Most patients infected with HIV-1 develop AIDS unless they receive antiretroviral medication. However, a small number of HIV-infected individuals with high viral titers remain disease free and do not experience progressive immunosuppression, even in the absence of therapy. Such individuals are labeled long-term nonprogressors (LTNPs) and are characterized by a series of laboratory parameters that are usually compromised in HIV-1 carriers who ultimately develop AIDS. In particular, LTNPs possess a high frequency of peripheral CD4plus; T cells, as well as a low level of spontaneous apoptosis, correlating with a normal mitochondrial transmembrane potential (Δψm) among circulating T cells. It has long been assumed that, as an experimentum naturae, LTNPs might furnish valuable clues for the identification of molecular determinants of HIV-1 pathogenesis. A study by Badley and colleagues involving LTNPs, reported in this issue of the JCI (1), strongly suggests that viral protein R (Vpr) is a major HIV-1 virulence factor.
Vpr – a cytocidal protein encoded by HIV-1
The HIV-1 genome encodes structural and enzymatic proteins common to all retroviruses, but also some accessory proteins that are not always required for the replication of HIV-1. One of these accessory proteins, Vpr, is found in virions, HIV-1–infected cells, and in the serum and cerebrospinal fluid of HIV-1 carriers. Vpr is a small (96 amino acids) soluble protein composed of three α-helical domains (Figure 1a). Vpr is dispensable for viral replication in T lymphocytes, but not in monocytes, and thus is rapidly lost among laboratory HIV-1 isolates. In contrast, Vpr is maintained in most HIV-1–infected patients, indicating that this protein may be important for the in vivo biology of HIV-1. In vitro, Vpr reduces the proliferation of CD4plus; lymphocytes and of various other cell types via a G2 cell cycle arrest. Moreover, Vpr induces cell death through the intrinsic pathway of apoptosis, which involves mitochondrial membrane permeabilization (MMP), the release of cytochrome c from mitochondria, and the cytochrome c–dependent activation of caspases (2, 3).
Figure 1.
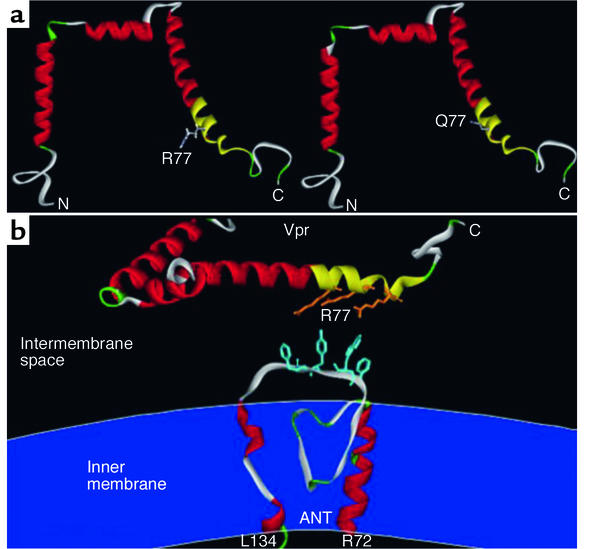
Impact of the R77Q mutation of Vpr on the conformation of Vpr and the interaction between Vpr and ANT. (a) Representation of wild-type (R77) Vpr and R77Q Vpr. Three amphipathic helices (shown in red and yellow) are linked by turns (green) and flexible loops (gray). The mitochondriotoxic domain of Vpr has been marked in yellow. Only the side chain of amino acid 77 is shown. (b) View of the arginine residues (R73, R77, R80) within the mitochondriotoxic domain of Vpr (yellow) and the aromatic residues of ANT (F109, W111, Y113 and/or F114) involved in cation-π interactions, as calculated by molecular modeling (10). The inner mitochondrial membrane (dark blue) is indicated to facilitate the topological orientation of the third loop (residues 104–116) of ANT. C, carboxy terminus; N, amino terminus; L134 and R72 are amino residues of ANT.
A fraction of Vpr transfected into cells can be found in the mitochondrial compartment (2). When added to purified mitochondria, recombinant or synthetic Vpr crosses the outer membrane through the voltage-dependent anion channel. It then interacts with the adenine nucleotide translocator (ANT) in the inner mitochondrial membrane to form a composite ion channel. This channel dissipates the Δψm and thus favors MMP and subsequent apoptosis (3). The physical interaction between Vpr and ANT has been determined by coimmunoprecipitation, electrophysiological measurements, and surface plasmon resonance (3). Vpr fails to kill ANT-deficient cells (3), which suggests that the Vpr/ANT interaction is central to the apoptosis-inducing properties of Vpr.
Vpr is mutated in LTNPs
In this issue of the JCI, Badley and colleagues (1) report that 80% of LTNPs manifest a point mutation in Vpr leading to the exchange of arginine 77 with a glutamine residue. This mutation (R77Q) is far less frequent (33%) among patients with progressive disease. Badley and coauthors’ analysis confirms previous studies (4) and supports the idea that Vpr may serve as a mediator of CD4plus; T cell depletion in HIV infection. In T cell infections utilizing vesicular stomatitis virus G protein–pseudotyped HIV-1 particles, the R77Q mutation had no impact on cell cycle arrest or viral replication, yet it led to a significant reduction in apoptosis (1). Compared to the wild-type Vpr peptide, a synthetic Vpr peptide carrying the R77Q mutation induced much less apoptosis, Δψm dissipation, cytochrome c release, and caspase-3 activation (1). This result has been demonstrated in vitro, when Vpr-derived peptides were added to human T lymphoma cells, and in vivo, when such peptides were injected into mice in which the depletion of CD4plus; and CD8plus; T cells was being monitored (1). Based on these findings, Badley and coauthors (1) propose that the R77Q mutation (and the subsequent reduction in Vpr-mediated apoptosis) might explain the LTNP phenotype.
How the Vpr mutation attenuates Vpr function
How is it possible that the R77Q mutation reduces Vpr-mediated apoptosis induction and HIV-1 virulence, even though computer-based calculations do not predict any major effect of this single amino acid substitution on the overall folding of the α-helical structure (5) of Vpr (Figure 1a)? The answer to this enigma may reside in the mechanism through which Vpr interacts with ANT. Indeed, the minimum unit of Vpr capable of inducing MMP in vitro in isolated mitochondria is a dodecapeptide (aa 72-83) located within the C-terminal α-helix (3). This mitochondriotoxic domain contains three arginine (R) residues (R73, R77, and R80), which asymmetrically distribute to just one side of the α-helix and participate in the physical interaction with one particular moiety of the six–transmembrane domain ANT, namely the first loop (amino acids 104–116) exposed to the mitochondrial intermembrane space. These positively charged R residues (e.g., R77, but also R73 or R80) within Vpr are likely to be involved in cation-π interactions with F109, W111, Y113 and/or F114 within ANT (Figure 1b). This type of noncovalent binding, which is as least as strong as a hydrogen bond, is increasingly considered for drug design and has been shown to be instrumental for antigen recognition by immune receptors, detection of DNA damage by repair enzymes, and ligand-elicited receptor signaling (6). Mutations of R77, as described by Badley and colleagues (1), may be expected to strongly reduce the affinity of Vpr for ANT. This would explain the reduced mitochondriotoxic and proapoptotic potential of Vpr R77Q.
Based on these findings, several tantalizing possibilities arise. It appears possible that R77Q could act as a dominant-negative Vpr mutant, based on the previous finding that R73-mutated Vpr reverses the wild-type Vpr-mediated block in host cell proliferation, presumably through the N-terminus–mediated oligomerization with wild-type Vpr (7). Such dominant-negative Vpr inhibitors, as well as synthetic blockers of the Vpr/ANT interaction, may be expected to have a positive impact on the prognosis of HIV-1 infection, provided that Vpr and the Vpr/ANT interaction are truly important for AIDS pathogenesis. Indeed, HIV protease inhibitors have already been shown to suppress the MMP-inducing effects of two ANT ligands, Vpr and atractyloside, as assessed using isolated mitochondria (8). This could explain why HIV-1 protease inhibitors can inhibit HIV-1–induced apoptosis, independently of their anti-retroviral effect.
It is also important to note that the proapoptotic Vpr protein is not the only viral mitochondrion-targeted protein to be identified as a virulence factor (9). It will be interesting to learn which other viruses produce similar mitochondrion-targeted, apoptosis-regulatory, disease-relevant proteins, all of which could constitute promising pharmacological targets.
Acknowledgments
Our work is supported by grants from the Centre National de la Recherche Scientifique, the Agence Nationale pour la Recherche sur le SIDA, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the European Commission (QLG1-CT-1999-00739), the Ligue Nationale contre le Cancer, and the Ministry of Science. The authors thank Aurélien Deniaud for assistance in molecular modeling.
Footnotes
See the related article beginning on page 1547.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: long-term nonprogressive/nonprogressor (LTNP); mitochondrial transmembrane potential (Δψm); viral protein R (Vpr); mitochondrial membrane permeabilization (MMP); adenine nucleotide translocator (ANT).
References
- 1.Lum JJ, et al. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J. Clin. Invest. 2003;111:1547–1554. doi:10.1172/JCI200316233. doi: 10.1172/JCI16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacotot E, et al. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 2000;191:33–46. doi: 10.1084/jem.191.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacotot E, et al. Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein R and Bcl-2. J. Exp. Med. 2001;193:509–520. doi: 10.1084/jem.193.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Huang Y, Yuan H, Tuttleton S, Ho D. Genetic characterization of vif, vpr, and vpu sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology. 1997;228:340–349. doi: 10.1006/viro.1996.8378. [DOI] [PubMed] [Google Scholar]
- 5.Morellet N, Bouaziz S, Petitjean P, Roques B. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 2003;327:215–227. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- 6.Zacharias N, Dougherty D. Cation-π interactions in ligand recognition and catalysis. Trends Pharmacol. Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 7.Sawaya B, et al. Transdominant activity of human immunodeficiency virus type 1 Vpr with a mutation at residue R73. J. Virol. 2000;74:4877–4881. doi: 10.1128/jvi.74.10.4877-4881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phenix B, Lum J, Nie Z, Sanchez-Dardon J, Badley A. Antiapoptotic mechanism of HIV protease inhibitors: preventing mitochondrial transmembrane potential loss. Blood. 2001;98:1078–1085. doi: 10.1182/blood.v98.4.1078. [DOI] [PubMed] [Google Scholar]
- 9.Castedo M, Perfettini JL, Kroemer G. Mitochondrial apoptosis and the peripheral benzodiazepine receptor: a novel target for viral and pharmacological manipulation. J. Exp. Med. 2002;196:1121–1125. doi: 10.1084/jem.20021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]