Abstract
Abstract Comparative genomic hybridization (CGH) studies have provided a wealth of information on common copy number aberrations in pancreatic cancer, but the genes affected by these aberrations are largely unknown. To identify putative amplification target genes in pancreatic cancer, we performed a parallel copy number and expression survey in 13 pancreatic cancer cell lines using a 12,232-clone cDNA microarray, providing an average resolution of 300 kb throughout the human genome. CGH on cDNA microarray allowed highly accurate mapping of copy number increases and resulted in identification of 24 independent amplicons, ranging in size from 130 kb to 11 Mb. Statistical evaluation of gene copy number and expression data across all 13 cell lines revealed a set of 105 genes whose elevated expression levels were directly attributable to increased copy number. These included genes previously reported to be amplified in cancer as well as several novel targets for copy number alterations, such as p21-activated kinase 4 (PAK4), which was previously shown to be involved in cell migration, cell adhesion, and anchorage-independent growth. In conclusion, our results implicate a set of 105 genes that is likely to be actively involved in the development and progression of pancreatic cancer.
Keywords: Pancreatic cancer, amplification, CGH microarray, cDNA microarray, overexpression
Introduction
Nearly 30,000 new pancreatic cancer cases were diagnosed in the United States in 2001, representing about 2.3% of all new cancer cases [1]. Despite this relatively low incidence, pancreatic cancer was the fourth common cause of cancer deaths in the United States, with 5-year survival rates of 3% to 5% [1,2]. The poor prognosis of pancreatic cancer is largely due to the fact that the first symptoms (e.g., jaundice caused by the growing tumor obstructing the common bile duct, or abdominal and back pain caused by perineural invasion in the celiac plexus) come typically rather late in the disease progression. Therefore, most pancreatic cancers have already metastasized and are inoperable at the time of diagnosis [3,4].
There are several well-documented risk factors for the development of pancreatic cancer, including smoking [5], chronic pancreatitis [6], and a family history of pancreatic cancer [7,8]. However, the genetic changes involved in the pathogenesis of pancreatic cancer are not fully understood, although the role of a few known oncogenes and tumorsuppressor genes has been well established (reviewed in Ref. [9]). Mutations leading to activation of the KRAS oncogene are the most common genetic changes in pancreatic cancer occurring in nearly all primary tumors [9,10]. KRAS mutations have been observed already in normal pancreas as well as in noninvasive neoplastic precursor lesions, indicating that they represent an early event in the pathogenesis of pancreatic cancer [11]. Furthermore, overexpression of EGFR and MYC, as well as inactivation of TP53 and MADH4, have been frequently detected in pancreatic cancer [10,12–14]. In addition to these alterations involving known oncogenes or tumor-suppressor genes, cytogenetic and molecular cytogenetic studies have revealed frequent structural and numeric chromosome abnormalities in pancreatic cancer. For example, comparative genomic hybridization (CGH) analyses have indicated common gains affecting chromosomes 7, 8q, 17q, 19q, and 20q, and losses of 6q, 8p, 9p, and 18q in pancreatic adenocarcinomas [15–19].
Recently, DNA microarray-based methods have been applied to the analysis of pancreatic cancer, mainly in an effort to identify new diagnostic markers or targets for the development of new therapies against pancreatic cancer [20–25]. These studies have revealed a considerable number of differentially expressed genes, the majority of which has not been previously implicated in pancreatic cancer. For example, Iacobuzio- Donahue et al. [24] identified a set of more than 400 genes that were differentially expressed in pancreatic cancer tissues and cell lines as compared to normal pancreas. These genes were linked to multiple cellular processes, such as cell-cell and cell-matrix interactions, cytoskeletal remodeling, proteolytic activity, and Ca2+ homeostasis. A set of 149 genes was more highly expressed in pancreatic cancers compared with normal pancreas and contained 103 genes that had not been previously reported in association with pancreatic cancer [24]. However, the exact role of these differentially expressed genes discovered by Iacobuzio- Donahue et al. [24] and others in pancreatic cancer pathogenesis remains to be elucidated.
In this study, we used a combination of CGH and cDNA microarray technologies to specifically identify those gene expression change events that were associated with gene copy number alterations. Analysis of 13 pancreatic cancer cell lines using a 12,232-clone cDNA microarray revealed 24 independent amplicons and a set of 105 genes whose expression levels were directly attributable to increased copy number. We hypothesize that these genes are likely to have a central role in the pathogenesis of pancreatic cancer.
Materials and Methods
Cell Lines
Thirteen established cell lines derived from pancreatic adenocarcinomas or their metastases (AsPC-1, BxPC-3, Capan-1, Capan-2, CFPAC-1, HPAC, HPAF-II, Hs 700T, Hs 766T, MIA PaCa-2, PANC-1, SU.86.86, and SW 1990) were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The CFPAC-1 cell line originated from a patient with cystic fibrosis. The cells were grown under recommended culture conditions. Genomic DNA was isolated using standard protocols and mRNA was extracted with FastTrack 2.0 Kit (Invitrogen, Carlsbad, CA). Both DNA and mRNA preparations were derived from a single harvest of cells.
cDNA Microarray-Based Copy Number and Expression Analyses
The overall experimental design consisted of two parallel microarray experiments where gene copy numbers and gene expression levels were measured in 13 pancreatic cancer cell lines by using an identical cDNA microarray. The cDNA microarray contained polymerase chain reaction (PCR)- amplified inserts derived from a total of 12,232 cDNA clones, representing 8881 different transcripts printed on poly-Llysine- coated glass slides as previously described [26,27] (full description of the clones and their locations on the array is provided at http://sigwww.cs.tut.fi/TICSP/Mahlamaki_ et_al_2003). All experiments were performed using slides from a single print set.
Copy number analyses were performed as previously reported [28,29]. Briefly, genomic DNA from cell lines and sex-matched normal human white blood cells were digested for 14 to 18 hours with AluI and RsaI restriction enzymes (Life Technologies, Inc., Rockville, MD) and purified by phenol/chloroform extraction. Six micrograms of digested cell line and normal DNA was labeled with Cy3-dUTP and Cy5-dUTP (Amersham Pharmacia, Piscataway, NJ), respectively, using Bioprime Labeling kit (Life Technologies, Inc.). The hybridization mixture, containing the labeled probes, 150 µg of CotI DNA (Life Technologies, Inc.), 300 µg of yeast tRNA (Gibco/BRL, Gaithersburg, MD), as well as 60 µg each of poly(dA) and poly(dT) in 3.4 x SSC/0.3% sodium dodecyl sulfate (SDS), was denatured at 100°C for 1.5 minutes, incubated for 30 minutes at 37°C, and hybridized to a microarray slide at 65°C for 16 to 24 hours in a sealed humidified chamber. The slides were then washed in 0.1% SDS, 0.5 x SSC/0.01% SDS, and 0.06 x SSC for 2 minutes each. For expression analyses, a pool of mRNA derived from all 13 cell lines was used as a common reference. Four micrograms of cell line mRNA was labeled with Cy3-dUTP and an equal amount of control mRNA was labeled with Cy5-dUTP by use of oligo(dT)-primed polymerization by Super-Script II reverse transcriptase (Life Technologies, Inc.). The labeled cDNA were hybridized on microarrays as described [27,29]. Briefly, they were combined with 12 µg of poly(dA) (Pharmacia, Bridgewater, NJ), 6 µg of tRNA, and 10 µg of CotI DNA (Life Technologies, Inc.) in 0.25% SDS/2 x SSC. The probe mix was incubated at 98°C for 2 minutes and at 4°C for 10 seconds, and hybridized on a cDNA microarray. Hybridization and subsequent washing were carried out as described above for copy number analyses.
For both copy number and expression analyses, a laser confocal scanner (Agilent Technologies, Palo Alto, CA) was used to measure the fluorescence intensities at the target locations. The fluorescent images from the test and control hybridizations were scanned separately, and image analysis was performed using the DeARRAY software [30]. After background subtraction, the average intensity at each clone in the test hybridization was divided by the average intensity of the corresponding clone in the control hybridization. Within-slide normalization for each cDNA and CGH microarray was performed using the Local Weighted Scatter Plot Smoother (LOWESS) method [31,32] for each print tip group. Fraction of data points used in local regression (f) was 0.4 and other parameters were adjusted as suggested by Cleveland [32]. After within-slide normalization, low quality measurements (i.e., copy number data with mean reference intensity less than 50 fluorescent units, and expression data with both test and reference intensity less than 100 fluorescent units and/or with spot size less than 50 units) were excluded from the analysis and were treated as missing values. Self versus self experiments were performed to ensure the performance of copy number and expression hybridizations. In addition, 15 clones were printed as triplicates on the microarray. Evaluation of data derived from these 15 clones showed highly consistent results for each of the 13 cell lines with standard deviations ranging between 0.01 and 0.346 and between 0.011 and 0.427, for copy number and expression data, respectively (all original data are available at http://sigwww.cs.tut.fi/TICSP/Mahlamaki_ et_al_2003).
Identification of Amplicons
The chromosome and base-pair positions for each cDNA clone on the array were determined by using information from the November 2002 freeze of the University of California Santa Cruz's GoldenPath database (www.genome.ucsc.edu) as described [33]. This information was available for 10,389 clones, providing an average spacing of 308 kb throughout the human genome. The CGH copy number data were ordered according to the location of the clones along chromosomes. Genes with copy number ratio >1.4 (representing the upper 5% of the CGH ratios across all experiments) were considered to be amplified. Two different criteria were used to define amplicons. First, based on the fact that typical amplicons span a large region of the genome, six (providing an average coverage of 1.5 Mb) or more adjacent clones were expected to show a copy number ratio >1.4. Secondly, to avoid missing small amplicons or amplicons located in regions of the genome with poorer-than-average clone coverage, regions with at least three adjacent clones with a copy number ratio >1.4 and no less than one clone with a ratio >2.0 were also considered as amplicons. The amplicon start and end positions were extended to include neighboring nonamplified clones (ratio <1.4). The amplicon size determination was dependent on the local clone density.
Statistical Analyses
The influence of gene copy number on gene expression level was evaluated as described [33,34]. Briefly, within-slide normalized CGH and cDNA ratios in each cell line were logtransformed and median-centered. Furthermore, cDNA data were median-centered using values across 13 cell lines. For each gene, the CGH data were represented by a vector that was labeled “1” for amplification ratio >1.4 and “0” for no amplification. Amplification was correlated with gene expression using the signal-to-noise statistics [33,34]. A weight wg was calculated for each gene:
where mg1,σg1 and mg0,σg0 denote the means and standard deviations for the expression levels for amplified and nonamplified cell lines, respectively. To assess the statistical significance of each weight, 10,000 random permutations of the label vector were performed. The probability that a gene had a larger or equal weight by random permutation than the original weight was denoted by α. A low α (<0.05) indicates a strong association between gene expression and amplification.
Results
A genome-wide copy number analysis was performed in 13 pancreatic cancer cell lines using CGH on a cDNA microarray containing 12,232 clones. Chromosomal and base-pair locations were obtained for 10,389 clones, providing an average spacing of 308 kb throughout the human genome. The clone density varied considerably from one region of the genome to another, with chromosome 19 showing the highest clone density (on average, one clone per 80 kb) and chromosome 13 having the poorest clone density (one clone per 1 Mb). Evaluation of copy number changes as a function of the genomic position of the clones revealed a total number of 24 independent amplicons in the 13 cell lines (Table 1). The number of amplicons varied from one cell line to another, with Capan1 cells showing the highest number of amplicons (8 of 24). The amplicons were located on 12 different chromosomes with multiple separate amplicons observed on chromosomes 15q, 17q, and 19. The use of base-pair information allowed exact determination of the locations and boundaries of each amplicon (Table 1). In several cases, the CGH microarray analysis resulted in high-resolution mapping of amplicons previously detected by chromosomal CGH (Figure 1A). The size of the amplicons ranged from 130 kb to 11 Mb, with an average of 2 Mb (Table 1), although it has to be noted that this size determination was dependent on the local clone density.
Table 1.
Summary of Independent Amplicons in 13 Pancreatic Cancer Cell Lines by CGH on cDNA Microarray.
| Location | Start (Mb) | End (Mb) | Size (Mb) |
| 3p21 | 39.73 | 42.01 | 2.28 |
| 3q29 | 198.68 | 199.49 | 0.81 |
| 5p15.3 | 0.19 | 0.48 | 0.29 |
| 6p21 | 29.97 | 30.91 | 0.94 |
| 6p21 | 30.94 | 32.48 | 1.55 |
| 7q21 | 90.70 | 94.85 | 4.15 |
| 7q22 | 95.13 | 99.58 | 4.44 |
| 8q24 | 133.88 | 135.60 | 1.72 |
| 11q13 | 70.15 | 71.17 | 1.02 |
| 14q21 | 44.90 | 48.66 | 3.76 |
| 15q22 | 66.15 | 69.29 | 3.14 |
| 15q24 | 74.74 | 76.26 | 1.52 |
| 15q25 | 83.12 | 84.32 | 1.20 |
| 15q26 | 84.98 | 96.03 | 11.05 |
| 17q21 | 39.56 | 40.31 | 0.75 |
| 17q21 | 42.27 | 42.71 | 0.44 |
| 17q21 | 44.46 | 44.60 | 0.14 |
| 17q25 | 82.61 | 83.27 | 0.66 |
| 19p13.3 | 1.97 | 2.11 | 0.13 |
| 19q13.1 | 39.26 | 42.12 | 2.86 |
| 19q13.3 | 50.85 | 51.24 | 0.39 |
| 20q13.3 | 62.13 | 62.28 | 0.15 |
| Xp22.3 | 2.72 | 11.76 | 9.05 |
| Xq28 | 149.31 | 149.66 | 0.35 |
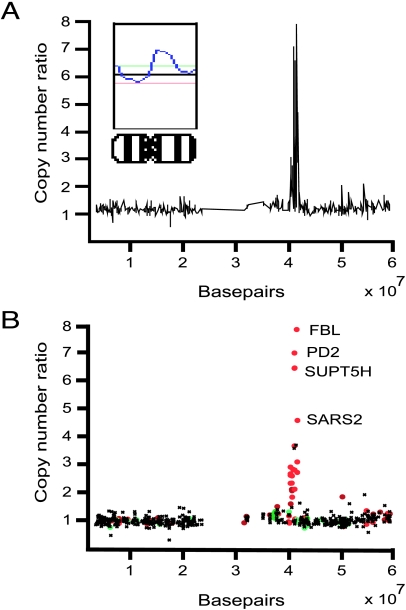
Figure 1.
Gene copy number and expression analysis for chromosome 19 in the Panc1 pancreatic cancer cell line by cDNA microarray. The copy number ratios were plotted as a function of the position of the cDNA clones along chromosome 19. In (A), individual data points were connected with a line. The inset shows chromosomal CGH copy number ratio profile. In (B), individual data points were color-coded according to cDNA expression ratios. The red dots indicate overexpressed genes (the upper 7% of expression ratios) and the green dots indicate underexpressed genes (lowest 7% of expression ratios) in Panc1 cells.
Next we analyzed the expression levels of the 12,232 cDNA clones in the 13 pancreatic cancer cell lines using an identical cDNA microarray. Direct comparison between copy number and expression levels in each cell line allowed unambiguous identification of genes whose expression levels were elevated due to increased copy number. For example, annotation of CGH copy number information with color-coded expression data revealed increased expression of several highly amplified genes, such as FBL, PD2, SUPT5H, and SARS2, located at the 19q13.1 region in the PANC-1 cell line (Figure 1B). Although such annotation is extremely effective in pinpointing putative amplification target genes in individual samples, it becomes progressively more complicated when the number of genes and samples to be analyzed increases. To facilitate the identification of possible amplification target genes, we applied a statistical approach with random permutation tests to explore the effects of gene copy number on gene expression levels across all 13 cell lines. This analysis revealed a set of 105 genes, including 70 known genes, whose expression levels were highly dependent on gene copy number (i.e., these genes were activated by increased copy number in the pancreatic cancer cell lines) (Table 2). The set included genes previously shown to be amplified in human cancer (e.g., MLN51) as well as known oncogenes (e.g., RAB4A, member of the RAS oncogene family). In addition, 11 (16%) of the 70 known genes (ARHGDIB, CLDN4, FBL, MCM7, PSMC4, RELA, SIRT2, ST14, STK15, SUPT5H, and TRIAD3) had been shown to be highly expressed in previous global gene expression surveys by either DNA microarrays [20–25] or serial analysis of gene expression (http://www.ncbi.nlm.nih.gov/SAGE).
Table 2.
List of 105 Genes with Statistically Significant Correlation (α < 0.05) between Gene Copy Number and Expression Level.
| Gene Name | Clone ID | Locus | α |
| EST | 4541713 | 0.0035 | |
| CDC42-binding protein kinase beta (DMPK-like) | 4582722 | 14q32.3 | 0.0045 |
| POP7 (processing of precursor, Saccharomyces cerevisiae) homolog | 3138336 | 7q22 | 0.0047 |
| TRIAD3 protein* | 3506949 | 7p22.2 | 0.0073 |
| Hypothetical protein F231491 | 3504227 | 19q13.1 | 0.0083 |
| Hypothetical protein DKFZp434H247 | 3010753 | 19q13.13 | 0.0104 |
| Hypothetical protein FLJ10055 | 37134 | 17q24 | 0.0107 |
| FK506-binding protein 4 (59 kDa) | 3940730 | 12p13.33 | 0.0109 |
| Hypothetical protein MGC3234 | 3504261 | 11p15.5 | 0.0111 |
| Opioid growth factor receptor | 3050950 | 20q13.3 | 0.0112 |
| Tubulin, gamma 1 | 108377 | 17q21 | 0.0117 |
| Mitochondrial ribosomal protein S12 | 2959394 | 19q13.1–q13.2 | 0.0118 |
| Ribosomal protein L8 | 3506015 | 8q24.3 | 0.012 |
| ATP-dependent RNA helicase | 3687438 | 17q21.1 | 0.0123 |
| EST | 3503397 | 0.0125 | |
| β-1,3-Glucuronyltransferase 3 (glucuronosyltransferase I) | 4299539 | 11q12.2 | 0.0127 |
| EST | 3543708 | 0.0129 | |
| p21 (CDKN1A)-activated kinase 4 | 3956856 | 19q13.1 | 0.0129 |
| seryl-tRNA synthetase 2 | 3627808 | 19q13.13 | 0.0129 |
| Metaxin 2 | 3542457 | 2q31.2 | 0.013 |
| Sirtuin (silent mating type information regulation 2 homolog) 2* | 2820929 | 19q13 | 0.013 |
| Hypothetical protein FLJ20721 | 3544728 | 17q25.1 | 0.0132 |
| Rho GDP dissociation inhibitor (GDI) β-* | 3621138 | 12p12.3 | 0.0133 |
| Ring finger protein 26 | 3028875 | 11q23 | 0.0133 |
| KIAA0182 protein | 2819273 | 16q24 | 0.014 |
| Ribosomal protein S16 | 4310307 | 19q13.1 | 0.0141 |
| MCM2 minichromosome maintenance deficient 2 | 3544403 | 3q21 | 0.0145 |
| Cyclin D-binding myb-like transcription factor 1 | 3010038 | 7q21 | 0.0146 |
| Hypothetical gene ZD52F10 | 3690018 | 19q13.11 | 0.015 |
| RNA-binding motif protein 10 | 3163064 | Xp11.23 | 0.015 |
| Cleavage and polyadenylation specific factor 4 (30 kDa) | 3356071 | 7q22.1 | 0.0152 |
| Fibrillarin* | 3504198 | 19q13.1 | 0.0153 |
| Hypothetical protein FLJ14888 | 3141437 | 15q25.1 | 0.0157 |
| Hypothetical protein MGC3040 | 3163990 | 3q21 | 0.0157 |
| Suppressor of Ty 5 homolog (S. cerevisiae)* | 3532191 | 19q13 | 0.0159 |
| Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 3051442 | 5p15 | 0.0164 |
| EST | 4543771 | 0.0167 | |
| suppression of tumorigenicity 14 (colon carcinoma)* | 2959447 | 11q24–q25 | 0.0173 |
| GTP-binding protein 1 | 4127961 | 22q13.1 | 0.0175 |
| Nuclear factor I/C (CCAAT-binding transcription factor) | 4128493 | 19p13.3 | 0.0183 |
| Similar to RIKEN cDNA 0610011N22 | 4334552 | 5p15.33 | 0.0185 |
| Similar to RIKEN cDNA 1810006A16 gene | 3942185 | 16q22.1 | 0.0186 |
| Syndecan 1 | 3347793 | 2p24.1 | 0.0187 |
| ADP ribosylation factor GTPase-activating protein 1 | 3162991 | 20q13.33 | 0.0191 |
| Likely ortholog of mouse another partner for ARF 1 | 2822464 | 14q24.3 | 0.0192 |
| Proteasome (prosome, macropain) 26S subunit, ATPase, 4* | 4046205 | 19q13.11–q13.13 | 0.0202 |
| EST | 3628799 | 0.0203 | |
| Hypothetical protein MGC45840 | 3941077 | 11p15.5 | 0.0205 |
| PCTAIRE protein kinase 1 | 3504276 | Xp11.3–p11.23 | 0.0205 |
| EST | 3958225 | 0.0209 | |
| Hypothetical protein BC011824 | 3830276 | 19p13.3 | 0.021 |
| DKFZP564B147 protein | 2821721 | Xq26.3 | 0.0213 |
| Claudin 4* | 3349211 | 7q11.23 | 0.022 |
| EST, weakly similar to IDN3 protein, isoform B | 3528352 | 0.0221 | |
| Major histocompatibility complex, class I, C | 4249847 | 6p21.3 | 0.0225 |
| Homo sapiens, clone IMAGE: 3029191, mRNA | 3029191 | 7 | 0.0228 |
| Inhibitor of kappa light polypeptide gene enhancer in B-cells | 4651870 | Xq28 | 0.0233 |
| EST | 3678374 | 0.0238 | |
| Proteasome (prosome, macropain) subunit, α type, 1 | 4105838 | 11p15.1 | 0.0241 |
| Splicing factor, arginine/serine-rich 8 | 3352093 | 12q24.33 | 0.0243 |
| EST | 3503182 | 0.0244 | |
| KIAA0632 protein | 3356434 | 7q22.1 | 0.0246 |
| Putative translation initiation factor | 2823520 | 17q21.2 | 0.0248 |
| Ribonuclease H2, large subunit | 3160621 | 19p13.12 | 0.0248 |
| Adaptor-related protein complex 3, sigma 2 subunit | 4139509 | 15q25.3 | 0.0254 |
| Regulator of G-protein signalling 14 | 4547415 | 5q35.3 | 0.0254 |
| KIAA1023 protein | 37440 | 7p22.3 | 0.0257 |
| Chromosome 20 open reading frame 126 | 4126826 | 20q11.21 | 0.0261 |
| MLN51 protein | 40515 | 17q11–q21.3 | 0.0271 |
| Likely ortholog of mouse synembryn | 3633778 | 11p15.5 | 0.0274 |
| Ubiquitin-specific protease 11 | 2961383 | Xp11.23 | 0.0276 |
| v-rel reticuloendotheliosis viral oncogene homolog A* | 4547184 | 11q13 | 0.029 |
| ATP synthase, H+ transporting, mitochondrial F0 complex | 3826469 | 7q22.1 | 0.0291 |
| Hypothetical protein MGC11242 | 3939938 | 17q21.32 | 0.0293 |
| SIN3 homolog A, transcriptional regulator (yeast) | 3604034 | 15q23 | 0.0293 |
| Dual specificity phosphatase 3 | 3605391 | 17q21 | 0.0298 |
| Asparagine synthetase | 3010719 | 7q21.3 | 0.03 |
| Copine I | 4074508 | 20q11.21 | 0.0305 |
| mitochondrial Ribosomal protein L45 | 3951804 | 17q21.2 | 0.0306 |
| RPA-binding transactivator | 3926937 | 19q13.13 | 0.0308 |
| Hexosaminidase A (α polypeptide) | 3353424 | 15q23–q24 | 0.0312 |
| SPRY domain-containing SOCS box protein SSB-3 | 3342825 | 16p13.3 | 0.0319 |
| Steroidogenic acute regulatory protein related | 2823277 | 17q11–q12 | 0.0319 |
| Chromosome 20 open reading frame 31 | 3051376 | 20q11.21 | 0.036 |
| Enoyl coenzyme A hydratase 1, peroxisomal | 4300082 | 19q13.1 | 0.036 |
| WD repeat domain 5B | 4097434 | 3q21.1 | 0.0367 |
| Actin-related protein 2/3 complex, subunit 1B (41 kDa) | 3138319 | 7q22.1 | 0.037 |
| Epoxide hydrolase 1, microsomal (xenobiotic) | 3547085 | 1q42.1 | 0.0371 |
| Alkaline phosphatase, placental-like 2 | 4652865 | 2q37 | 0.0376 |
| FtsJ homolog 2 (Escherichia coli) | 3343278 | 7p22 | 0.0379 |
| GTP-binding protein 5 (putative) | 4564032 | 20q13.33 | 0.0386 |
| Inactive progesterone receptor (23 kDa) | 3353485 | 12q13.13 | 0.0387 |
| EST | 4634558 | 0.0401 | |
| FK506-binding protein 8 (38 kDa) | 3543042 | 19p12 | 0.0421 |
| Thyroid hormone receptor interactor 6 | 3610413 | 7q22 | 0.0421 |
| Tripartite motif-containing 8 | 4561807 | 10q24.3 | 0.0444 |
| RAB4A, member RAS oncogene family | 3346455 | 1q42–q43 | 0.0455 |
| Effector cell protease receptor 1 | 3351130 | 17q25 | 0.0459 |
| MCM7 minichromosome maintenance deficient 7 (S. cerevisiae)* | 4134871 | 7q21.3–q22.1 | 0.0462 |
| ADP ribosylation factor-related protein 1 | 2989010 | 20q13.3 | 0.047 |
| Chromosome 20 open reading frame 35 | 2959575 | 20q13.11 | 0.0473 |
| KIAA0943 protein | 3543516 | 2q37.3 | 0.0479 |
| Serine/threonine kinase 15* | 3532438 | 20q13.2–q13.3 | 0.048 |
| Protein kinase, cAMP-dependent, regulatory, type I, β | 3677877 | 7pter-p22 | 0.0485 |
| Hypothetical protein FLJ10468 | 2822261 | 1p34.2 | 0.0495 |
Genes previously shown to be overexpressed in pancreatic cancer by global expression profiling [20–25].
To obtain further information on the possible role of the 105 genes in the pathogenesis of pancreatic cancer, we explored their functional characteristics by using the SOURCE database (http://source.stanford.edu). According to the SOURCE, 17 of 105 genes represented hypothetical proteins and 21 were known genes or ESTs with no functional annotation. A large fraction (78%) of the remaining 67 genes was involved in key cellular processes including signal transduction (17 genes), protein processing (11), metabolism (8), RNA processing (7), transcription (5), and DNA replication (4).
Discussion
Pancreatic ductal adenocarcinoma is one of the top 10 causes of cancer death in industrialized countries, with over 40,000 deaths/year in Europe [35] and nearly 30,000 deaths/year in the United States [1]. New methods for early detection, a better understanding of the biologic mechanisms underlying cancer progression, and cancer-targeted treatment modalities are urgently needed to reduce the mortality from this lethal disease. The availability of global expression and copy number platforms, such as cDNA microarrays, has greatly facilitated the identification of novel tumor markers in many cancer types. Here we used a 12,232-clone cDNA microarray to search for genes that are activated by increased copy number in pancreatic cancer. The main focus of this study was to specifically identify genes whose overexpression is caused by increased copy number (i.e., those genes that are overexpressed only in cell lines with amplification). Gene copy numbers were first determined by performing CGH on the cDNA microarray, and subsequent expression analysis using an identical cDNA microarray permitted direct correlation between copy numbers and expression levels on a gene-by-gene basis.
The high-resolution copy number analysis by CGH microarray revealed a total of 24 independent regions of copy number increase in the 13 pancreatic cancer cell lines. These included several chromosomal segments, such as 3q, 5p, 7q, 8q24, 11q13, 15q, 17q, 19q, and 20q, previously implicated to be gained or amplified by chromosomal CGH or fluorescence in situ hybridization in pancreatic cancer [17,36,37]. In addition to validating data from these previous studies, the CGH microarray analysis permitted mapping of these copy number increases in much higher accuracy and determination of the exact base-pair boundaries for each aberration. For example, frequent copy number increases were observed at three separate regions on chromosome 19 (19p13.3, 19q13.1, and 19q13.3). Although gains and amplifications of 19q have been frequently reported both in pancreatic cancer cell lines and primary pancreatic carcinomas [17,18,36], the CGH microarray analysis was able to narrow down the affected regions to sizes of 130 kb, 2.9 Mb, and 390 kb, respectively (Table 1). However, it has to be noted that the size determination was fully dependent on the local clone density on the microarray and therefore does not necessarily correspond to the actual size of the amplicon.
The CGH microarray results obtained in this study considerably advance our knowledge on common copy number increases in pancreatic cancer and provide an improved starting point for the identification of genes affected by such copy number alterations. The subsequent expression survey performed using an identical cDNA microarray revealed that several genes located within the regions of increased copy number also showed elevated expression levels in the pancreatic cancer cell lines. For example, several genes at the 19q13.1 amplicon, such as FBL, PD2, SUPT5H, and SARS2, were highly expressed in cell lines with increased copy number. None of these genes has been previously linked to cancer, but based on their function, FBL, which is known to act in ribosomal RNA processing [38], and SUPT5H, a putative modulator of chromatin structure [39], might be implicated in cancer formation.
To evaluate the contribution of gene copy number to gene expression level, a statistical procedure was applied to systematically identify genes whose expression levels were attributable to copy number increase across all 13 cell lines. This analysis revealed 105 genes and included previously described amplified genes as well as known oncogenes, such as STK15, MLN51, RAB4A, and RELA. For example, the STK15 gene (also called AURKA, aurora kinase A, and BTAK) located at 20q13 is a putative oncogene that has been shown to be amplified and overexpressed in many human cancers [40–42]. STK15 is essential for normal centrosome function and chromosome segregation [43] and, recently, STK15 overexpression was shown to be involved in degradation of the p53 tumor-suppressor protein, leading to loss of p53 function [44].
Although gene copy number has been shown to be a major determinant of gene expression level, a large fraction of genes shows elevated expression levels without any increase in gene copy number [33,45]. It was therefore expected that our statistical approach identified an overlapping but a clearly separate set of genes than those reported in studies where only expression levels were evaluated. A direct comparison between our data and previously published global gene expression analyses [20–25] (http://www.ncbi.nlm.nih.gov/SAGE) revealed that 16% of the 70 known genes identified here have been previously reported to be highly expressed in pancreatic cancer.
Most of the genes identified by the statistical approach in this study had not been previously linked to pancreatic cancer. However, some of them have been implicated in tumor development. For example, p21-activated kinase 4 (PAK4) has been shown to regulate cell migration, cell adhesion, and anchorage-independent growth both in human cancer cell lines and in fibroblasts, suggesting a central role in oncogenic transformation and tumorigenesis [46–48]. To further clarify the possible roles of the 105 genes in pancreatic cancer, we explored their functional characteristics by using information from the SOURCE database (http://source.stanford.edu). This analysis showed that 78% of these genes were involved in functions, such as signal transduction, transcription, and DNA replication, which are essential for normal cellular processes but could also be envisioned to have an impact on cancer development. Taken together, the set of 105 genes identified in this study is likely to represent genes that are activated by increased copy number and might be actively involved in the pathogenesis of pancreatic cancer. However, it has to noted that it will be essential to validate the results obtained in this study by using uncultured primary pancreatic tumors. Such validation will also be essential to assess the possible clinical significance of the genes implicated here.
In summary, our microarray-based genome-wide copy number survey resulted in the identification of 24 independent regions of copy number increase in 13 pancreatic cancer cell lines. This approach allowed highly accurate mapping of copy number increases with an average resolution of about 300 kb throughout the genome. Parallel expression analysis using an identical cDNA microarray permitted direct comparison between gene copy numbers and expression levels. A statistical evaluation disclosed a set of 105 genes whose expression levels were directly linked to copy number increase, indicating that they are activated through amplification in pancreatic cancer. These results implicate a set of genes that are likely to have an important role in pancreatic cancer pathogenesis.
Acknowledgement
We thank Kati Rouhento for excellent technical assistance.
Abbreviations
- CGH
comparative genomic hybridization
Footnotes
This study was financially supported by the Academy of Finland, the Medical Research Fund of the Tampere University Hospital, the Finnish Medical Society (Duodecim), and the Pirkanmaa Cultural Foundation.
References
- 1.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Greenlee RT, Hill-Harmon MB, Murray T, Thun MJ. Cancer statistics. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Schnall SF, Macdonald JS. Chemotherapy of adenocarcinoma of the pancreas. Semin Oncol. 1996;23:220–228. [PubMed] [Google Scholar]
- 4.Lowenfels AB, Maisonneuve P. Pancreatico-biliary malignancy: prevalence and risk factors. Ann Oncol. 1999;10:1–3. [PubMed] [Google Scholar]
- 5.Schuller HM. Mechanisms of smoking-related lung and pancreatic adenocarcinoma development. Nat Rev Cancer. 2002;2:455–463. doi: 10.1038/nrc824. [DOI] [PubMed] [Google Scholar]
- 6.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, Levy P, Ruszniewski P. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenk M, Schwartz AG, O'Neal E, Kinnard M, Greenson JK, Fryzek JP, Ying GS, Garabrant DH. Familial risk of pancreatic cancer. J Natl Cancer Inst. 2001;93:640–644. doi: 10.1093/jnci/93.8.640. [DOI] [PubMed] [Google Scholar]
- 8.Tersmette AC, Petersen GM, Offerhaus GJ, Falatko FC, Brune KA, Goggins M, Rozenblum E, Wilentz RE, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Increased risk of incident pancreatic cancer among first-degree relatives of patients with familial pancreatic cancer. Clin Cancer Res. 2001;7:738–744. [PubMed] [Google Scholar]
- 9.Bardeesy N, De Pinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 10.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ, Kern SE. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 11.Luttges J, Reinecke-Luthge A, Mollmann B, Menke MA, Clemens A, Klimpfinger M, Sipos B, Kloppel G. Duct changes and K-ras mutations in the disease-free pancreas: analysis of type, age relation and spatial distribution. Virchows Arch. 1999;435:461–468. doi: 10.1007/s004280050428. [DOI] [PubMed] [Google Scholar]
- 12.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, daCosta LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 13.Schleger C, Verbeke C, Hildenbrand R, Zentgraf H, Bleyl U. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol. 2002;15:462–469. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- 14.Tobita K, Kijima H, Dowaki S, Kashiwagi H, Ohtani Y, Oida Y, Makuuchi H, Nakamura M, Ueyama Y, Tanaka M, Inokuchi S, Makuuchi H. Epidermal growth factor receptor expression in human pancreatic cancer: significance for liver metastasis. Int J Mol Med. 2003;11:305–309. [PubMed] [Google Scholar]
- 15.Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. 1996;56:3803–3807. [PubMed] [Google Scholar]
- 16.Mahlamäki EH, Höglund M, Gorunova L, Karhu R, Dawiskiba S, Andrén-Sandberg A, Kallioniemi OP, Johansson B. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q, and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes Cancer. 1997;20:383–391. doi: 10.1002/(sici)1098-2264(199712)20:4<383::aid-gcc10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Schleger C, Arens N, Zentgraf H, Bleyl U, Verbeke C. Identification of frequent chromosomal aberrations in ductal adenocarcinoma of the pancreas by comparative genomic hybridization (CGH) J Pathol. 2000;191:27–32. doi: 10.1002/(SICI)1096-9896(200005)191:1<27::AID-PATH582>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 18.Shiraishi K, Okita K, Kusano N, Harada T, Kondoh S, Okita S, Ryozawa S, Ohmura R, Noguchi T, Iida Y, Akiyama T, Oga A, Fukumoto Y, Furuya T, Kawauchi S, Sasaki K. A comparison of DNA copy number changes detected by comparative genomic hybridization in malignancies of the liver, biliary tract and pancreas. Oncology. 2001;60:151–161. doi: 10.1159/000055313. [DOI] [PubMed] [Google Scholar]
- 19.Harada T, Okita K, Shiraishi K, Kusano N, Kondoh S, Sasaki K. Interglandular cytogenetic heterogeneity detected by comparative genomic hybridization in pancreatic cancer. Cancer Res. 2002;62:835–839. [PubMed] [Google Scholar]
- 20.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7446. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- 21.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 22.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 23.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, Van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 26.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 27.Mousses S, Bittner ML, Chen Y, Dougherty ER, Baxevanis A, Meltzer PS, Trent JM. Gene expression analysis by cDNA microarrays. In: Livesey FJ, Hunt SP, editors. Functional Genomics. Oxford: Oxford University Press; 2000. pp. 113–137. [Google Scholar]
- 28.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genomewide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 29.Monni O, Bärlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Dougherty ER, Bittner ML. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J Biomed Opt. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Dudoit S, Luu P, Speed T. Normalization for cDNA microarray data. In: Bittner M, Chen Y, Dorsel A, Dougherty E, editors. Microarrays: Optical Technologies and Informatics SPIE. San Jose: Society for Optical Engineering; 2001. pp. 141–152. [Google Scholar]
- 32.Cleveland W. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- 33.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 34.Hautaniemi S, Ringner M, Kauraniemi P, Autio R, Edgren H, Yli-Harja O, Astola J, Kallioniemi A, Kallioniemi OP. A strategy for identifying putative causes of gene expression variation in human cancers. J Franklin Inst. 2004;341:77–88. [Google Scholar]
- 35.Fernandez E, La Vecchia C, Porta M, Negri E, Lucchini F, Levi F. Trends in pancreatic cancer mortality in Europe, 1955–1989. Int J Cancer. 1994;57:786–792. doi: 10.1002/ijc.2910570605. [DOI] [PubMed] [Google Scholar]
- 36.Höglund M, Gorunova L, Andrén-Sandberg A, Dawiskiba S, Mitelman F, Johansson B. Cytogenetic and fluorescence in situ hybridization analyses of chromosome 19 aberrations in pancreatic carcinomas: frequent loss of 19p13.3 and gain of 19q13.1–13.2. Genes Chromosomes Cancer. 1998;21:8–16. [PubMed] [Google Scholar]
- 37.Mahlamäki EH, Bärlund M, Tanner M, Gorunova L, Höglund M, Karhu R, Kallioniemi A. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 38.Jansen RP, Hurt EC, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stachora AA, Schafer RE, Pohlmeier M, Maier G, Ponstingl H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Lett. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- 40.Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 41.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 43.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 44.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 45.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu J, Cammarano MS, Shi Q, Ha KC, de Lanerolle P, Minden A. Activated PAK4 regulates cell adhesion and anchorageindependent growth. Mol Cell Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, Jallal B, Smeal T. Requirement for PAK4 in the anchorageindependent growth of human cancer cell lines. J Biol Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Li Z, Viklund EK, Stromblad S. p21-activated kinase 4 interacts with integrin alpha v beta 5 and regulates alpha v beta 5-mediated cell migration. J Cell Biol. 2002;158:1287–1297. doi: 10.1083/jcb.200207008. [DOI] [PMC free article] [PubMed] [Google Scholar]