Abstract
The intronic Ig heavy chain (IgH) enhancer, which consists of the core enhancer flanked by 5′ and 3′ matrix attachment regions, has been implicated in control of IgH locus recombination and transcription. To elucidate the regulatory functions of the core enhancer and its associated matrix attachment regions in the endogenous IgH locus, we have introduced targeted deletions of these elements, both individually and in combination, into an IgHa/b-heterozygous embryonic stem cell line. These embryonic stem cells were used to generate chimeric mice by recombination activating gene-2 (Rag-2)-deficient blastocyst complementation, and the effects of the introduced mutations were assayed in mutant B cells. We find that the core enhancer is necessary and sufficient to promote normal variable (V), diversity (D), and joining (J) segment recombination in developing B lineage cells and IgH locus transcription in mature B cells. Surprisingly, the 5′ and 3′ matrix attachment regions were dispensable for these processes.
The Ig heavy chain (IgH) locus is an intriguing model for studying gene regulation because of the functional interplay between transcription and recombination in the context of cell lineage and developmental stage-specific expression patterns (1, 2). The IgH locus comprises a 5′ region that harbors variable, diversity, and joining (VH, DH, and JH) segments and a 3′ region that harbors the constant region exons (Cμ-Cδ-Cγ3-Cγ1-Cγ2b-Cγ2a-Cɛ-Cα); each region spans several hundred kb. The VH-DH-JH locus undergoes V(D)J recombination during early B cell development. V(D)J recombination initiates at the pro-B cell stage and is ordered with DH to JH rearrangement preceding VH to DJH rearrangement. Generation of a μ IgH chain from a productive V(D)J segment results in differentiation to the precursor-B cell stage in which most Ig light (L) chain variable region genes are assembled, eventually producing the complete (H and L chains) surface Ig complexes. (reviewed in refs. 1 and 2).
Multiple enhancer elements have been identified within the IgH locus, including the intronic enhancer (Eμ) between JH and Cμ (3, 4) and a series of enhancers (collectively referred to as the 3′ IgH regulatory region) that lie downstream of Cα (reviewed in ref. 5). Extensive transfection and transgenic studies delineated the Eμ sequences based on ability to direct lymphoid-specific expression (3, 4, 6–8; reviewed in refs. 9–11). Studies of cell lines with spontaneous Eμ region deletions suggested this enhancer was necessary for IgH expression in precursor-B cells (12, 13), but dispensable for expression in terminally differentiated B cell lines (14–17).
Transfection assays defined a small 220-bp core element (hereafter referred as cEμ) within Eμ, which is necessary and sufficient for transcriptional stimulation; cEμ contains multiple binding sites for both ubiquitous and cell-specific factors with negative and positive activity (reviewed in ref. 18). Biochemical assays further identified two AT-rich nuclear matrix attachment regions (MARs) flanking cEμ (19). MARs are generally defined by the ability to bind to the nuclear matrix, which is a rather poorly defined protein fraction containing factors important for regulation of gene expression in addition to structural scaffold components (reviewed in refs. 20–25). Despite a strictly biochemical definition, several functions for MARs have been proposed (reviewed in refs. 20, 23–27). For example, MARs have been implicated in defining physical boundaries between genes (27, 28). In addition, MARs often are found in close association with active elements such as enhancers (19, 27, 28), promoters (29, 30), and putative replication origins (31, 32), potentially serving to anchor these elements to specific nuclear matrix sites. MARs have also been described as regions susceptible to histone H1 displacement and, therefore, chromatin “opening” by way of the interaction of minor groove binding proteins like HMG-I/Y (33).
The Eμ-associated MARs initially were implicated as a negative regulator in non-B cells (34–39). The Eμ MARs also contain topoisomerase II cleavage consensus sequences and regions susceptible to unpairing under negative supercoiling, which are speculated to control chromatin superhelicity (26). More recently, it has been concluded that the Eμ MARs contribute positively to Eμ function based on ability to promote position-independent expression of VH promoter-driven transgenes (40). The MARs also increased the distance from Eμ at which a prokaryotic promoter was accessible to its specific polymerase (41, 42). Together, the latter two findings suggested that cEμ can induce local chromatin unwinding, but that the MARs are necessary to increase the spatial range of this effect (42).
Our earlier studies indicated that Eμ may be positively involved in regulating V(D)J rearrangement. In this context, we found that Eμ could drive V(D)J and DJ rearrangement of a T cell antigen receptor (TCR)β/IgH hybrid minilocus in normal developing lymphocytes (43–45) and in a B-lineage cell line (43–45), and also could replace the TCRβ enhancer in driving endogenous TCRβ locus rearrangement in T lineage cells (46). We further used gene-targeted mutation in embryonic stem (ES) cells to show that recombination of the JH locus was greatly inhibited by replacement of the entire core/MARs complex (Eμ) with a phosphoglycerate-kinase (pgk)-neomycin-resistance (neor) gene cassette (47). However, others showed that D to JH rearrangement occurred relatively normally when the same region was replaced with a short oligonucleotide sequence, although VH to DJH rearrangement was substantially inhibited by this mutation (48).
To more clearly delineate contributions of cEμ and the MARs in the physiologic context of the native IgH locus, we have now introduced targeted deletions of each of these elements, both individually and in combination, and examined their effect on V(D)J rearrangement and expression of the endogenous locus in vivo.
MATERIALS AND METHODS
Targeting Vectors.
A 3.1-kb partial HindIII genomic 129/sv (IgHa allotype) DNA fragment containing the JH-Eμ-Iμ region (for Δ5′MAR and ΔcEμ constructs) and a 3.8-kb partial HindIII–PvuII fragment with an additional 0.7 kb at the 3′ end (for other constructs) was cloned into the HindIII site of pBluescript II (Stratagene). The thymidine kinase gene driven by pgk promoter was cloned into the EcoR V site in the 5′ homology arm. Each region to be deleted was first replaced with a NotI site; for the ΔMARS construct, the site was 3′ of the core element. Finally, a pgk-neor gene flanked by two loxP sites (49) was cloned into the introduced NotI site. For the ΔMARS construct, an additional copy of the 1-kb XbaI–XbaI enhancer fragment was inserted between the 5′ loxP site and the pgk promoter, and an additional pgk-thymidine kinase gene was inserted downstream of the 3′ homology arm.
Generation of Mutant Chimeric Mice.
Transfection of vectors into F1/1 ES cells (50) and isolation of drug-resistant clones was carried out as described (47). Targeting efficiencies for each replacement (R) mutation with neor gene were: REμ, 2/129; RcEμ, 2/404; R5′MAR, 1/327; R3′MAR; 2/182; RMARS, 2/384. The homozygous replacement of both MARs (R/R MARS) was obtained by high G418 selection of heterozygous mutant clones (1.2–1.6 mg/ml) (51). Positive clones were subjected to Cre recombinase-mediated deletion (49) to generate the heterozygous (ΔEμ, ΔcEμ, Δ5′MAR, Δ3′MAR, and ΔMARS) and homozygous (Δ/ΔMARS) deletion mutants. Clones were injected into blastocysts from recombination activating gene-2 (Rag-2)-deficient mice (CBA × C57BL6 mixed background) and transferred into foster mothers as described (52). Chimeras were analyzed at 5 weeks to 4 months of age. Nonlymphoid tissue chimerism was 10–30%, as assessed by Southern blotting of kidney DNA.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
Single-cell suspensions from spleens (5 × 105 cells/100 μl) were stained by using 0.5 μg of phycoerythrin-labeled anti-mouse IgMa and fluorescein isothiocyanate -labeled anti-mouse IgMb antibodies (PharMingen) and analyzed by using a FACStarplus flow cytometer (Becton Dickinson).
Southern Blot Analysis.
IgM-positive B cells were purified from spleen cells by panning using goat anti-mouse IgM antibody (Southern Biotech.). Recovered cells were >80% B220 positive. For Southern blotting, genomic DNA from 106 cells was digested with appropriate restriction enzymes. For analyses of DH to JH rearrangements, SacI- or PvuII-digested DNA was hybridized with probe A (a 0.38-kb SacI–ApaI fragment between DQ52 and JH1 segments). For VH to DJH rearrangements, BamHI and EcoR V-digested DNA was assayed for hybridization with probe C (a 0.18-kb EcoRI–SacI fragment upstream of DFL16.1). To normalize for DNA content, blots were reprobed with a c-myc cDNA fragment (XhoI–KpnI 1.6-kb fragment). To estimate DNA contribution from Rag-2-deficient cells, as opposed to ES cells, in chimeric tissues, DNA samples were separately digested with PvuII and assayed for hybridization to a Rag-2 cDNA probe (1.0-kb PstI–EcoR V fragment) that distinguishes wild-type vs. mutant Rag-2 genes (53). Quantitation was carried out with a Storm 860 PhosphorImager (Molecular Dynamics). V(D)J rearrangements in hybridomas were similarly analyzed.
RNA Analysis.
Pooled spleen cells from two mutant chimeras were stained with fluorescein isothiocyanate-labeled anti-mouse IgM antibody and phycoerythrin-labeled anti-mouse B220 antibody (PharMingen), and the double positive cells were isolated by sorting (FACStar, Becton Dickinson). Purity was >96% for wild-type and >90% for chimera cells. Total RNA was isolated by using Trizol (Gibco) and assayed by Northern blotting for hybridization to: a Cμ fragment (probe B), a 0.5-kb BglII–HpaI fragment from the Cκ gene, a 0.3-kb EcoRI–PstI fragment from VHNP.B4 (54) (J558 family), and a 0.28-kb EcoRI–PstI fragment from VH81X (55) (7183 family).
Hybridomas.
Single-cell suspensions were prepared from the spleens of Rag-2 chimeras, cultured for 5 days in the presence of 15 μg/ml of lipopolysaccharide, and used for making IgG-secreting hybridomas, as described (56). Clonality was confirmed by Southern blotting for V(D)J rearrangements.
RESULTS
Generation of Mutant cEμ/MAR ES Cells.
The cEμ is contained within a 220-bp HinfI fragment, whereas the entire 5′MAR/cEμ/3′MAR region (hereafter called Eμ) spans a 1-kb XbaI fragment (Fig. 1A). Targeting constructs designed to replace Eμ, the 5′MAR region, the 3′ MAR region, and cEμ with a loxP-flanked neor gene cassette were transfected into F1/1 IgHa/b heterozygous ES cells. The use of the F1/1 ES cells (50), which were generated from C57B6/CBA mice, facilitates analyses of the effects of the mutations through the use of a vs. b allele restriction fragment-length polymorphism and the use of monoclonal antibodies that specifically recognize allele-specific IgH constant region gene products. The neor cassette was removed from specifically targeted ES cells to generate specific deletion mutants by transfection of a Cre-recombinase expression construct (Fig. 1B). ES cells that contained deletions of both MARs (ΔMARS), but that retained cEμ, were generated by targeting cEμ into an ES cell line that harbored the previously targeted ΔEμ mutation (Fig. 1B).
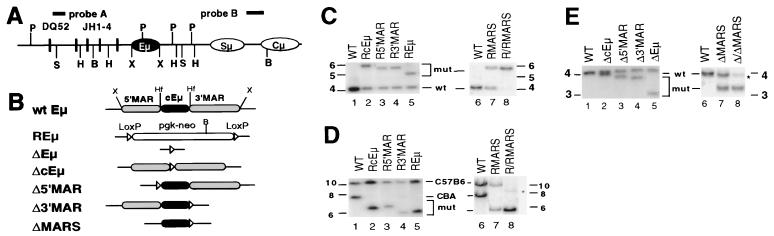
Figure 1.
Targeted mutations of cEμ and MAR elements. (A) Schematic of the IgH locus showing DQ52, JH gene segments, intronic enhancer (Eμ), switch μ region (Sμ), and the μ constant region (Cμ). Also indicated are BamHI (B), HindIII (H), HinfI (Hf), PvuII (P), SacI (S), and XbaI (X) restriction sites, and probe A (380-bp SacI–ApaI fragment) and probe B (0.9-kb XbaI–BamHI fragment). The schematic is not to scale. (B) Schematic diagram of targeted mutations of cEμ and MAR elements. Shaded and filled boxes represent MARs (5′MAR, 350-bp XbaI–HinfI fragment, and 3′MAR, 430-bp HinfI–XbaI fragment), and cEμ (220-bp HinfI–HinfI fragment) elements, respectively. The loxP sites are indicated by open triangles. The structures of individual mutations are shown below. (C) Southern blot analysis of ES cells with neor gene replacement mutations. Genomic DNA from parental ES cells and ES cells in which the 5′MAR (R5′MAR), 3′MAR (R3′MAR), Eμ (REμ), cEμ (RcEμ), and both MARs (RMARS) are replaced with the neor gene heterozygously and both MARs homozygously (R/R MARS), was analyzed by SacI digestion and probe A. The fragment size detected is as follows: 4.8 kb (REμ), 5.6 kb (RcEμ), 5.4 kb (R5′MAR), 5.3 kb (R3′MAR) 5.9 kb (RMARS and R/R MARS), and 4.0 kb (wild type). (D) Identification of the targeted CBA alleles. The same set of DNA as in C was digested with BamHI and subjected to Southern blot analysis by using probe B. The targeted CBA allele gives fragments of 5.6 kb (REμ), 6.0 kb (RcEμ), 6.2 kb (R5′MAR), 5.6 kb (R3′MAR), and 5.6 kb (RMARS and R/R MARS) for individual mutants, as opposed to a wild-type 7.8-kb fragment. (E) Southern blot analysis of mutant ES cells after Cre-mediated deletion of neor gene. SacI-digested genomic DNA was probed with probe A. The size of DNA fragment detected is: 3.1 kb (ΔEμ), 3.9 kb (ΔcEμ), 3.8 kb (Δ5′MAR), 3.7 kb (Δ3′MAR) 3.3 kb (ΔMARS and Δ/ΔMARS), and 4.0 kb (wild type). The asterisk indicates bands derived from feeder cells.
Targeted ES cells were initially identified by Southern blot analyses by using SacI-digested DNA and probe A (Fig. 1 A and C, lanes 1–7). Subsequent analyses using probe B and BamHI-digested DNA revealed that all mutant ES cells had been targeted on the CBA (IgHa) allele (Fig. 1 A and D, lanes 1–7). The mutation on CBA allele was demonstrated by the retention of a 10.5-kb probe B hybridizing band from the C57B6 allele and the loss of the 7.8-kb probe B hybridizing band from the CBA allele in each of the targeted ES cells (Fig. 1D, lanes 1–7). ES cells homozygous for the replacement mutation of both MARs (R/R MARS) were isolated by selecting heterozygous mutants under increasing G418 concentration (51) (Fig. 1C, lane 8; Fig. 1D, lane 8). The double mutants had not only lost the MARs on both alleles, but also lost other IgHb-specific restriction polymorphisms in the Sμ-Cμ region as well as in regions upstream of DH segments and downstream of CH exons (data not shown), suggesting that generation of homozygous knockouts is likely to involve a long-range gene conversion or, possibly, loss of the IgHb allele and duplication of the IgHa allele. Cre recombinase-mediated deletion of the neor gene was carried out on all targeted ES cell clones. Clones that had undergone Cre-mediated deletion of the neor gene were identified by assaying SacI digested DNA with probe A (Fig. 1E, lanes 1–8). Alleles in which the neor gene had been deleted (leaving a single loxP sequence in place of the targeted sequence) generated probe A-hybridizing bands that were distinct from those generated from the wild-type and neor gene targeted alleles (Fig. 1 C and E). The various clones in which the inserted neor gene was deleted, along with one carrying a neor replacement of the Eμ (REμ), were used to generate mice by Rag-2 deficient complementation (52).
Eμ Regulates Recombinational Accessibility of the IgH Locus.
To assess the role of Eμ in regulating IgH locus expression, spleen cells from REμ and ΔEμ mice were stained for surface IgM by using anti-IgMa (CBA allele) and anti-IgMb (C57B6 allele) monoclonal antibodies (Fig. 2A) and assayed by flow cytometry. This analysis revealed significantly reduced numbers of IgMa-expressing B cells in the periphery of REμ and ΔEμ mice (Fig. 2A). To quantitate the level of V(D)J recombination on the mutant alleles of REμ and ΔEμ mice, splenic IgM+ B cells were isolated by panning, yielding >80% pure B cell samples, as determined by flow cytometric analysis (data not shown). SacI-digested genomic DNA from purified B cells was analyzed by Southern blotting for hybridization to probe A (to quantify JH rearrangements) as well as for hybridization of a c-myc probe (to control for DNA loading in the various lanes). These analyses revealed a significant block at the D to JH rearrangement stage on the targeted allele in the REμ mutants, as evidenced by substantial retention of the targeted allele as opposed to the nontargeted allele (Fig. 2B). In fact, the faint germline band from the wild-type allele is likely because of low-level contamination by nonlymphoid cells. The latter conclusion is confirmed by the presence of the Rag-2-targeted allele in these samples; this allele can be derived only from nonlymphoid cells that originated from the Rag-2−/− blastocyst (Fig. 2B, lane 4). In contrast, ΔEμ B cell samples showed levels of D to JH rearrangement on the targeted allele that approached those of the normal allele (Fig. 2B).
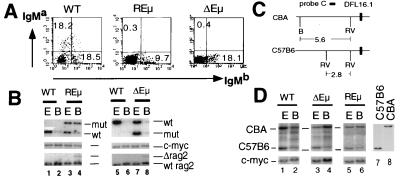
Figure 2.
Analysis of REμ and ΔEμ mice. (A) Flow cytometric analysis of surface IgMa and IgMb expression by spleen cells from wild-type, REμ, and ΔEμ mice. Percentages shown are of lymphocytes gated by forward and side scatter. (B) D to J rearrangement in REμ and ΔEμ B cell. Genomic DNA from ES cells (E) and purified IgM+ splenic B cells (B) of wild type and mutants was digested with SacI and subjected to Southern blot analysis by using probe A and then the c-myc probe. The level of Rag-2−/− cells in the DNA samples are estimated by using PvuII-digested genomic DNA and Rag-2 probe. (C) V to DJ rearrangement in REμ and ΔEμ B cells. The polymorphic restriction enzyme fragments of the region upstream of DFL16.1, the most 5′ D segment, are schematically shown in the left. BamHI (B) and EcoR V (RV) enzyme sites are shown. Genomic DNA samples from ES cells and purified IgM+ splenic B cells of wild type and mutants were digested with BamHI and EcoR V and subjected to Southern blot analysis by using probe C and c-myc probe.
To assay for VH to D rearrangements, EcoR V/BamHI-digested genomic DNA was assayed by Southern blotting for hybridization to probe C (Fig. 2D). VH to DJH rearrangements result in deletion of the IgH region from which probe C is derived and, therefore, generate a decrease in intensity of the hybridizing band in proportion to the extent of rearrangement. In wild-type B cells, we observed a decrease, but not complete loss, of the band from both the CBA and C57B6 alleles. This level of reduction is expected as IgH VH to DJH rearrangement is regulated in the context of allelic exclusion with the residual band being caused by alleles that are retained in the DJH configuration. In contrast, both REμ and ΔEμ B cells exhibited a substantial retention of the germline band from the targeted allele as compared with the wild-type allele (Fig. 2D), demonstrating that VH to DJH rearrangement is significantly decreased by both mutations.
Together, these results confirm that Eμ is required in cis to promote accessibility of the IgH locus; however, additional elements, capable of promoting D to JH accessibility, must exist in the IgH locus, and their activity appears to be hampered by the presence of a pgk-neor gene.
The MARs Do Not Function Independently to Mediate Recombinational Accessibility of the IgH Locus.
Previous studies have established a role for the Eμ-associated MARs in regulating transcription of IgH transgenes (40). To assess for potentially independent roles of these sequences in modulating accessibility to V(D)J recombinase, we analyzed the effect of the Δ5′MAR, Δ3′MAR, and ΔcEμ mutations on V(D)J recombination at the JH locus. Chimeric mice generated with Δ5′MAR and Δ3′MAR ES cell lines exhibited a normal ratio of IgMa- vs. IgMb-expressing B cell numbers in the periphery (Fig. 3A). Purified peripheral IgM-expressing B cells from Δ5′MAR and Δ3′MAR mice were used directly for Southern analyses and also to make B cell hybridomas that were subsequently analyzed for DH to JH and VH to DJH rearrangements.
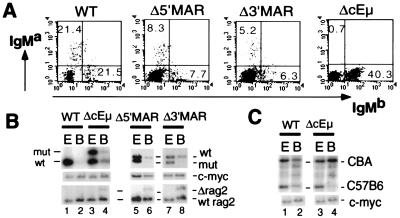
Figure 3.
Analysis of Δ5′MAR, Δ3′MAR, and ΔcEμ mice. (A) Flow cytometric analysis of surface IgMa and IgMb expression by spleen cells from wild-type, Δ5′MAR, Δ3′MAR, and ΔcEμ mice. (B) D to J rearrangement in Δ5′MAR, Δ3′MAR, and ΔcEμ B cells. Genomic DNA from ES cells and purified IgM+ splenic B cells of wild type and mutants was digested with SacI (Δ5′MAR and Δ3′MAR) or Pvu II (ΔcEμ) and subjected to Southern blot analysis as in Fig. 2B. (C) V to DJ rearrangement in Δ5′MAR, Δ3′MAR, and ΔcEμ B cells. Genomic DNA samples from ES cells and purified IgM+ splenic B cells of wild type and mutants was analyzed by Southern blotting as in Fig. 2C.
DH to JH rearrangements occurred at normal levels in Δ5′MAR and Δ3′MAR mice, as demonstrated by the loss of the probe A hybridizing band generated from the mutant and wild-type alleles in B cell populations isolated from Δ5′MAR and Δ3′MAR mice (Fig. 3B, lanes 5–8). The residual wild-type germline band is likely caused by contamination by nonlymphoid cells, as demonstrated by the presence of a Rag-2− hybridizing band (Fig. 3B, lanes 5–8). In addition, B cell hybridomas derived from these mice exhibited equivalent levels of DH to JH rearrangement on the targeted and nontargeted alleles (Table 1). VH to DJH rearrangements also occurred at normal levels on the Δ5′MAR and Δ3′MAR alleles, as demonstrated by Southern blot analysis of purified B cells (data not shown) and by analysis of B cell hybridomas derived from Δ5′MAR and Δ3′MAR mice (Table 1). Together, these analyses demonstrated that the Δ5′MAR and Δ3′MAR alleles undergo normal levels of DH to JH and VH to DJH rearrangement; therefore, we conclude that the MAR sequences do not have essential independent functions in mediating these processes.
Table 1.
V(D)J rearrangement status of mutant hybridomas
| Total number | Allele | Germline | DJ | V(D)J | |
|---|---|---|---|---|---|
| ΔcEμ | 21 | wt | 0 | 0 | 21 |
| mutant | 4 (19%) | 15 (71%) | 2 (10%) | ||
| Δ5′MAR | 33 | wt | 2 (6%) | 10 (30%) | 21 (64%) |
| mutant | 2 (6%) | 9 (27%) | 22 (66%) | ||
| Δ3′MAR | 51 | wt | 2 (4%) | 14 (27%) | 35 (69%) |
| mutant | 1 (2%) | 17 (33%) | 33 (65%) | ||
| ΔMARS | 48 | wt | 4 (8%) | 13 (27%) | 31 (65%) |
| mutant | 1 (2%) | 12 (25%) | 35 (73%) |
The rearrangement status of wild type (wt) and mutant alleles was assayed by Southern blot analysis, as described in the legend to Fig. 3.
We then assayed for the potential of the 5′ and 3′ MAR elements to mediate IgH accessibility in B cells in the absence of cEμ. Flow cytometric analyses revealed a significant reduction in IgMa-expressing splenic B cells with a ΔcEμ-mutated IgHa allele (Fig. 3A). Southern blot analyses of genomic DNA isolated from purified peripheral B cells demonstrated significant levels of DH to JH rearrangement on the targeted allele (Fig. 3B; lanes 1–4). However, VH to DJH rearrangement was dramatically reduced in these mice to levels similar to those observed in ΔEμ mice (Fig. 3C; lanes 1–4). Moreover, hybridoma analyses showed that only 2/21 ΔcEμ alleles, as opposed to all 21 wild-type alleles in these cells, had VHDJH rearrangements, confirming the dramatic decrease in VH to DJH recombination. Therefore, the 5′ and 3′ MAR elements are not able to mediate VH to DJH recombination in the absence of cEμ.
The cEμ Is Sufficient to Mediate Recombinational Accessibility and Transcription of the IgH L.
To determine whether cEμ alone could provide Eμ-associated function within the endogenous IgH locus, chimeric mice were generated from ΔMARS ES cells. Flow cytometric analysis of splenic B cells revealed normal ratio of the numbers of IgMa- and IgMb-expressing B cells, demonstrating that cEμ is capable of promoting rearrangement and expression of the IgH locus at a level similar to that of the intact IgHb locus in the same cell, even in the absence of its associated MAR elements (Fig. 4A). Southern blot analyses of genomic DNA isolated from peripheral B cells further demonstrated that both DH to JH and VH to DJH rearrangements occurred at normal levels on the ΔMARS allele (Fig. 4B). Analysis of B cell hybridomas with this mutation independently confirmed equivalent levels of D to JH and VH to DJH rearrangement on the targeted and wild-type alleles (Table 1). These data demonstrate that cEμ can promote normal levels of V(D)J rearrangement of the IgH locus in the absence of its associated MARs.
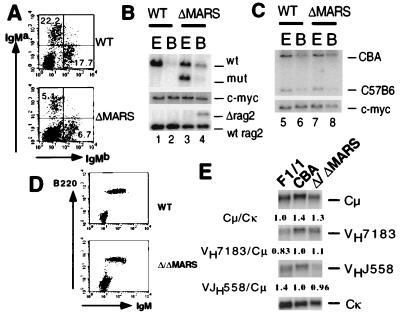
Figure 4.
Analysis of ΔMARS and Δ/ΔMARS mice. (A) Flow cytometric analysis of surface IgMa and IgMb expression by spleen cells from wild-type and ΔMARS mice. (B) D to J rearrangement in ΔMARS B cells. Genomic DNA from ES cells and purified IgM+ splenic B cells of wild type and mutant was digested with SacI and subjected to Southern blot analysis as in Figs. 2B and 3B. (C) V to DJ rearrangement in ΔMARS B cells. Genomic DNA samples from ES cells and purified IgM+ splenic B cells of wild type and mutants were analyzed by Southern blotting as in Figs. 2C and 3C. (D) Flow cytometric analysis of surface IgM and B220 expression by spleen cells from wild-type and Δ/ΔMARS mice. (E) Northern blot analysis of μ chain and VDJH expression by B220+IgM+ spleen B cells from wild-type F1/1 and CBA and Δ/ΔMARS mice. The Cμ/Cκ signal intensity (parental F1/1 cells normalized to 1.0) and the VH7183/Cμ and VHJ558/Cμ intensities (CBA cells to 1.0) are shown below each panel.
In the ΔMARS mutant, the relative numbers of IgMa vs. IgMb B cells, as well as the level of surface expression of the targeted allotype, were identical to controls (Fig. 4A). This observation indicates that the expression of the rearranged μ locus is not affected by the deletion of the two MAR sequences. To confirm this notion, chimeric mice were generated from Δ/ΔMARS ES cells. Flow cytometric analysis confirmed that B220+ IgM+ spleen cells in the mutants express similar levels of surface μ chain as wild-type B cells (Fig. 4D). To rule out secondary compensatory mechanisms in Ig surface expression in these mutants, RNA was prepared from FACS-sorted B cells and subjected to Northern blot analysis using a Cμ region probe (Fig. 4E). Both loading amount and purity were normalized by reprobing the same blot with a Cκ probe. Signal intensities with the two probes were compared in parental F1/1 cells and mutants, as well as in normal CBA B cells to correct for potential differences in expression between the CBA and C57B6 alleles. The Cμ/Cκ signal ratio, normalized to 1.0 for parental F1/1 cells, was 1.4 for the wild-type CBA allele and 1.3 for the mutant allele, indicating that, despite potential background effects on IgH expression, steady-state μ chain transcripts are certainly not diminished in Δ/ΔMARS-mutant mature B cells.
To test the possibility that MARs may control accessibility to the distal VH segments, the usage level of two major VH families, VHJ558 and VH7183, in rearranged transcripts was examined by assaying Δ/ΔMARS and control B cell RNA for hybridization to probes specific for these two families (Fig. 4E). VHJ558 and VH7183 represent the most JH-distal and JH-proximal VH families (reviewed in ref. 2). Signal intensities were normalized to the Cμ RNA expression level and compared between mutant and wild-type CBA to standardize the number of VH segments that belong to individual VH families (given that the Δ/ΔMARS cells contain two IgHa loci). The signal ratios of both VH7183/Cμ and VHJ558/Cμ ratio were quite similar in Δ/ΔMARS vs. wild-type CBA B cell RNA, indicating that VH segments are accessible throughout the VH locus in the absence of the 5′ and 3′ Eμ MARS.
DISCUSSION
The Role of Eμ Components in V(D)J Recombination and IgH Expression.
Previous targeted mutagenesis studies have indicated that sequences within the Eμ enhancer region are important for V(D)J recombination (47, 48). Our current studies show, for the first time, that the core region is the only critical component within Eμ necessary for allowing V(D)J recombination across the IgH locus. Quite unexpectedly, we found that the 5′ and 3′ MAR sequences of this enhancer region are essentially dispensable for V(D)J recombination of the IgH locus, as well as for normal expression of rearranged IgH alleles in mature B cells.
Differential Control of D to JH vs. VH to DJH Rearrangement.
Our studies confirm and extend two separate previous studies that found different effects of a full Eμ deletion (48) vs. replacement of this region with a pgk-neor cassette (47). Our observation that D to JH rearrangement was only minimally impaired by cEμ or Eμ deletions, in contrast to the nearly complete inhibition of VH to DJH rearrangements, demonstrates that additional elements, distinct from Eμ, can function to enhance D to JH recombination. Such elements might promote D to JH rearrangement either specifically or by general opening of the locus. In this regard, it is notable that the D to JH rearrangement step is severely inhibited by the insertion of a pgk-neor cassette in place of the Eμ region (Fig. 2B), indicating that the additional element(s) is negatively influenced by the pgk-neor cassette.
The 3′ IgH regulatory locus is one candidate for a D to JH promoting element, as we have previously shown that this regulatory region can act over large chromosomal distances and also is markedly affected by insertion of pgk-neor cassettes at various positions 5′ to the region (58, 59, 69). Furthermore, insertion of a pgk-neor cassette within the 3′ IgH regulatory region in cells lacking the Eμ region negatively affected IgH expression (59). However, similar mutations of the 3′IgH locus had no influence on V(D)J recombination or expression of the IgH locus with Eμ intact (57), showing that any role for the 3′IgH region in these processes must be redundant with Eμ. Similarly, our findings do not rule out the possibility that Eμ also could influence D to JH recombination; such activities may only become apparent in the absence of potential redundant cis-regulatory elements (e.g., 3′IgH regulatory locus).
The Eμ Core Promotes V(D)J Rearrangement in the Absence of Flanking MARs.
Recent studies of transgenic constructs have shown that the Eμ core element induces local accessibility, but that extension of such chromatin remodeling activity over longer distances requires the flanking MARs (41, 42). However, we show that rearrangement of endogenous VH, D, and JH segments occurs in the presence of just the Eμ core without the MARs, despite the fact that these segments span an extremely large genomic region. Likewise, we show expression of rearranged IgH genes occurs at normal levels in B cells that lack both MARs. Whereas our current findings are consistent with the possibility that cEμ may promote V(D)J rearrangement by means of functions other than alteration of chromatin structure (e.g., by directly recruiting factors required for recombination), the differences between the transgenic and endogenous mutation studies require further explanation.
There are several developmental scenarios by which the apparently contradictory findings regarding the requirement for the 5′ and 3′ MAR sequences in endogenous loci vs. transgenes might be rationalized in the context of a putative required endogenous function. The first would involve the unanticipated possibility that MAR function for B cell specific gene expression is established in very early development and not in B lineage cells. Thus, our MAR mutations were passed on to B cells from ES cells, whereas the B cell transgenes were passed on from the germline. In addition, pgk-neor insertion into the IgH locus in ES cells might somehow mimic the putative role of MAR elements in normal pro-B cells, but this possibility would also require that the putative ES cell effect would be imprinted after the neor gene was deleted. Finally, the MARs could theoretically exert their function only during early B cell development as IgH transgene accessibility was assayed in precursor-B cells, as opposed to the mature B cell stages which we assayed. However, such an effect could not be absolute, as our studies clearly showed comparable IgMa to IgMb B cell numbers in ΔMARS mutants, indicating that mutant allele transcription occurred during the precursor-B cell stage at levels sufficient to support further development.
Overall, our current findings are reminiscent of other recent studies that found different requirements for specific elements within their endogenous vs. transgenic settings including MAR sequences (60–66). For example, the HS2 element in the β-globin locus control region, which had been shown to maintain erythroid-lineage specific expression of transgenes (60), has been found to be dispensable in the endogenous locus (61). Similarly, the Igκ 3′ enhancer had been reported to determine tissue- and stage-specific rearrangement of transgene substrates (62), but clearly was not required for this level of control with respect to the endogenous locus (63, 64). Therefore, it appears that endogenous loci often have a complex potentially redundant regulation that cannot necessarily be linearly dissected with smaller transgenes. In this context, transgenes contain a limited array of components (basically consisting of V(D)J, Eμ, Sμ, and Cμ), whereas the endogenous IgH locus harbors additional known MARs, potentially compensatory in function to the Eμ between downstream CH genes (67) and upstream of the VH107 segment (30). The lack of such elements could well render transgenes more susceptible to negative effects of surrounding foreign genomic regions and potentially repressive influences of tandemly arranged transgenes (68).
Acknowledgments
We thank Bill Forrester and Jianzhu Chen for critical reading of the manuscript and Roger Ferrini for technical assistance. The work was supported by the Howard Hughes Medical Institute and by the National Institutes of Health Grants A.I.20047 and A.I.35714 (to F.W.A.) and A.I.01297–01 (B.P.S.). E.S. was supported in part by the Japan Society for Promotion of Science. B.P.S. is a recipient of a Career Development Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
ABBREVIATIONS
- IgH
Ig heavy chain
- Eμ
intronic enhancer
- V
variable
- D
diversity
- J
joining
- MARs
matrix attachment regions
- C
constant
- H
heavy
- L
light
- cEμ
core enhancer
- TCR
T cell antigen receptor
- ES
embryonic stem
- pgk
phosphoglycerate kinase
- neor
neomycin-resistance
- R
replacement
- FACS
fluorescence-activated cell sorter
- Rag-2
recombination activating gene-2
References
- 1.Bottaro A, Alt F W. In: Local and General Control Elements of Ig Class Switching. Vercelli D, editor. Chichester, U.K.: Wiley; 1997. pp. 155–177. [Google Scholar]
- 2.Lansford R, Okada A, Chen J, Oltz E, Blackwell T, Alt F, Rathburn G. In: Mechanism and Control of Ig Gene Rearrangement. Hames B D, Glover D M, editors. Oxford: Oxford Univ. Press; 1996. pp. 248–282. [Google Scholar]
- 3.Banerji J, Olson L, Schaffner W. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 4.Gillies S D, Morrison S L, Oi V T, Tonegawa S. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 5.Birshtein B K, Chen C, Saleque S, Michaelson J S, Singh M, Little R D. Curr Top Microbiol Immunol. 1997;224:73–80. doi: 10.1007/978-3-642-60801-8_7. [DOI] [PubMed] [Google Scholar]
- 6.Garcia J V, Bich-Thuy L T, Stafford J, Queen C. Nature (London) 1986;322:383–385. doi: 10.1038/322383a0. [DOI] [PubMed] [Google Scholar]
- 7.Grosschedl R, Weaver D, Baltimore D, Costantini F. Cell. 1984;38:647–658. doi: 10.1016/0092-8674(84)90259-9. [DOI] [PubMed] [Google Scholar]
- 8.Grosschedl R, Baltimore D. Cell. 1985;41:885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- 9.Calame K, Ghosh S. In: Factors Regulating Ig Transcription. Honjo T, Alt F W, Rabbits T H, editors. London: Academic; 1995. [Google Scholar]
- 10.Nelsen B, Sen R. Int Rev Cytol. 1992;133:121–149. doi: 10.1016/s0074-7696(08)61859-8. [DOI] [PubMed] [Google Scholar]
- 11.Staudt L M, Lenardo M J. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 12.Alt F W, Rosenberg N, Casanova R J, Thomas E, Baltimore D. Nature (London) 1982;296:325–331. doi: 10.1038/296325a0. [DOI] [PubMed] [Google Scholar]
- 13.Burrows P D, Beck-Engeser G B, Wabl M R. Nature (London) 1983;306:243–246. doi: 10.1038/306243a0. [DOI] [PubMed] [Google Scholar]
- 14.Aguilera R J, Hope T J, Sakano H. EMBO J. 1985;4:3689–3693. doi: 10.1002/j.1460-2075.1985.tb04136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckhardt L A, Birshtein B K. Mol Cell Biol. 1985;5:856–868. doi: 10.1128/mcb.5.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein S, Sablitzky F, Radbruch A. EMBO J. 1984;3:2473–2476. doi: 10.1002/j.1460-2075.1984.tb02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaller D M, Eckhardt L A. Proc Natl Acad Sci USA. 1985;82:5088–5092. doi: 10.1073/pnas.82.15.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst P, Smale S T. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 19.Cockerill P N, Yuen M H, Garrard W T. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- 20.Bode J, Stengert-Iber M, Kay V, Schlake T, Dietz-Pfeilstetter A. Crit Rev Eukaryotic Gene Expression. 1996;6:115–138. doi: 10.1615/critreveukargeneexpr.v6.i2-3.20. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K, Kas E, Poljak L, Adachi Y. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 22.Berezney R, Mortillaro M J, Ma H, Wei X, Samarabandu J. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 23.Boulikas T. Int Rev Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- 24.Nickerson J A, Blencowe B J, Penman S. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 25.Stein G S, van Wijnen A J, Stein J, Lian J B, Montecino M. Int Rev Cytol. 1995;162A:251–278. doi: 10.1016/s0074-7696(08)61233-4. [DOI] [PubMed] [Google Scholar]
- 26.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 27.Cockerill P N, Garrard W T. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 28.Mirkovitch J, Mirault M E, Laemmli U K. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 29.Lauber A H, Barrett T J, Subramaniam M, Schuchard M, Spelsberg T C. J Biol Chem. 1997;272:24657–24665. doi: 10.1074/jbc.272.39.24657. [DOI] [PubMed] [Google Scholar]
- 30.Webb C F, Das C, Eneff K L, Tucker P W. Mol Cell Biol. 1991;11:5206–5211. doi: 10.1128/mcb.11.10.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ariizumi K, Wang Z, Tucker P W. Proc Natl Acad Sci USA. 1993;90:3695–3699. doi: 10.1073/pnas.90.8.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkwel P A, Hamlin J L. Mol Cell Biol. 1988;8:5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K, Kas E, Gonzalez E, Laemmli U K. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadesch T, Zervos P, Ruezinsky D. Nucleic Acids Res. 1986;14:8209–8221. doi: 10.1093/nar/14.20.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasylyk C, Wasylyk B. EMBO J. 1986;5:553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheuermann R H, Chen U. Genes Dev. 1989;3:1255–1266. doi: 10.1101/gad.3.8.1255. [DOI] [PubMed] [Google Scholar]
- 37.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 38.Kohwi-Shigematsu T, Maass K, Bode J. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S R, Dudley J P. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forrester W C, van Genderen C, Jenuwein T, Grosschedl R. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T, Forrester W C, Qiu R G, Grosschedl R. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 42.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Nature (London) 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 43.Okada A, Mendelsohn M, Alt F. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oltz E M, Alt F W, Lin W C, Chen J, Taccioli G, Desiderio S, Rathbun G. Mol Cell Biol. 1993;13:6223–6230. doi: 10.1128/mcb.13.10.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrier P, Krippl B, Blackwell T K, Furley A J, Suh H, Winoto A, Cook W D, Hood L, Costantini F, Alt F W. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bories J C, Demengeot J, Davidson L, Alt F W. Proc Natl Acad Sci USA. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Young F, Bottaro A, Stewart V, Smith R K, Alt F W. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serwe M, Sablitzky F. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu H, Zou Y R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga T, Tsunoda Y. Dev Growth Differ. 1992;34:561–566. doi: 10.1111/j.1440-169X.1992.00561.x. [DOI] [PubMed] [Google Scholar]
- 51.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A, Seidman J G. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Lansford R, Stewart V, Young F, Alt F W. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 54.Yancopoulos G D, Alt F W. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 55.Yancopoulos G D, Desiderio S V, Paskind M, Kearney J F, Baltimore D, Alt F W. Nature (London) 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 56.Bottaro A, Young F, Chen J, Serwe M, Sablitzky F, Alt F W. Int Immunol. 1998;10:799–806. doi: 10.1093/intimm/10.6.799. [DOI] [PubMed] [Google Scholar]
- 57.Manis J P, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt F W. J Exp Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng H L, Alt F W. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 59.Lieberson R, Ong J, Shi X, Eckhardt L A. EMBO J. 1995;14:6229–6238. doi: 10.1002/j.1460-2075.1995.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraser P, Pruzina S, Antoniou M, Grosveld F. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 61.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 62.Hiramatsu R, Akagi K, Matsuoka M, Sakumi K, Nakamura H, Kingsbury L, David C, Hardy R R, Yamamura K, Sakano H. Cell. 1995;83:1113–1123. doi: 10.1016/0092-8674(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 63.Gorman J R, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt F W. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 64.van der Stoep N, Gorman J R, Alt F W. Immunity. 1998;8:743–750. doi: 10.1016/s1074-7613(00)80579-8. [DOI] [PubMed] [Google Scholar]
- 65.Chattopadhyay S, Whitehurst C E, Chen J. J Biol Chem. 1998;273:29838–29846. doi: 10.1074/jbc.273.45.29838. [DOI] [PubMed] [Google Scholar]
- 66.Chattopadhyay S, Whitehurst C E, Schwenk F, Chen J. J Immunol. 1998;160:1256–1267. [PubMed] [Google Scholar]
- 67.Cockerill P N. Nucleic Acids Res. 1990;18:2643–2648. doi: 10.1093/nar/18.9.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garrick D, Fiering S, Martin D I, Whitelaw E. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 69.Seidl, K. J., Manis, J. P., Bottaro, A., Zhang, J., Davidson, L., Kisselgof, A., Oettinger, H. & Alt, F. W. (1999) Proc. Natl. Acad. Sci. USA 96, in press. [DOI] [PMC free article] [PubMed]