissue, 72% of the normal tissues expressed higher levels of c-jun [38]. In light of these observations, additional studies will need to be performed to define the role of c-kit, c-fgr, c-src, and c-jun in tumorigenesis.
Growth Factors and Related Genes
Several growth factors and related genes were also expressed differentially in the NSCLC subtypes compared to normal lung tissues. Hepatocyte growth factor (HGF) is a potent inducer of cell proliferation, motility, and morphogenesis [39]. A probable autocrine loop involving HGF and its receptor c-met exists in NSCLC [6,40]. HGF is secreted in an inactive form and must be proteolytically activated by factors such as HGF activator (HGFA) [41]. Although HGFA levels have not been examined in many tissues (including lung), significantly higher HGFA levels were reported in breast cancer tissue than in normal control tissues [39]. Increased HGFA expression was found in two of seven squamous cell carcinomas and five of seven adenocarcinomas.
Another differentially expressed gene in this study was AR. It is a strong growth promoter for breast, colon, lung, and ovarian epithelial cells and may function in autocrine and paracrine loops with the epidermal growth factor receptor (EGFR) to promote tumorigenesis [42,43]. However, it has also been reported to inhibit growth of carcinoma cell lines [44,45]. Both reduced (63%) and increased (11%) AR expression levels have been reported in NSCLC tumors compared to uninvolved lung tissue [46]. Most of the NSCLC samples in our study exhibited reduced AR expression compared to normal lung tissue. Only two of seven (28%) adenocarcinomas showed increased expression of AR compared to normal lung counterparts.
Intracellular Signal Transducers
Several modulators, effectors, and intracellular transducers showed increased expression in the human lung tumors. The receptor tyrosine kinase erythropoietin-producing hepatocellular (eph) family transduces signals to proteins involved in cytoskeletal organization, adhesion, migration, and proliferation [47,48] and may also be involved in angiogenesis [49–51]. Overexpression of eph receptors has been detected in gastric carcinomas, malignant melanomas, and hepatomas [52]. Overexpression of ephA1 receptor was able to transform NIH3T3 cells [48]. We found frequent overexpression of both ephA1 receptor and ephrin A3 ligand in both squamous cell carcinomas and adenocarcinomas compared to matching normal tissues.
LIM domain kinase (LIMK-1) is a serine/threonine kinase involved in the Rho GTPase signaling pathway that leads to actin cytoskeleton reorganization and the formation of lamellipodia [53–55]. Although LIMK-1 has not previously been implicated in cancer, it was overexpressed in two of seven squamous cell carcinomas and four of seven adenocarcinomas in this study.
Apoptotic Factors
Another component of neoplastic transformation is the prevention of apoptosis. Death receptor-dependent and -independent activation of caspase 8 leads to cleavage and activation of effector caspases 3, 6, and 7 and other intermediate apoptotic signaling proteins [56,57] that then initiate cell death. In neuroblastoma, caspase 8 was often inactivated by gene deletion and/or DNA methylation in cells with N-myc amplification [58]. However, in several other cancers including high-grade non-Hodgkins lymphoma, breast carcinoma, pancreatic carcinoma, and gallbladder carcinoma, caspase 8 gene expression was upregulated [59–62] and was strongly associated with the extent of apoptosis [58,60,62]. In this study, caspase 8 was down-regulated in several squamous cell carcinomas (3/7) but primarily upregulated in adenocarcinomas (3/7).
Using the rapid, high-throughput human Atlas cDNA expression array system, we examined the gene expression profiles of 14 NSCLCs and their matching normal tissues. Numerous cell surface receptors, proto-oncogenes, growth factors, and signal transduction molecules were differentially expressed in these NSCLC subtypes and may have significant roles in lung tumorigenesis. The divergent expression patterns of c-src, caspase 8, cdc25A, rab2, and ICAM1 (see Table 2) suggest subtle but probable molecular differences between human lung adenocarcinomas and squamous cell carcinomas. These differences may ultimately play a role in determining the best treatments for these two NSCLC subtypes. Differential gene expression in tumor and normal lung tissues does not necessarily suggest tumor promoter or tumor inhibitory function. These differences may be a consequence of other biochemical changes that occur in a cell on its path to neoplastic transformation. Nevertheless, the changes in gene expression may serve as diagnostic or prognostic tumor biomarkers. Others may prove to be modulated by therapeutic or preventative agents and, thus, serve as surrogate endpoint biomarkers.
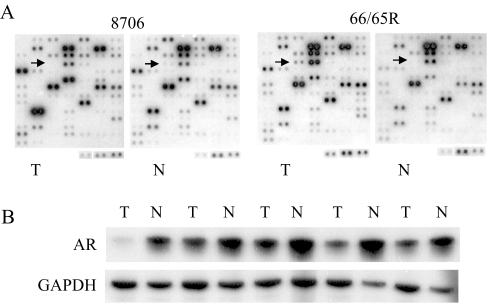
Figure 3.
Differential expression of AR. (A) cDNA expression array hybridization with the squamous cell carcinoma 8706 and its matching normal tissue and the adenocarcinoma 94-11-C066R (66R) and its matching normal tissue 94-11-C065R (65R). (B) RT-PCR confirmation. The first two pairs of tumor/normal tissues from the left are squamous cell carcinomas of the lung. The remaining three pairs of tumor/normal tissues are lung adenocarcinomas.
Abbreviations
- AR
amphiregulin
- eph
erythropoietin-producing hepatocellular
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HGF
hepatocyte growth factor
- ICAM1
intercellular adhesion molecule 1
- IL2Rα
interleukin 2 receptor alpha
- NSCLC
non-small cell lung cancer
- RT-PCR
reverse-transcriptase polymerase chain reaction
- SCLC
small cell lung cancer
Footnotes
This work was supported by NIH grants CN05113, R01CA58554, R01CA78797, and P30CA16058.
References
- 1.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer Statistics, 2001. CA-Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg RA. Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res. 1989;49:3713–3721. [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 4.Pitterle DM, Jolicoeur EM, Bepler G. Hot spots for molecular genetic alterations in lung cancer. In Vivo. 1998;12:643–658. [PubMed] [Google Scholar]
- 5.Wang XW. Role of p53 and apoptosis in carcinogenesis. Anticancer Res. 1999;19:4759–4771. [PubMed] [Google Scholar]
- 6.Sekido Y, Fong KM, Minna JD. Progress in understanding the molecular pathogenesis of human lung cancer. Biochim Biophys Acta. 1998;1378:F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 7.Robbins SL, Kumar V, Cotran RS. In: Lung Cancer. Saunders WB, editor. Philadelphia, PA: 1994. pp. 673–734. [Google Scholar]
- 8.Linnoila RI, Aisner SC. Pathology of Lung cancer: an Exercise in Classification. New York, NY: Wiley-Liss; 1995. pp. 73–98. [Google Scholar]
- 9.Jiang SX, Sato Y, Kuwao S, Kameya T. Expression of Bcl-2 oncogene protein is prevalent in small cell lung carcinomas. J Pathol. 1995;177:135–138. doi: 10.1002/path.1711770206. [DOI] [PubMed] [Google Scholar]
- 10.Jiang SX, Kameya T, Sato Y, Yanase N, Yoshimura H, Kodama T. Bcl-2 protein expression in lung cancer and close correlation with neuroendocrine differentiation. Am J Pathol. 1996;148:837–846. [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser U, Schilli M, Haag U, Neumann K, Kreipe H, Kogan E, Havemann K. Expression of Bcl-2-protein in small cell lung cancer. Lung Cancer. 1996;15:31–40. doi: 10.1016/0169-5002(96)00568-5. [DOI] [PubMed] [Google Scholar]
- 12.Pezzella F, Turley H, Kuzu I, Tungekar MF, Dunnill MS, Pierce CB, Harris A, Gatter KC, Mason DY. Bcl-2 protein in non-small-cell lung carcinoma. N Engl J Med. 1993;329:690–694. doi: 10.1056/NEJM199309023291003. [DOI] [PubMed] [Google Scholar]
- 13.Higashiyama M, Doi O, Kodama K, Yokouchi H, Nakamori S, Tateishi R. Bcl-2 oncoprotein in surgically resected non-small cell lung cancer: possibly favorable prognostic factor in association with low incidence of distant metastasis. J Surg Oncol. 1997;64:48–54. doi: 10.1002/(sici)1096-9098(199701)64:1<48::aid-jso10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 14.Reissmann PT, Koga H, Takahashi R, Figlin RA, Holmes EC, Piantadosi S, Cordon-Cardo C, Slamon DJ. Inactivation of the retinoblastoma susceptibility gene in non-small-cell lung cancer. The Lung Cancer Study Group. Oncogene. 1993;8:1913–1919. [PubMed] [Google Scholar]
- 15.Cagle PT, el-Naggar AK, Xu HJ, Hu SX, Benedict WF. Differential retinoblastoma protein expression in neuroendocrine tumors of the lung. Potential diagnostic implications. Am J Pathol. 1997;150:393–400. [PMC free article] [PubMed] [Google Scholar]
- 16.Dosaka-Akita H, Hu SX, Fujino M, Harada M, Kinoshita I, Xu HJ, Kuzumaki N, Kawakami Y, Benedict WF. Altered retinoblastoma protein expression in nonsmall cell lung cancer: its synergistic effects with altered ras and p53 protein status on prognosis. Cancer. 1997;79:1329–1337. [PubMed] [Google Scholar]
- 17.Washimi O, Nagatake M, Osada H, Ueda R, Koshikawa T, Seki T, Takahashi T. In vivo occurrence of p16 (MTS1) and p15 (MTS2) alterations preferentially in non-small cell lung cancers. Cancer Res. 1995;55:514–517. [PubMed] [Google Scholar]
- 18.Shapiro GI, Park JE, Edwards CD, Mao L, Merlo A, Sidransky D, Ewen ME, Rollins BJ. Multiple mechanisms of p16INK4A inactivation in non-small cell lung cancer cell lines. Cancer Res. 1995;55:6200–6209. [PubMed] [Google Scholar]
- 19.Richardson GE, Johnson BE. The biology of lung cancer. Semin Oncol. 1993;20:105–127. [PubMed] [Google Scholar]
- 20.Eerola AK, Tormanen U, Rainio P, Sormunen R, Bloigu R, Vahakangas K, Lehto VP, Soini Y, Paakko P. Apoptosis in operated small cell lung carcinoma is inversely related to tumour necrosis and p53 immunoreactivity. J Pathol. 1997;181:172–177. doi: 10.1002/(SICI)1096-9896(199702)181:2<172::AID-PATH715>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Nishio M, Koshikawa T, Kuroishi T, Suyama M, Uchida K, Takagi Y, Washimi O, Sugiura T, Ariyoshi Y, Takahashi T, Ueda R. Prognostic significance of abnormal p53 accumulation in primary, resected non-small-cell lung cancers. J Clin Oncol. 1996;14:497–502. doi: 10.1200/JCO.1996.14.2.497. [DOI] [PubMed] [Google Scholar]
- 22.Konishi T, Lin Z, Fujino S, Kato H, Mori A. Association of p53 protein expression in stage I lung adenocarcinoma with reference to cytological subtypes. Hum Pathol. 1997;28:544–548. doi: 10.1016/s0046-8177(97)90076-9. [DOI] [PubMed] [Google Scholar]
- 23.Kohno T, Yokota J. How many tumor suppressor genes are involved in human lung carcinogenesis? Carcinogenesis. 1999;20:1403–1410. doi: 10.1093/carcin/20.8.1403. [DOI] [PubMed] [Google Scholar]
- 24.Schwendel A, Langreck H, Reichel M, Schrock E, Ried T, Dietel M, Petersen I. Primary small-cell lung carcinomas and their metastases are characterized by a recurrent pattern of genetic alterations. Int J Cancer. 1997;74:86–93. doi: 10.1002/(sici)1097-0215(19970220)74:1<86::aid-ijc15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 25.Testa JR, Liu Z, Feder M, Bell DW, Balsara B, Cheng JQ, Taguchi T. Advances in the analysis of chromosome alterations in human lung carcinomas. Cancer Genet Cytogenet. 1997;95:20–32. doi: 10.1016/s0165-4608(96)00337-8. [DOI] [PubMed] [Google Scholar]
- 26.Clontech, author. I. Atlas cDNA Expression Arrays User Manual. Palo Alto, CA: Clontech Laboratories, Inc.; 1999. pp. 4–12. [Google Scholar]
- 27.Ashman LK. The biology of stem cell factor and its receptor C-kit. Int J Biochem Cell Biol. 1999;31:1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- 28.Sekido Y, Takahashi T, Ueda R, Takahashi M, Suzuki H, Nishida K, Tsukamoto T, Hida T, Shimokata K, Zsebo KM, et al. Recombinant human stem cell factor mediates chemotaxis of small-cell lung cancer cell lines aberrantly expressing the c-kit protooncogene. Cancer Res. 1993;53:1709–1714. [PubMed] [Google Scholar]
- 29.Krystal GW, Hines SJ, Organ CP. Autocrine growth of small cell lung cancer mediated by coexpression of c-kit and stem cell factor. Cancer Res. 1996;56:370–376. [PubMed] [Google Scholar]
- 30.Bredin CG, Liu Z, Hauzenberger D, Klominek J. Growth-factor-dependent migration of human lung-cancer cells. Int J Cancer. 1999;82:338–345. doi: 10.1002/(sici)1097-0215(19990730)82:3<338::aid-ijc6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 31.Tonary AM, Macdonald EA, Faught W, Senterman MK, Vanderhyden BC. Lack of expression of c-KIT in ovarian cancers is associated with poor prognosis. Int J Cancer. 2000;89:242–250. doi: 10.1002/1097-0215(20000520)89:3<242::aid-ijc6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Biscardi JS, Tice DA, Parsons SJ. c-Src, receptor tyrosine kinases, and human cancer. Adv Cancer Res. 1999;76:61–119. doi: 10.1016/s0065-230x(08)60774-5. [DOI] [PubMed] [Google Scholar]
- 33.Mazurenko NN, Kogan EA, Zborovskaia IB, Sukhova NM, Kiselev FL. The detection of the c-src protein gene product in human lung tumors. Vopr Onkol. 1991;37:683–690. [PubMed] [Google Scholar]
- 34.Mazurenko NN, Kogan EA, Sukhova NM, Zborovskaia IB. Synthesis and distribution of oncoproteins in tumor tissue. Vopr Med Khim. 1991;37:53–59. [PubMed] [Google Scholar]
- 35.Budde RJ, Ke S, Levin VA. Activity of pp60c-src in 60 different cell lines derived from human tumors. Cancer Biochem Biophys. 1994;14:171–175. [PubMed] [Google Scholar]
- 36.Leppa S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. 1999;18:6158–6162. doi: 10.1038/sj.onc.1203173. [DOI] [PubMed] [Google Scholar]
- 37.Koomagi R, Stammler G, Manegold C, Mattern J, Volm M. Expression of resistance-related proteins in tumoral and peritumoral tissues of patients with lung cancer. Cancer Lett. 1996;110:129–136. doi: 10.1016/s0304-3835(96)04471-0. [DOI] [PubMed] [Google Scholar]
- 38.Levin WJ, Press MF, Gaynor RB, Sukhatme VP, Boone TC, Reissmann PT, Figlin RA, Holmes EC, Souza LM, Slamon DJ. Expression patterns of immediate early transcription factors in human non-small cell lung cancer. The Lung Cancer Study Group. Oncogene. 1995;11:1261–1269. [PubMed] [Google Scholar]
- 39.Jiang W, Hiscox S, Matsumoto K, Nakamura T. Hepatocyte growth factor/scatter factor, its molecular, cellular and clinical implications in cancer. Crit Rev Oncol Hematol. 1999;29:209–248. doi: 10.1016/s1040-8428(98)00019-5. [DOI] [PubMed] [Google Scholar]
- 40.Olivero M, Rizzo M, Madeddu R, Casadio C, Pennacchietti S, Nicotra MR, Prat M, Maggi G, Arena N, Natali PG, Comoglio PM, Di Renzo MF. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br J Cancer. 1996;74:1862–1868. doi: 10.1038/bjc.1996.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazawa K, Shimomura T, Kitamura N. Activation of hepatocyte growth factor in the injured tissues is mediated by hepatocyte growth factor activator. J Biol Chem. 1996;271:3615–3618. doi: 10.1074/jbc.271.7.3615. [DOI] [PubMed] [Google Scholar]
- 42.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 43.Fontanini G, De Laurentiis M, Vignati S, Chine S, Lucchi M, Silvestri V, Mussi A, De Placido S, Tortora G, Bianco AR, Gullick W, Angeletti CA, Bevilacqua G, Ciardiello F. Evaluation of epidermal growth factor-related growth factors and receptors and of neoangiogenesis in completely resected stage I-IIIA non-small-cell lung cancer: amphiregulin and microvessel count are independent prognostic indicators of survival. Clin Cancer Res. 1998;4:241–249. [PubMed] [Google Scholar]
- 44.Plowman GD, Green JM, McDonald VL, Neubauer MG, Disteche CM, Todaro GJ, Shoyab M. The amphiregulin gene encodes a novel epidermal growth factor-related protein with tumor-inhibitory activity. Mol Cell Biol. 1990;10:1969–1981. doi: 10.1128/mcb.10.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoyab M, McDonald VL, Bradley JG, Todaro GJ. Amphiregulin: a bifunctional growth-modulating glycoprotein produced by the phorbol 12-myristate 13-acetate-treated human breast adenocarcinoma cell line MCF-7. Proc Natl Acad Sci USA. 1988;85:6528–6532. doi: 10.1073/pnas.85.17.6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusch V, Baselga J, Cordon-Cardo C, Orazem J, Zaman M, Hoda S, McIntosh J, Kurie J, Dmitrovsky E. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res. 1993;53:2379–2385. [PubMed] [Google Scholar]
- 47.Kalo MS, Pasquale EB. Signal transfer by Eph receptors. Cell Tissue Res. 1999;298:1–9. [PubMed] [Google Scholar]
- 48.Nakamoto M. Eph receptors and ephrins. Int J Biochem Cell Biol. 2000;32:7–12. doi: 10.1016/s1357-2725(99)00096-5. [DOI] [PubMed] [Google Scholar]
- 49.Pandey A, Shao H, Marks RM, Polverini PJ, Dixit VM. Role of B61, the ligand for the Eck receptor tyrosine kinase, in TNF-alpha-induced angiogenesis. Science. 1995;268:567–569. doi: 10.1126/science.7536959. [DOI] [PubMed] [Google Scholar]
- 50.Daniel TO, Stein E, Cerretti DP, St John PL, Robert B, Abrahamson DR. ELK and LERK-2 in developing kidney and microvascular endothelial assembly. Kidney Int Suppl. 1996;57:S73–S81. [PubMed] [Google Scholar]
- 51.Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 52.Xu Q, Wilkinson DG. Eph-related receptors and their ligands: mediators of contact dependent cell interactions. J Mol Med. 1997;75:576–586. doi: 10.1007/s001090050142. [DOI] [PubMed] [Google Scholar]
- 53.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 54.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 55.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 56.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri ES. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bratton SB, MacFarlane M, Cain K, Cohen GM. Protein complexes activate distinct caspase cascades in death receptor and stress-induced apoptosis. Exp Cell Res. 2000;256:27–33. doi: 10.1006/excr.2000.4835. [DOI] [PubMed] [Google Scholar]
- 58.Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 59.Soini Y, Paakko P. Apoptosis and expression of caspases 3, 6 and 8 in malignant non-Hodgkin's lymphomas. APMIS. 1999;107:1043–1050. doi: 10.1111/j.1699-0463.1999.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 60.Vakkala M, Paakko P, Soini Y. Expression of caspases 3, 6 and 8 is increased in parallel with apoptosis and histological aggressiveness of the breast lesion. Br J Cancer. 1999;81:592–599. doi: 10.1038/sj.bjc.6690735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virkajarvi N, Paakko P, Soini Y. Apoptotic index and apoptosis influencing proteins Bcl-2, mcl-1, bax and caspases 3, 6 and 8 in pancreatic carcinoma. Histopathology. 1998;33:432–439. doi: 10.1046/j.1365-2559.1998.00553.x. [DOI] [PubMed] [Google Scholar]
- 62.Turunen N, Paakko P, Soini Y. Apoptosis in gallbladder carcinomas and dysplasias, its relation to the expression of caspases 3, 6 and 8 and apoptosis regulating proteins Bcl-2, mcl-1 and bax. Histol Histopathol. 2000;15:53–60. doi: 10.14670/HH-15.53. [DOI] [PubMed] [Google Scholar]