Abstract
Previous studies in our laboratory have shown that the elevation of reactive oxygen species levels and the repression of the antioxidant enzyme, catalase, played a critical role in the in vitro progression of benign papilloma cells to malignant carcinoma cells. Catalase message, protein levels, and activity levels were found to be downregulated in the malignantly progressed cells. The goal of this study is to further characterize the repression of catalase in malignant progression of mouse skin tumors. To validate the in vitro observations, we examined catalase expression in tumor samples generated by the multistep chemical carcinogenesis protocol. Higher levels of catalase mRNA and protein were observed in benign papillomas versus malignant carcinomas. Nuclear run-on analysis showed that catalase repression in the cultured malignant cells was transcription-dependent. Results from luciferase reporter assays indicated that malignant cells have lower catalase promoter activities than benign papilloma cells, in part through the Wilm's tumor suppressor 1 (WT1) binding site within the proximal promoter region. The WT1 protein levels were found to be inversely correlated with the observed catalase promoter activities, with higher levels observed in the malignant cells versus the benign cells. These results led us to conclude that WT1 is acting as a transcription repressor in catalase gene regulation during tumor progression.
Keywords: Catalase, Wilm's tumor suppressor 1, antioxidant, transcriptional regulation, tumor progression and mouse skin carcinogenesis
Abbreviations: ROS, reactive oxygen species; SCC, squamous cell carcinoma; DMBA, 7,12-dimethylbenzanthracene; TPA, 12-O-tetradecanoyl-phorbol-13-acetate; MNNG, N-methyl-N-Vnitro-N-nitrosoguanidine; WT1, Wilm's tumor suppressor 1
Introduction
Nonmelanoma skin cancer is the most common form of malignancy in the United States. It is estimated that over 1 million new cases are diagnosed every year [1]. About 20% of those cases are categorized as squamous cell carcinomas (SCCs) [2]. The multistep mouse skin carcinogenesis protocol has provided researchers a model system to study and identify key events in the development of such cancers. The protocol is divided into three major steps, including initiation, promotion, and progression [3]. Initiation involves the application of a carcinogen such as 7,12-dimethylbenzanthracene (DMBA) to normal mouse skin to induce genetic mutations in the epidermal cells. In the promotion step, a tumor promoter such as 12-O-tetradecanoyl-phorbol-13-acetate (TPA) is applied to cause a selective outgrowth of initiated cells that carry activating Ha-ras gene mutations [4]. The hyperproliferation of these cells leads to the development of benign papillomas. In the progression step, additional genetic and epigenetic changes are acquired by the tumor cells to convert from benign papillomas to malignant SCCs. In a previous study, we developed an in vitro model to study the molecular mechanisms involved in the malignant progression process [5]. The benign papilloma-producing mouse keratinocyte cell line 308, containing a Ha-ras mutation [6], was subjected to repeated treatments with N-methyl-N-nitro-N-nitrosoguanidine (MNNG) or ionizing radiation to introduce DNA damage, thus enhancing their conversion into malignant cells. This protocol produced two malignantly transformed cell lines: 6M90 (for-MNNG treated) and 6R90 (for ionizing radiation-treated). The 6M90 and 6R90 cells were able to develop into SCCs upon subcutaneous injection into athymic nude mice. In comparison, the parental 308 cells were unable to form tumors when injected subcutaneously. We reported previously that the malignant variants had elevated levels of steady-state reactive oxygen species (ROS) in comparison to the parental 308 cells, and this elevation of ROS played a functional role in maintaining the malignant phenotypes of the 6M90 and 6R90 cells [5].
ROS, which include superoxide anion, hydroxyl radical, hydrogen peroxide, and singlet oxygen, are continuously produced by the cell in a variety of normal cellular functions. The majority of ROS are generated by the mitochondria as unwanted by-products during cellular respiration. The cell employs a host of antioxidant defense mechanisms to maintain the steady-state level of ROS. In a previous study, we showed that attenuation of the antioxidant defense enzyme, catalase, was mainly responsible for the elevated ROS levels found in our malignantly transformed cell lines [7]. We showed that catalase message, protein levels, and activity levels, in both 6M90 and 6R90 cells, were decreased compared to the parent 308 cells. The catalase activities in the malignant cell lines were found to decrease from 21.5 ± 1.63 µmol/min per milligram of protein in 308 cells to 10.96 ± 1.73 and 7.23 ± 0.57 µmol/min per milligram of protein in 6M90 and 6R90 cells, respectively. We were unable to detect any significant changes in the expression and activity levels of Mn or Cu/Zn superoxide dismutase in the malignant cells. The levels of glutathione (both oxidized and reduced form) were decreased in 6M90 and increased in 6R90 compared to the 308 cells [7]. These data indicated to us that the changes in glutathione might not be as critical or common as the repression of catalase in the progression of cancer cells. In addition, expression of an exogenous rat catalase gene in the 6M90 and 6R90 cells resulted in an increase in catalase activities and a reduction of ROS [8]. Taken together, these observations demonstrated that downregulation of catalase expression plays a crucial role in the elevation of ROS in the malignantly transformed 6M90 and 6R90 cells.
Catalase is a heme-containing enzyme found mainly in the peroxisomes and cytosol, and is responsible for detoxifying hydrogen peroxide by catalyzing its degradation to water and oxygen. We hypothesized that catalase functions as a tumor-suppressor gene in the malignant progression of mouse skin tumors. Repression of catalase causes the elevation of hydrogen peroxide, which acts as a secondary messenger to activate signal transduction pathways, leading to the formation of SCC. Our goal in this study was to determine the mechanism(s) behind catalase repression during tumor progression.
Materials and Methods
In Vivo Tumor Progression
A single initiating dose of 100 µg of DMBA (Sigma, St. Louis, MO) in 0.2 ml of acetone was applied topically to the shaved backs of female ICR mice (Harlan Sprague Dawley, Indianapolis, IN). It was followed by twice-weekly application of 5 nmol of TPA (Alexis Corp., San Diego, CA) in 0.2 ml of acetone for 21 weeks. On week 22, TPA applications were stopped and replaced with weekly application of 120 µg of MNNG in 0.2 ml of acetone for a total of 40 weeks.
Histology and Immunohistochemistry
Formalin-fixed and paraffin-embedded tumor samples were cut into 5-µm serial sections. The sections were hematoxylin and eosin (H&E)-stained and used for pathology to determine tumor classifications. Immunohistostainings were performed on the remaining sections. Briefly, the sections were deparaffinized with xylene and rehydrated with ethanol. The sections were rinsed in PBS and incubated with the following primary antibodies: catalase (Rockland, Gilbertsville, PA), keratin 1, and keratin 10 (Berkeley Antibody Company, Richmond, CA). The pan-keratin antibody, 18A, was given by Dr. Raymond Nagle (Department of Pathology, University of Arizona, Tuczon, AZ) and had been shown to stain high-molecular-weight cytokeratins from the epidermis. The Vectastain ABC kit (Vector Laboratories, Burlingame, CA) was used to perform the secondary antibody incubations. The staining was visualized with diaminobenzidene (DAB) and counterstained with hematoxylin.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR) Assay
Total RNA were isolated from the cultured cells or frozen tumor sample using the RNeasy Mini Kit (Qiagen, Chatsworth, CA) and quantitated by spectrophotometry. Two hundred nanograms of total RNA was used to generate random hexamer primed cDNA with the Omniscript Reverse Transcription kit (Qiagen). Ten percent of the cDNA product was used to perform real-time PCR for the quantitation of the catalase and GAPDH message levels. The primers and Taqman probes for the assay were purchased from the Assay-by Design Service (Applied Biosystems, Foster City, CA). The sequences of the primers and probe used for detecting the catalase message were as follows: mCas1 forward primer (5′-CCGACCAGGGCATCAAAA-3′), mCas1 reverse primer (5′-GAGGCCATAATCCGGATCTTC-3′), and mCas1 probe (5′-6FAM-CAGGAAGGCTTGCTC-3′). The sequences of the primers and probe used for detecting GAPDH message were as follows: mGAPDH forward primer (5′-GTCCTGAGTATGTCGTGGAGTCTAC-3′), mGAPDH reverse primer (5′-GGCCCCGGCCTTCTC-3′), and mGAPDH probe (5′-6FAM-ATGGTGGTGAAGACAC-3′). The real-time PCR assay was carried out with the Taqman Universal PCR Master Mix and the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) under the follow conditions: 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute. The Relative CT Method (described in the User Bulletin No. 2, ABI PRISM 7700 Sequence Detection System, pp. 11–15) was used to calculate the relative mRNA level of catalase (normalized to GAPDH) in each sample.
Cells and Cell Culture
Papilloma-derived mouse skin keratinocyte 308 cells were kindly provided by Dr. Stuart H. Yuspa [6]. The progressed variants, 6M90 and 6R90 cells, were generated as described in previous publications [5,7]. The cells were cultured in minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 7.5% fetal bovine serum, 2.5% calf serum (Gemini Bioproducts, Woodland, CA), 500 U/ml penicillin, and 5000 U/ml streptomycin (Invitrogen) at 37°C in a 7% CO2 humidified environment.
Southern Blot Analysis
Genomic DNA was isolated from the cultured cells as previously described [9] and quantitated by spectrophotometry. The DNA was digested overnight with 10 U/µg restriction enzymes (New England Biolaboratories, Beverly, MA) at 37°C. Twenty-five micrograms of digested DNA was eletrophoresed on a 1% agarose gel and transferred onto a GeneScreen membrane (NEN Life Science Products, Boston, MA) with the Pressure Blotter (Stratagene, La Jolla, CA). The membrane was hybridized with 32P-labeled cDNA probes according to the manufacturer's protocol. The hybridized bands were visualized and quantitated with the MD Storm Phosphorimaging system (Molecular Dynamics, Sunnyvale, CA).
Catalase Promoter Sequencing
Genomic DNA was isolated as described above for Southern blot analysis. The DNA was subjected to two rounds of PCR with nested primers for amplification. The outside flanking primers for the first round of PCR are as follows: -567 to -546 (5′-CCTCTCCCCACAAACAAAGAGC-3′) and +110 to +93 (5′-GCTGGGTCCCGACTGTCC-3′). The internal nested primers for the second round of PCR are as follows: -557 to -530 (5′-CAAACAAAGAGCTTTTCAGTATTCTTCC-3′) and +89 to +70 (5′-ATGGTGTAGGATTGCGGAGC-3′). The PCR reaction was performed with the Pfx polymerase (Invitrogen) to increase the fidelity of the product. PCR was performed under the follow conditions: 94°C for 3 minutes followed by 25 cycles of 94°C for 1 minute, 58°C for 1 minute, and 68°C for 1.5 minutes, ending with a final extension at 68°C for 5 minutes. The PCR product was diluted 1:300 to be used as template for the second round of PCR, which was performed under identical conditions. The 646-bp product after two rounds of PCR was cloned into pCR 2.1 and sequenced as described for bisulfite sequencing.
Nuclear Run-On
A novel nonradioactive-based nuclear run-on method [10] was used in conjunction with real-time RT-PCR to assay for catalase transcription rate in our cell lines. Briefly, 50 x 106 cells were trypsinized and washed twice in Ca2+- and Mg2+- free PBS. The cell pellet was resuspended in cell lysis buffer containing 10 mM Tris-HCl, pH 7.4, 3 mM MgCl2, 10 mM NaCl, 150 mM sucrose, and 0.5% NP-40. The nuclei were collected by centrifugation and stored frozen in 40% glycerol at -70°C until used. The run-on reaction was carried out in a 2 x transcription buffer containing 200 mM KCl; 20 mM Tris-HCl, pH 8.0; 5 mM MgCl2; 5 mM dithiothreitol (DTT); 4 mM each of ATP, GTP, and CTP; 200 mM sucrose; and 20% glycerol. The reaction was initiated with the addition of 4 mM biotin-16-UTP (Roche Applied Science, Indianapolis, IN) and incubated for 1 hour at 29°C. The RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's protocol. Dynabeads M-280 (Dynal, Oslo, Norway), a magnetic bead covalently linked to streptavidin, was used to isolate the biotin-labeled run-on RNA according to the manufacturer's protocol. The beads were resuspended in RNase-free water and stored at -20°C until used. Reverse transcription and real-time PCR were performed as described above.
Sodium Bisulfite Genomic Sequencing
Genomic DNA was isolated as described above for Southern blot analysis. Five micrograms of DNA was modified with sodium bisulfite under the conditions described [11]. Briefly, DNA was denatured with 0.3 M NaOH and treated with sodium bisulfite at 55°C for 14 hours. The DNA were desalted by the Wizard Prep kit (Promega, Madison, WI) and desulfonated with 0.3 M NaOH. The DNA was then purified with standard ethanol precipitation and subjected to two rounds of PCR with nested primers for amplification. The nested primers were designed specifically to the bisulfite-modified DNA sequence for the mouse catalase promoter region. The outside flanking primers for the first round of PCR are as follows: -527 to -507 (5′-GGGAGAAAATGGAGGAGTTTG-3′) and +140 to +119 to (5′-CTACTCCTTCCACTACTTCATC-3′). The internal nested primers for the second round of PCR are as follows: -268 to -248 (5′-GAGGAATTGGATTTTGAGTTG-3′) and +112 to +92 (5′-CTAACTAAATCCCAACTATCC-3′). PCR was performed under the following conditions: 95°C for 1 minute followed by 35 cycles of 92°C for 1 minute, 55°C for 3 minutes, and 72°C for 1 minute, ending with a final extension at 72°C for 5 minutes. Twenty percent of the PCR product was used as the template for the second round of nested PCR. Identical conditions were used for the second round of PCR. After two rounds of amplifications, the PCR product was subjected to 1% agarose gel electrophoresis for separation. The 380-bp product was excised from the gel and purified with the Qiaex II kit (Qiagen). The purified product was cloned into the pCR 2.1 TA cloning vector according to the protocol provided by Invitrogen. Ten positive recombinant clones were isolated from each cell line using the QIAprep Spin Plasmid Miniprep kit (Qiagen) and sequenced by an ABI automated DNA sequencer (University of Arizona Sequencing Facility). The methylation status of CpG islands on the catalase promoter was determined by comparing the bisulfite-modified sequence to the published GeneBank sequence (accession no. L25070).
Transient Transfection and Luciferase Assay
The series of catalase promoter luciferase reporter constructs was a kind gift from Dr. Thomas A. Rando (Stanford University) [12]. About 1.5 x 105 cells were plated into sixwell plates the day before the experiment. They were transiently transfected with 0.5 µg of plasmid DNA using the DOTAP liposomal transfection reagent (Avanti Polar Lipids, Alabaster, AL) for 12 hours. The transfectant was then removed and the cells were left to recover for an additional 12 hours in full serum-containing medium before harvesting. The cells were harvested in Cell Culture Lysis Reagent (Promega). The protein concentrations of the samples were determined by the DC protein assay (Bio-Rad Laboratories, Hercules, CA). Five micrograms of protein was used for each reaction. The luciferase activity was measured with the single luciferase kit (Promega) on a TD-20/20 luminometer (Turner Designs, Sunnydale, CA). The relative activity was calculated by dividing the raw light units of each sample by the raw light units of control samples that had been transfected with pGL3 control plasmid (Promega).
Site-Directed Mutagenesis of Wilm's Tumor Suppressor (WT1) Consensus Binding Site
The QuikChange II Site-Directed Mutagenesis Kit from Stratagene was used in accordance with the manufacturer's protocol. The following mutational primers were used: 5′-GGCGGCCGTCCCAAGTTATGGGGGCGGTGCTGATTG-3′ and 5′-CAATCAGCACCGCCCCCATAACTTGGGACGGCCGCC-3′. The underlined bases represent the WT1 binding site that was mutated in the -191/+68 construct.
Results
Catalase Expression In Vivo
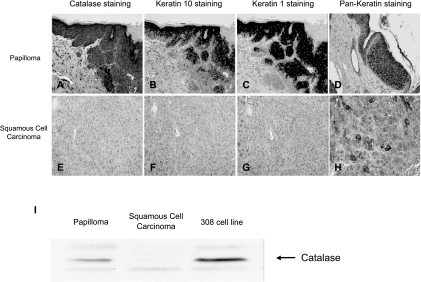
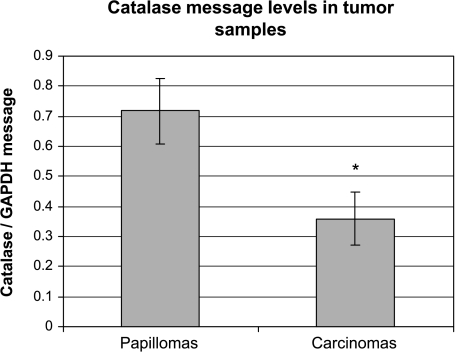
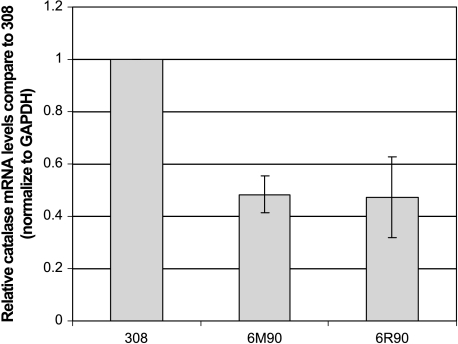
We have shown that catalase expression was attenuated in the in vitro progression model [7]. To validate that observation, we decided to examine catalase expression in tumors generated in vivo. A multistep chemical carcinogenesis protocol with DMBA (initiating agent) and TPA (promoting agent) was used to generate papillomas on the mouse skin [3]. A third stage carcinogen, MNNG, was applied to increase the rate of conversion into SCCs. A total of 12 tumors was harvested and examined for catalase expression. The tumors were classified into six papillomas and six SCCs based on gross anatomy. Immunostaining was used to detect the expression of catalase on the formalin-fixed tumor sections. Detailed histologic examination on H&E-stained sections revealed that most tumors were heterogeneous in their pathology. At the end of experiment, most of the “papillomas” have started the progression process and contained small areas that were classified as SCCs. However, most “carcinomas” retained small areas that have not undergone the progression process and those sections were classified as papillomas in nature. As a result, we performed additional immunostaining with keratinocyte differentiation markers, keratins 1 and 10, to confirm with the pathology of particular sections. Keratins 1 and 10 are known to be highly expressed in well-differentiated keratinocytes found in normal skin and papillomas, but lost in dedifferentiated SCCs [13]. High catalase expression was detected in all the well-differentiated papilloma sections examined (Figure 1A). Conversely, very low to no catalase expression was detected in the dedifferentiated carcinoma sections examined (Figure 1E). In addition, we wanted to determine if the loss of catalase expression correlates to a decrease in catalase mRNA level in the tumors. Real-time RT-PCR analysis was performed with RNA isolated from the tumor samples. The average catalase message levels of five papillomas and six SCCs are shown in Figure 2. The results indicated that carcinomas have reduced catalase message and protein expression compared to papillomas.
Figure 1.
Catalase, keratin 1, and keratin 10 expression patterns in benign papillomas and SCCs. Tumors were excised from mice and fixed in 10% formalin. The samples were paraffin-embedded, sectioned, and mounted on glass slides. Immunohistochemistry was performed with anti-bovine catalase (A and E), anti-mouse keratin 10 (B and F), and anti-mouse keratin 1 (C and G) antibodies at 1:500 dilutions. The Vectastain ABC kit at 1:200 dilution was used to perform the secondary antibody incubations. Catalase, keratin 10, and keratin 1 stainings were visualized with 5-, 2 1/2-, and 2 1/2-minute incubation of DAB, respectively (brown staining). Hematoxylin was used to counterstain the nucleus. The pan-keratin antibody specific for high-molecular-weight epidermal cytokeratins (D and H) was used as a positive control for staining in the samples. (I) Western blot analysis for catalase expression in tumor samples. Lysate from the 308 keratinocyte cell line was included as a positive control.
Figure 2.
Depression of catalase mRNA levels in SCCs. Total RNA was isolated from 12 tumors at the end of the 40-week experiment. The tumors were divided into six papillomas and six carcinomas based on gross pathology. Real-time RT-PCR was performed to determine the relative level of catalase mRNA in each tumor. The relative level of GAPDH mRNA in each tumor sample was used as an internal standard to normalize loading for the real-time RT-PCR reaction. The error bars represent ± SEM deviation within the each sample group (*P < .05 compared to the papilloma group).
No Gross Deletion or Rearrangement of the Catalase Gene Was Detected
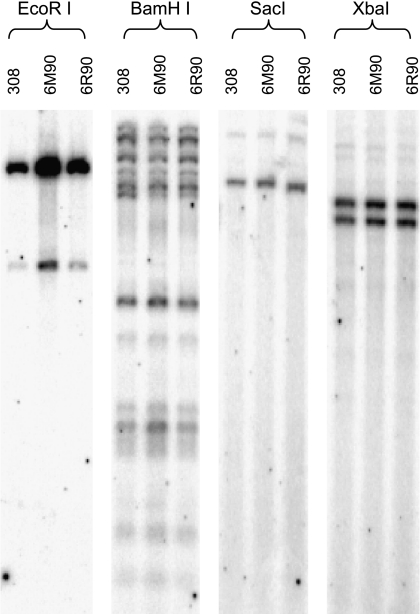
We previously observed that malignant variants of 6M90 and 6R90 cells had decreased catalase expression compared to the benign 308 cells [7]. One possibility was that chemical (MNNG) and physical (γ-ray irradiation) mutagens used to create the malignant phenotypes had caused mutations in the catalase gene. Any deletions and/or rearrangements of the catalase genomic sequence could affect the expression levels. Southern blot analysis was performed to detect any gross changes that might have occurred. Genomic DNA from 308, 6M90, and 6R90 cells were digested with four unique restriction enzymes and eletrophoresed on a 1% agarose gel. The blot was probed with a 1.8-kb full-length rat catalase cDNA probe (the coding sequences of rat and mouse catalase gene are 93% identical) to provide a complete coverage of the catalase gene. As shown in Figure 3, banding patterns created by all four restriction enzymes were identical between the three cell lines. These results indicated that no deletions or rearrangements had taken place in genomic sequence encoding for the catalase mRNA transcript and suggested that an alternate mechanism(s) was responsible for the decreased expression in the 6M90 and 6R90 cells.
Figure 3.
Southern blot analysis of catalase genomic structures in 308, 6M90, and 6R90 cells. Twenty-five micrograms of genomic DNA was digested with the appropriate restriction enzyme overnight and loaded onto a 1% agarose gel for Southern blotting. The blot was hybridized with a 32P-labeled rat catalase cDNA probe.
Catalase Message Stabilities Were Not Reduced in the Malignant Variants
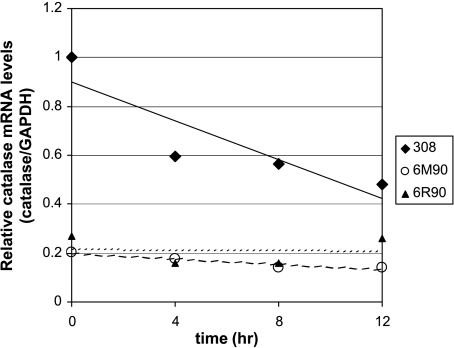
Steady-state message levels represent a balance between the rate of transcription and the rate of degradation. Studies in different cell types have shown that catalase expression can be regulated at the level of message stability [14,15]. To address the question of whether our malignant cell lines had decreased catalase message stability, we performed a time course experiment to measure the message half-life of catalase in our three cell lines. We treated the cells with 1 µg/ml actinomycin D to block transcription and harvested cellular RNA at time points indicated on Figure 4. Real-time RT-PCR was used to measure the degradation in catalase message level after transcription blockage. We did not find any decrease in message stability in the 6M90 and 6R90 cells compared to the parent 308 cells. In fact, the 6M90 and 6R90 cells' catalase mRNA appeared to have a longer half life (t1/2 6M90 ∼51 hours and t1/2 6R90 ∼91 hours) than the 308 cells (t1/2 308 ∼17 hours).
Figure 4.
Catalase message stabilities in 308, 6M90, and 6R90 cells. The cells were treated with 1 µg/ml actinomycin D in serum-free medium at time zero. RNA was isolated at the time points indicated and analyzed with real-time RT-PCR. To compare the differences between cell lines, catalase mRNA levels of 308 cells at the zero time point were set to one.
Reduced Catalase Transcription Rates in Both 6M90 and 6R90 Cells
There is evidence in the literature to suggest that catalase expression can be regulated at the transcription level [16,17]. Here, we used nuclear run-on analysis to determine if decreases in steady-state catalase message levels found in the malignant variants were due to a lower transcription rate. Nuclei isolated from 308, 6M90, and 6R90 cells were incubated with biotin-16-UTP for the run-on reaction. The newly synthesized biotin-labeled RNA transcripts were captured by streptavidin-coated magnetic beads. The RNAbound beads were used for real-time RT-PCR to evaluate differences in catalase transcription between cell lines. The results showed that malignant variants 6M90 and 6R90 cells have approximately a 50% decrease in catalase transcription rates compared to the benign 308 cells (Figure 5). This suggested that downregulation of catalase expression observed in our in vitro progression model was due to transcriptional depression.
Figure 5.
Decreased catalase transcription rates in malignant variants. Nuclear run-on assays were performed as described in Materials and Methods section. The results shown here represent the average ± SD of three experiments. To compare the differences between cell lines, the level of run-on catalase message produced by the 308 cells was set as one in each experiment.
Catalase Promoter Analysis
To determine if there were genetic changes associated with transcriptional repression, we sequenced the -557 to +89 region of the catalase promoter in the three cell lines. Ten individual recombinant clones from each cell lines were analyzed. We detected a single nucleotide polymorphism (SNP) in all three cell lines when compared to the published sequence submitted by Reimer et al. [18]. However, we failed to detect any sequence differences between the benign 308 and the malignant 6M90 and 6R90 cells. Next, we examined epigenetic alterations that might have occurred in the malignant progression process. Aberrant cytosine methylation of a tumor-suppressor gene promoter is a mechanism by which tumor cells can downregulate its transcription [19,20]. Sun et al. [21] have shown, by methylation-sensitive Southern blot analysis, that catalase depression in transformed mouse liver cells was due, in part, to an increase in cytosine methylation. The mouse catalase promoter sequence published by Reimer et al. [18] showed 21 potential methylation-sensitive CpG sites, making it a likely candidate as a CpG island that could be targeted for gene regulation. We performed bisulfite sequencing to examine the methylation status of 17 individual CpG sites on the -248 to +92 region of the catalase promoter. None of the cytosines in the catalase promoter of 308, 6M90, and 6R90 cells was found to be methylated (Table 1). This led us to conclude that methylation did not play a role in the repression of catalase in our system.
Table 1.
Sodium Bisulfite Sequencing of the Catalase Promoter Regions in 308, 6M90, and 6R90 Cells.
| Methylation Status of the Catalase Promoter | |
| Methylated | CpG/Total CpG Cell Line |
| 308 | 0/17 |
| 6M90 | 0/17 |
| 6R90 | 0/17 |
Genomic DNA from the three cells lines were bisulfite-modified and subjected to two rounds of nested PCR to amplify the -248 to +92 catalase promoter region. Ten clones of each cell line were sequenced and analyzed for CpG dinucleotide methylations.
Catalase Promoter Transactivation
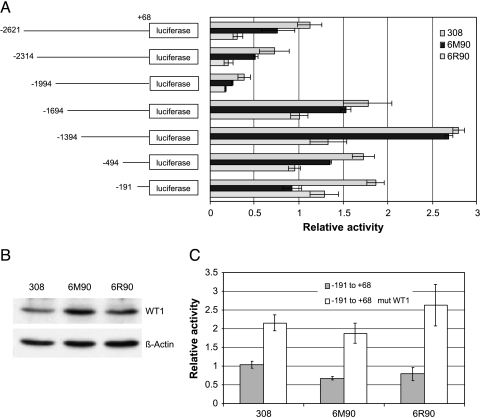
The results from the promoter analysis suggested that the same catalase promoter might have different transactivation activities depending on the cell lines. To test this idea, we employed a series of luciferase reporter constructs driven by different lengths of the 5′-flanking region of the mouse catalase gene [2]. The reporter constructs were transiently transfected into 308, 6M90, and 6R90 cells to assay for catalase promoter transactivation. The luciferase activities of 308 cells were higher than for 6M90 and 6R90 cells, using all the fragments tested, from -2621/+68 to -191/+68 (Figure 6A). This indicates that the same catalase promoter has a higher transactivation potential in 308 cells than its malignant counterparts 6M90 and 6R90 cells. The deletion series also allowed us to identify potential regulatory elements that could explain the repression of promoter activities in the 6M90 and 6R90 cells. For all three cell lines, there was a gradual decrease in luciferase activities from the full-length -2621 to -1994, indicating the presence of positive regulatory elements that were removed as the fragment was shortened. A dramatic increase in luciferase activity was then observed in the next shorter fragment (-1694/+68), indicating the presence of a strong negative regulatory element(s) in the 300-bp sequence between -1994/+68 and -1694/+68. The constructs with fragments from -1694/+68 to -191/+68 show similar luciferase activities, indicating a lack of major regulatory element in those regions. The most significant finding was the fact that the shortest fragment (-191/+68) retains the repression of catalase promoter transactivation in the 6M90 and 6R90 cells. These results suggested that a regulatory element(s) that is responsible for transcriptional repression resides within the -191/+68 sequence of the catalase promoter.
Figure 6.
Catalase promoter transactivation. (A) 308, 6M90, and 6R90 cells were transfected with a luciferase reporter construct driven by the catalase promoters ranging in size from -2621/+68 to -191/+68. Luciferase assays were performed as described in Materials and Methods section. (B) Expression levels of WT1 protein in 308, 6M90, and 6R90 cells. Western blot analysis was performed with 20 µg of total protein in each lane and probed with anti-WT1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The blot was then stripped and reprobed with anti-β-actin antibody (Sigma) for loading control. (C) Increase in catalase promoter activity by abrogation of WT1 binding on the -191/+68 reporter construct. Results of the luciferase reporter assays shown are representative of three independent experiments performed in triplicate ± SD.
Sequence analysis revealed multiple regulatory elements within the -191/+68 region. They include NF-κB, CAAT boxes, and GC boxes [12]. Nenoi et al. had reported that GC box binding proteins play a critical role in the repression of catalase transcription. The positive transcription regulator, stimulating protein 1 (SP1), and the negative transcription regulator, WT1, compete for the GC box binding site within the human catalase gene promoter to differentially regulate transcription in leukemia cells [19]. Western blot analysis of total cellular extracts from our three cell lines showed that the WT1 protein was expressed at higher levels in the malignant cells. Densitometry of the WT1 band normalized to β-actin loading control band revealed an 84% increase in 6M90 and a 23% increase in 6R90 compared to 308 cells (Figure 6B). We hypothesized that the increased expression of WT1 protein, a negative transcription regulator, in the malignant cell lines could lead to the repression of catalase promoter activity. To test this hypothesis, we performed site-directed mutagenesis to mutate the WT1 consensus binding site within the GC box, while retaining the SP1 consensus binding site in the -191/+68 reporter construct. Luciferase activity of the WT1 mutated construct was significantly higher than the wild type, indicating that WT1 protein maybe acting as a transcription repressor in this system (Figure 6C). Moreover, the mutation causes the luciferase activities of both 6M90 and 6R90 to increase to a level similar to the 308 levels, demonstrating that WT1 binding site plays a significant role in the repression of catalase transcription in 6M90 and 6R90 cells.
Discussion
Based on our previous in vitro observations concerning catalase repression, we hypothesized that catalase functions as a tumor-suppressor gene to impede malignant transformation of mouse skin tumors. In these studies, we presented in vivo evidence to support the role of catalase repression in tumor progression. We showed by immunohistochemistry that papilloma samples have a robust catalase expression, whereas the SCCs have low to undetectable levels of catalase expression. We also showed that catalase mRNA levels correlate with the levels of protein expression in tumors. We believed that the variations in message levels (Figure 2) were due to the heterogeneous nature of these tumors. Total RNA was harvested from frozen tumor samples that might contain a mixture of tumor cells (papilloma and SCC) as well as other nonkeratinocytes such as fibroblasts and endothelial cells. The classification of these tumors into papilloma or SCC was based on gross pathology, which reflected the majority of the tumor cells in a given sample but failed to take into account other tissue types present in the samples. These factors contributed to the variations in catalase message levels found in the tumors. The results from the in vivo tumor progression experiment recapitulated our previous observation that catalase was downregulated at both mRNA and protein levels during in vitro tumor progression [7].
Catalase expression is known to be regulated at multiple levels, including the message, protein, and activity levels [22,18]. Reports in the literature have shown that cells have evolved different strategies to downregulate the expression of catalase. In both mouse and rat hepatoma cells, catalase is reported to be repressed at the transcriptional level [23,17,21,24]. In neonatal rat lung and human breast cancer cells, catalase expression is believed to be downregulated by decreases in message stability [25,26]. Reimer et al. have reported that the 3′ untranslated region of mouse catalase mRNA might be used to regulate translation rates in a tissuespecific manner during development [27]. In addition, the acatalasemic mouse strain, Csb, was found to contain a glutamine-to-histidine transversion mutation in the coding region of catalase that rendered the protein unstable under physiological conditions [28]. We have ruled out translational repression as a possible mechanism in our system because, in such a scenario, we would have expected the steady-state mRNA levels of 308, 6M90, and 6R90 cells to be very similar. Instead, 308 cells have approximately four-fold higher catalase mRNA than 6M90 and 6R90 cells (Figure 4, 0 hour time point). Southern blot analysis revealed that the catalase genomic structures were identical between the three cell lines. It indicated that no rearrangement or deletion had taken place in the 6M90 or 6R90 cells that could account for the reduction in steady-state mRNA levels. Next, we examined the message half-life of catalase in these cell lines using actinomycin D treatment. We failed to detect any decreases in 6M90 and 6R90 message stability that would lead to lower steady-state levels. In fact, we found that messages in 6M90 and 6R90 cells were more stable than those in 308 cells. The only other way to decrease steadystate message levels would be a transcriptional repression in the 6M90 and 6R90 cells. Results from the nuclear run-on experiments provided strong evidence to support that theory. The 6M90 and 6R90 cells' rates of catalase transcription were reduced in half compared to the 308 cells. We believe that this was the mechanism that accounts for the reduction in steady-state mRNA levels. To further dissect the mechanisms behind the transcriptional repression, we examined both genetic and epigenetic changes in the catalase promoter found in the 6M90 and 6R90 cells. Genomic sequencing of the catalase promoter (-557 to +89) did not reveal any differences between the three cell lines. Sun et al. [21] have shown in rat hepatoma cells that aberrant methylation in the catalase promoter led to downregulation of transcription. We failed to find any evidence for methylated CpG sites in the catalase promoter using bisulfite sequencing (Table 1) and methylation-sensitive Southern blot analysis (data not shown). It had recently been suggested by Georgel et al. [29] that chromatin condensation and gene silencing by the transcription repressor, MeCP2, could occur independent of the DNA methylation status. We have tested this potential explanation for catalase gene silencing by treating the three cell lines with histone deacetylase inhibitor, trichostatin A, but we failed to detect any change in the steady-state level of catalase mRNA (data not shown). Because the mechanism of action for trichostatin A is specific for histone deacetylase and is DNA methylation-independent, we concluded that MeCP2 or chromatin remodeling does not play an important role in the silencing of catalase in the malignant cells.
Based on these findings, we believe that the attenuation of catalase expression in the tumor cells is due to a dysregulation of catalase promoter activity. The catalase promoter in mice, rats, and humans have been documented to contain multiple enhancer and silencer regions [12,16,17,30,31]. The transcriptional repression observed in the malignant cells can be the result of increases in silencer elements, decreases in enhancer elements, or a combination of both. Sato et al. [17] and Takeuchi et al. [30] discovered two distinct silencer elements that were responsible for the negative regulation of catalase transcription in rat hepatoma cells. The silencer elements were located between 3.4 and 1.7 kb upstream of the rat catalase transcription start site. They determined that nuclear proteins, which are abundantly expressed in dedifferentiated hepatoma cells but scarcely expressed in well-differentiated liver cells, bound specifically to these silencer elements, leading to the downregulation of transcription. Based on our results from the transactivation reporter assays, we have determined that a potential silencer element exists in the -1994 to -1694 region. However, this silencer region does not seem to play a role in the differential transcription rate between the 308, 6M90, and 6R90 cells. The promoter region of the catalase gene shared a high degree of homology between species (mouse [18], rat [32], and human [33]). All three promoters lack TATA boxes but contain binding sites for CCAAT box and GC box proteins. It is likely that all three species share some common mechanisms for transcriptional regulation of catalase. Nenoi et al. had reported that hydrogen peroxide-resistant human leukemia HP100 cells have constitutively elevated catalase promoter activity. The promoter activity was reduced following ionizing radiation exposure [16]. They concluded that the interaction between multiple transcription factors in the -176 to +16 promoter region in the human catalase gene was responsible for the regulation of catalase transcription in the HP100 cells. These factors include SP1, NF-Y, and WT1. The elevation of catalase transcription was the result of elevated expression of SP1 and NF-Y in the HP100 cells. They theorized that ionizing radiation induced the association of WT1 to its response element and inhibited SP1 binding through steric hindrance. We speculate that a similar mechanism is at work in our model system. The increased expression of WT1 protein in our malignant cells could lead to increased binding to the GC box, thus reducing the binding of SP1 to the same element, leading to the lost of catalase promoter transactivation.
Our study has shown for the first time that catalase expression is downregulated in the multistep mouse skin carcinogenesis model in vivo. In the literature, MnSOD had been reported as the major antioxidant enzyme involved in the mouse skin carcinogenesis model by Zhao et al. [34]. That study focused on the role of MnSOD as a mediator of ROS during the promotion step of carcinogenesis. The results from our laboratory suggested that catalase plays an important role as an antioxidant in maintaining the redox state during the progression step of carcinogenesis. Furthermore, we were able to show that catalase repression occurred at the transcriptional level in both chemical and physical carcinogen-derived cell lines, thus highlighting its potential role as a key tumor-suppressor gene in the etiology of SCCs in general.
Acknowledgements
We thank our pathologist, Raymond B. Nagle, for his assistance in analyzing the histology samples.
Footnotes
This work was supported, in part, by National Institutes of Health grants CA40584 and CA23074, and T32 training grant CA0921.
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Stratton SP. Prevention of non-melanoma skin cancer. Curr Oncol Rep. 2001;3:295–300. doi: 10.1007/s11912-001-0080-x. [DOI] [PubMed] [Google Scholar]
- 3.Boutwell RK. The function and mechanism of promoters of carcinogenesis. CRC Crit Rev Toxicol. 1974;2:419–443. doi: 10.3109/10408447309025704. [DOI] [PubMed] [Google Scholar]
- 4.Balmain A, Ramsden M, Bowden GT, Smith J. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–660. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- 5.Gupta A, Rosenberger SF, Bowden GT. Increased ROS levels contribute to elevated transcription factor and MAP kinase activities in malignantly progressed mouse keratinocyte cell lines. Carcinogenesis. 1999;20:2063–2073. doi: 10.1093/carcin/20.11.2063. [DOI] [PubMed] [Google Scholar]
- 6.Strickland JE, Greenhalgh DA, Koceva-Chyla A, Hennings H, Restrepo C, Balaschak M, Yuspa SH. Development of murine epidermal cell lines which contain an activated rasHa oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 1988;48:165–169. [PubMed] [Google Scholar]
- 7.Gupta A, Butts B, Kwei KA, Dvorakova K, Stratton SP, Briehl MM, Bowden GT. Attenuation of catalase activity in the malignant phenotype plays a functional role in an in vitro model for tumor progression. Cancer Lett. 2001;173:115–125. doi: 10.1016/s0304-3835(01)00656-5. [DOI] [PubMed] [Google Scholar]
- 8.Butts BD, Kwei KA, Bowden GT, Briehl MM. Elevated basal reactive oxygen species and phospho-Akt in murine keratinocytes resistant to ultraviolet B-induced apoptosis. Mol Carcinog. 2003;37:149–157. doi: 10.1002/mc.10131. [DOI] [PubMed] [Google Scholar]
- 9.Futscher BW, Pieper RO, Dalton WS, Erickson LC. Genespecific DNA interstrand cross-links produced by nitrogen mustard in the human tumor cell-line Colo320HSR. Cell Growth Differ. 1992;3:217–223. [PubMed] [Google Scholar]
- 10.Patrone G, Puppo F, Ceccherine I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. BioTechniques. 2000;29:1012–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 11.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo D, Rando TA. The regulation of catalase gene expression in mouse muscle cells is dependent on the CCAAT-binding factor NF-Y. Biochem Biophys Res Commun. 2003;303:609–618. doi: 10.1016/s0006-291x(03)00397-8. [DOI] [PubMed] [Google Scholar]
- 13.Nelson KG, Slaga TJ. Keratin modifications in epidermis, papillomas, and carcinomas during two-stage carcinogenesis in the SENCAR mouse. Cancer Res. 1982;42:4176–4181. [PubMed] [Google Scholar]
- 14.Rohrdanz E, Kahl R. Alterations of antioxidant enzyme expression in response to hydrogen peroxide. Free Radic Biol Med. 1998;24:27–38. doi: 10.1016/s0891-5849(97)00159-7. [DOI] [PubMed] [Google Scholar]
- 15.Sampath D, Perez-Polo R. Regulation of antioxidant enzyme expression by NGF. Neurochem Res. 1997;22:351–362. doi: 10.1023/a:1027387105882. [DOI] [PubMed] [Google Scholar]
- 16.Nenoi M, Ichimura S, Mita K, Yukawa O, Cartwright IL. Regulation of the catalase gene promoter by Sp1, CCAAT-recognizing factors, and a WT1/Egr-related factor in hydrogen peroxide-resistant HP100 cells. Cancer Res. 2001;61:5885–5894. [PubMed] [Google Scholar]
- 17.Sato K, Ito K, Kohara H, Yamaguchi Y, Adachi K, Endo H. Negative regulation of catalase gene expression in hepatoma cells. Mol Cell Biol. 1992;12:2525–2533. doi: 10.1128/mcb.12.6.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimer DL, Bailley J, Singh SM. Complete cDNA and 5′ genomic sequences and multilevel regulation of the mouse catalase gene. Genomics. 1994;21:325–336. doi: 10.1006/geno.1994.1273. [DOI] [PubMed] [Google Scholar]
- 19.Greger V, Debus N, Lohmann D, Hopping W, Passarge E, Horsthemke B. Frequency and parental origin of hypermethylated RB1 alleles in retinoblastoma. Hum Genet. 1994;94:491–496. doi: 10.1007/BF00211013. [DOI] [PubMed] [Google Scholar]
- 20.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylationassociated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 21.Sun Y, Colburn NH, Oberley LW. Depression of catalase gene expression after immortalization and transformation of mouse liver cells. Carcinogenesis. 1993;14:1505–1510. doi: 10.1093/carcin/14.8.1505. [DOI] [PubMed] [Google Scholar]
- 22.el-Hage S, Singh SM. Regulation of catalase-specific mRNA and its processing during development in mice. Dev Genet. 1989;10:339–344. doi: 10.1002/dvg.1020100410. [DOI] [PubMed] [Google Scholar]
- 23.Kim HS, Lee TB, Choi CH. Down-regulation of catalase gene expression in the doxorubicin-resistant AML subline AML-2/DX100. Biochem Biophys Res Commun. 2001;281:109–114. doi: 10.1006/bbrc.2001.4324. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi Y, Sato K, Endo H. Depression of catalase gene expression in the liver of tumor bearing nude mice. Biochem Biophys Res Commun. 1992;189:1084–1089. doi: 10.1016/0006-291x(92)92315-o. [DOI] [PubMed] [Google Scholar]
- 25.Akman SA, Forrest G, Chu FF, Doroshow JH. Resistance to hydrogen peroxide associated with altered catalase mRNA stability in MCF7 breast cancer cells. Biochim Biophys Acta. 1989;930:140–144. doi: 10.1016/0167-4781(89)90080-8. [DOI] [PubMed] [Google Scholar]
- 26.Clerch LB, Iqbal J, Massaro D. Perinatal rat lung catalase gene expression: influence of corticosteroid and hyperoxia. Am J Physiol. 1991;260:L428–433. doi: 10.1152/ajplung.1991.260.6.L428. [DOI] [PubMed] [Google Scholar]
- 27.Reimer DL, Singh SM. Distinct mRNA-binding protein interacting with short repeat sequences of the 3′ UTR may be involved in the post-transcriptional regulation of the mouse catalase gene Cas-1. DNA Cell Biol. 1996;15:317–328. doi: 10.1089/dna.1996.15.317. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer JB, Preston KE. Molecular analysis of an acatalasemic mouse mutant. Biochem Biophys Res Commun. 1990;173:1043–1050. doi: 10.1016/s0006-291x(05)80891-5. [DOI] [PubMed] [Google Scholar]
- 29.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi T, Nakamura S, Kayasuga A, Isa S, Sato K. Multiple elements for negative regulation of the rat catalase gene expression in dedifferentiated hepatoma cells. J Biochem (Tokyo) 2000;128:1025–1031. doi: 10.1093/oxfordjournals.jbchem.a022830. [DOI] [PubMed] [Google Scholar]
- 31.Van Remmen H, Williams MD, Yang H, Walter CA, Richardson A. Analysis of the transcriptional activity of the 5′-flanking region of the rat catalase gene in transiently transfected cells and in transgenic mice. J Cell Physiol. 1998;174:18–26. doi: 10.1002/(SICI)1097-4652(199801)174:1<18::AID-JCP3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima H, Yamamoto M, Goto K, Osumi T, Hashimoto T, Endo H. Isolation and characterization of the rat catalase-encoding gene. Gene. 1989;79:279–288. doi: 10.1016/0378-1119(89)90210-2. [DOI] [PubMed] [Google Scholar]
- 33.Quan F, Korneluk RG, Tropak MB, Gravel RA. Isolation and characterization of the human catalase gene. Nucleic Acids Res. 1986;14:5321–5335. doi: 10.1093/nar/14.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Xue Y, Oberley TD, Kiningham KK, Lin SM, Yen HC, Majima H, Hines J, Clair D. Overexpression of manganese superoxide dismutase suppresses tumor formation by modulation of activator protein-1 signaling in a multistage skin carcinogenesis model. Cancer Res. 2001;61:6082–6088. [PubMed] [Google Scholar]