Abstract
The periodontal pathogen Fusobacterium nucleatum induces apoptosis in lymphocytes. We previously identified the autotransporter protein Fap2 in F. nucleatum strain PK1594 that induced apoptosis in lymphocytes when expressed in Escherichia coli. In this study, we identified protein homologs of Fap2 in the transformable F. nucleatum strain ATCC 23726, to determine their role in the induction of apoptosis in lymphocytes. We used a new gene-inactivation vector conferring thiamphenicol resistance (pHS31) to construct a mutant deficient in one of the homologs, aim1. Transcriptional analyses demonstrated disruption of aim1 expression, and phenotypic analyses revealed a 41% decrease in the ability of the mutant to induce apoptosis in Jurkat cells, as compared with the parental strain. These studies demonstrate, in the native host cell background, the contribution of aim1 to F. nucleatum induction of apoptosis and, to the best of our knowledge, represent the first report of a genetically defined and phenotypically characterized mutation in F. nucleatum.
Keywords: Fusobacterium nucleatum, apoptosis, autotransporter, aim1, mutant
INTRODUCTION
Fusobacterium nucleatum is a Gram-negative, anaerobic fusiform bacterium implicated as one of the causative agents of periodontal disease (Bolstad et al., 1996; Moore et al., 1982a,b, 1984, 1985), a chronic polymicrobial infection affecting the supporting tissues of teeth. Bacteria present in dental plaque advance the disease process by irritating tissues, leading to bone and attachment loss. The pathogenesis of F. nucleatum is not limited to the oral cavity; it is associated with a variety of systemic conditions, giving its pathogenesis broader relevance to the medical community (Sabiston and Gold, 1974; Sundqvist et al., 1979; Ribot et al., 1981; Edson et al., 1982; Truant et al., 1983; Gonzalez-Gay et al., 1993; Jousimies-Somer et al., 1993; Talan et al., 1999; Heckmann et al., 2003). F. nucleatum pathogenesis is still poorly understood, but the development of a genetic system (Haake et al., 2000) and sequencing of 3 strains (Kapatral et al., 2002, 2003) now enables the underlying mechanisms to be investigated at a molecular level.
Our previous research shows that F. nucleatum induces apoptosis in lymphocytes, an ability mediated by heat-labile outer membrane protein(s) that may allow the micro-organism to evade the immune system (Jewett et al., 2000). To identify the protein(s) involved, we expressed an F. nucleatum PK1594 gene library in E. coli and subjected the clones to cell death assays with lymphocytes. Five genes that mediated increased apoptosis in lymphocytes were found (unpublished observations). One of these genes, Fap2, showed homology to an autotransporter protein family. This was particularly interesting, since similar proteins have been identified as virulence factors in other bacteria (Henderson and Nataro, 2001).
In this study, we sought to establish the role of Fap2 in inducing apoptosis in lymphocytes. A genetic system to transform F. nucleatum was available, but strain PK1594 remained refractory to genetic manipulation. An integration vector (pHS31) possessing a catP gene capable of conferring thiamphenicol resistance in F. nucleatum was constructed. We used pHS31 to create a mutation in a Fap2 homolog in the transformable F. nucleatum strain ATCC 23726, and designated it aim1 (apoptosis inducing membrane protein gene 1), since transcriptional and phenotypic analyses demonstrated a role for this gene in inducing apoptosis in Jurkat cells. These data constitute, to the best of our knowledge, the first report of a genetically defined and phenotypically characterized mutation in F. nucleatum, and a genetic basis for its ability to induce apoptosis in lymphocytes.
MATERIALS & METHODS
Bacterial Strains, Cell Lines, and Culture Conditions
F. nucleatum strains were cultivated on Columbia agar or in Columbia broth (Difco, Detroit, MI, USA). Thiamphenicol (MP Biomedicals, Irvine, CA, USA) at 5 μg/mL was used for selection and maintenance of strains possessing the catP determinant. Jurkat cells were maintained in RPMI 1640 supplemented with 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 10 U/mL penicillin, 10 mg/mL streptomycin (Life Technologies, Grand Island, NY, USA), and 10% fetal calf serum (Irvine Scientific, Santa Ana, CA, USA).
Protein Analyses and Construction of a Fusobacterium nucleatum Mutant
Protein sequence homology was determined by use of the BLAST 2 program on the NCBI Web site (Tatusova and Madden, 1999). Aim1 mutants were created by single homologous recombination with the integration plasmid pHS31. Plasmid pHS31 was constructed from pJIR750, a clostridial shuttle plasmid possessing the catP gene conferring thiamphenicol resistance in F. nucleatum. Plasmid pJIR750 was digested with XbaI and SpeI, followed by isolation of the DNA fragment possessing the ColE1 origin and catP gene, and re-ligated to generate pHS31. The aim1 gene fragment, designated aim1′, was amplified with Taq DNA polymerase with use of the primers aim1F′ and aim1R′ (Table 1), and cloned into the vector pCR2.1 (Invitrogen, Carlsbad, CA, USA). The fragment was excised from the pCR2.1 vector by EcoRI, blunted, gel-purified, and ligated into a blunt-ended, dephosphorylated pHS31 to generate pIP-aim1. E. coli was transformed with the ligation reaction, and the resulting integration plasmid was confirmed by restriction analysis and PCR. F. nucleatum transformation was performed as previously described (Haake et al., 2000).
Table 1.
Primers Used in This Study
| Primer | Sequence |
|---|---|
| catP-F | 5′-TTAGGACGGCAATCAATCAA-3′ |
| catP-R | 5′-AAACGGCAAATGTGAAATCC-3′ |
| ColE1 | 5′-GCAGAGCGAGGTATGTAGGC-3′ |
| aim1′F | 5′-CTGTTGGGAAAGAAGGAGTTG-3′ |
| aim1′R | 5′-TTGAATAAAGGGCTGCTGTG-3′ |
| aim1-5′F | 5′-GTTTGGAGCAGGAGGTTCAA-3′ |
| aim1-3′R | 5′-CCTTGGCATCATTTTCAATAGTT-3′ |
| 2057rt-F | 5′-AAAAGAAGCCGATAAAAATGG-3′ |
| 2057rt-R | 5′-ATCACTGGGTATTGTCCTTGTTC-3′ |
| DSrt-F | 5′-CCATCTGCGTCCGTTATTGT-3′ |
| DSrt-R | 5′-GGAACTGCTGGTGGAGATAAG-3′ |
| DS2rt-F | 5′-TTGTCCTGGATCAGTAACAGAAAG-3′ |
| DS2rt-R | 5′-CCTAAAGGTGGAAGAACGAAGA-3′ |
| USrt-F | 5′-CCTGCTCCAAACATTCCAAC-3′ |
| USrt-R | 5′-AGTGAAACAACAACAGCAATGG-3′ |
| Control rt-F | 5′-GGTTAAGTCCCGCAACGA-3′ |
| Control rt-R | 5′-CATCCCCACCTTCCTCCTAC-3′ |
Transcriptional Analyses
Genomic DNA was extracted from stationary-phase cells following standard protocols.
Total RNA was extracted from mid-log-phase cells according to a hot-phenol protocol (Merritt et al., 2005). A 3-μg quantity of total RNA was used for cDNA synthesis by Stratascript RT (Stratagene, La Jolla, CA, USA), according to the manufacturer's protocol. For real-time RT-PCR, SYBR green (Bio-Rad, Hercules, CA, USA) was used for fluorescence detection with the iCycler real-time PCR system (Bio-Rad), according to the manufacturer's protocol. Statistical analysis was performed by analysis of variance (ANOVA) with SPLUS 6.0 (Insightful, Seattle, WA, USA).
DNA Staining and Apoptosis Assay
We assessed apoptosis, based on DNA staining, by labeling the cells with propidium iodide as described previously (Jewett et al., 2000). Briefly, samples of 2 × 105 cells were washed twice with PBS and incubated in 70% ethanol on ice. After 30 min of incubation, the cells were washed twice with PBS and 70 μL of RNase (1 mg/mL; Sigma), and a 140-μL quantity of propidium iodide (100 mg/mL; Sigma) was added to each sample. After incubation for 1 hr, analysis was performed with the use of a flow cytometer (Beckman Coulter, Fullerton, CA, USA). Each assay was run in 3 independent experiments to confirm reproducibility. Statistical analysis was performed as stated above.
RESULTS
Identification of Fap2 Homologs in F. nucleatum ATCC 23726
We previously identified the apoptosis-inducing autotransporter protein, Fap2, in F. nucleatum PK1594, based on its ability to induce apoptosis in lymphocytes when expressed in E. coli (unpublished observations). Because PK1594 is not amenable to genetic manipulation, Fap2 homologs were identified in the transformable F. nucleatum strain ATCC 23726. Since the F. nucleatum ATCC 23726 genome has not been sequenced, thus precluding direct analysis, homologs were first identified in the sequenced strain ATCC 25586, which is related to ATCC 23726 at the subspecies level. We used a local BLAST search and identified 8 predicted outer-membrane proteins with homology to Fap2 (Table 2). We then confirmed that the Fap2 homologs encoded by the ATCC 25586 genome were equally present in ATCC 23726 by partially sequencing each of the genes and comparing them with the ATCC 25586 sequences by a pairwise BLAST search (Table 2). The homologs in ATCC 23726 showed a high degree of similarity (97–100%) to ATCC 25586 homologs. The homologs identified in ATCC 25586 were considerably shorter than Fap2, with greatest similarity at the C-terminus, where the pore-forming transmembrane β-barrel is located in autotransporters. The C-terminus of each homolog showed similarity to the conserved autotransporter β-domain pfam03797.
Table 2.
Predicted Outer Membrane Protein Homology to Fap2 and F. nucleatum ATCC 23726 Homologs
| PK1594 Fap2 (3692 aa) | ATCC 23726 Sequenced Fragment | |||||
|---|---|---|---|---|---|---|
| ATCC 25586 Fap2 Homologs | Genome Annotation | Predicted Protein Length (aa) | Identities | Positives | Identities | Positives |
| Fn0254 | outer membrane protein | 1677 | 675/1852 (36%) | 930/1852 (49%) | 322/322 (100%) | 322/322 (100%) |
| Fn0387 | probable outer membrane protein | 1724 | 615/1879 (32%) | 900/1879 (47%) | 251/251 (100%) | 251/251 (100%) |
| Fn1449 | outer membrane protein | 3165 | 2025/3347 (60%) | 2407/3347 (71%) | 442/453 (97%) | 446/453 (97%) |
| Fn1526 | predicted outer membrane protein | 2143 | 597/2411 (24%) | 981/2411 (39%) | 317/322 (98%) | 319/322 (98%) |
| Fn1554 | predicted outer membrane protein | 1582 | 652/1815 (35%) | 909/1815 (49%) | 292/292 (100%) | 292/292 (100%) |
| Fn1893 | predicted outer membrane protein | 1361 | 546/1418 (38%) | 760/1418 (53%) | 287/287 (100%) | 287/287 (100%) |
| Fn2047 | predicted outer membrane protein | 1630 | 638/1736 (36%) | 890/1736 (50%) | 264/270 (97%) | 266/270 (97%) |
| Fn2058 (aim 1) | predicted outer membrane protein | 1794 | 677/1975 (34%) | 970/1975 (48%) | 1765/1791 (98%) | 1772/1791 (98%) |
Construction of the aim1 Mutant, aimless1
Using the gene sequence from F. nucleatum ATCC 25586 (Fn2058), we created primers to amplify a fragment of aim1. The amplified fragment was ligated into pHS31, creating integration plasmid pIP-aim1 (Fig. 1A). Transformation experiments yielded 16 thiamphenicol-resistant mutants (designated 'aimless1'), whose growth was indistinguishable from that of the wild-type parent strain in normal culture conditions.
Figure 1.
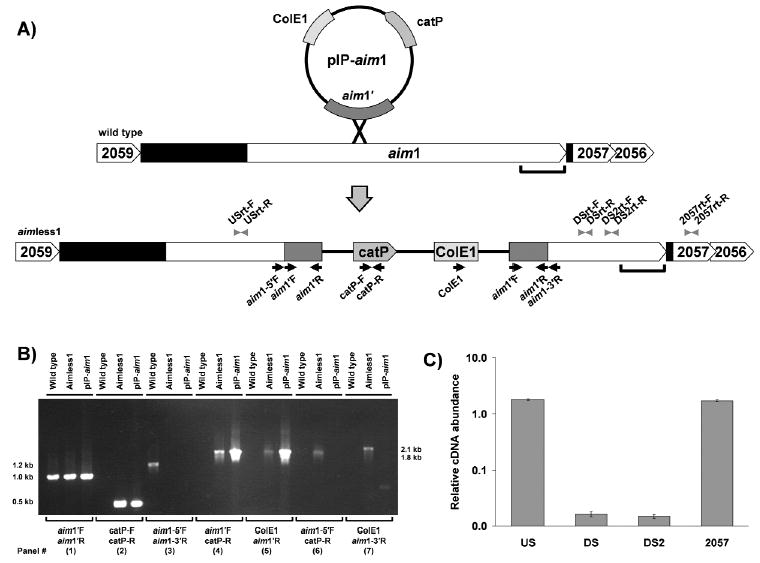
Structural and transcriptional analyses of wild-type and aimless1 mutant strains. (A) Schematic illustration of mutagenesis: organization of aim1 and flanking genes on the F. nucleatum ATCC 23726 wild-type and aimless1 mutant chromosome. Arrows indicate PCR primers used for construction and analysis of the aim1 mutant; arrowheads indicate real-time PCR primers; black shaded area indicates intergenic region; bracket indicates location of autotransporter domain. Full-length aim1 (5625 bases) was submitted to GenBank and has been assigned accession number DQ020251. The Fig. is not to scale. (B) PCR analysis confirming insertion into aim1. PCR analysis of chromosomal DNA from ATCC 23726 and the aimless1 mutant. Plasmid pIP-aim1, the construct used to create the inactivation in aim1, was used as the control DNA template. (C) Transcriptional analysis of aim1 and the downstream gene Fn2057. The relative RNA abundance of F. nucleatum wild-type and aimless1 was determined by real-time PCR. Results are expressed as relative means (mutant/wt) ± SEMs (error bars) of triplicate experiments.
PCR analyses confirmed that pIP-aim1, as well as wild-type and the aimless1 genomic DNA, harbored the aim1 gene fragment (aim1′; Fig. 1B, panel 1). The catP determinant was evident by PCR analyses in pIP-aim1 and in aimless1 genomic DNA, but, as predicted, was not present in the wild-type genome (Fig. 1B, panel 2). PCR analysis also confirmed the structure and site of vector integration, and ruled out the presence of a replicating plasmid carrying the catP gene. Primers flanking aim1′ (aim1-5′F and aim1-3′R) amplified a 1.2-kb product from the wild-type chromosome, but, as expected, yielded no product from pIP-aim1 or aimless1 (Fig. 1B, panel 3). Flanking primers were used in combination with pHS31-specific primers (catP-R and ColE1) to confirm the site of chromosomal integration. Expected 1.8-kb and 2.1-kb fragments were amplified for each primer pair (Fig. 1B, panels 6 and 7) from aimless1, but not from wild-type or pIP-aim1 DNA. This demonstrated specific integration of pIP-aim1 into the chromosome at the aim1 site, as predicted for a homologous recombination event.
Two genes downstream of aim1 could potentially be disrupted by plasmid integration (Fig. 1A). Real-time PCR transcriptional analyses of aim1 and downstream gene Fn2057 were conducted to confirm disruption of aim1 expression in aimless1, and to rule out the possibility of a polar effect on downstream genes. Two real-time PCR primer sets for the 3′ portion of aim1 showed that transcription levels of this region were at background levels in aimless1, 61-fold lower than in wild-type, indicating that the insertion disrupted transcription. Real-time PCR with primers for the 5′ portion of aim1 showed transcription levels similar to those of the wild type, indicating that this region was transcribed at normal levels. Importantly, primers for the downstream gene Fn2057 showed similar transcription levels in aimless1 and wild-type strains, indicating that expression of Fn2057 was not affected by vector integration (Fig. 1C).
Reduced Apoptosis-inducing Ability of the aim1 Mutant
We were interested in determining if aim1 contributed to the apoptosis phenotype. Incubation of Jurkat cells with F. nucleatum ATCC 23726 resulted in increased levels of apoptosis compared with that of Jurkat cells incubated alone. Our apoptosis assays showed that the number of apoptotic Jurkat cells present when incubated with wild-type F. nucleatum (14.2%) was significantly higher (p < 0.0001) than the number of apoptotic cells present when the cells were incubated with aimless1 mutant (11.2%), and both groups were significantly different from Jurkat cells incubated alone (6.9%). Thus, aimless1 demonstrated a 41.2% decrease in the number of apoptotic cells, indicating that aim1 plays a significant role in the induction of apoptosis (Fig. 2).
Figure 2.
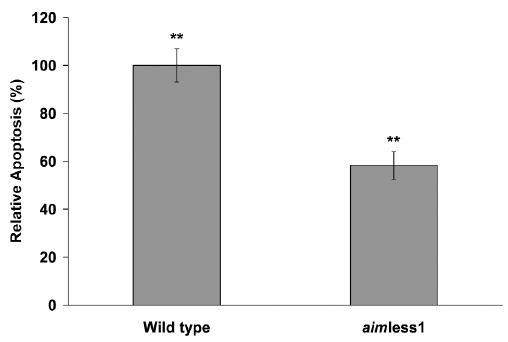
Induction of apoptosis in Jurkat cells after incubation with F. nucleatum wild-type or with aimless1. Jurkat cells were incubated with wild-type or aimless1 for 18 hrs before assay (n = 3). Uninfected Jurkat cells were incubated alone as a control. Values shown are relative to wild-type apoptosis levels after background subtraction. Error bars represent SD. **p < 0.0001.
DISCUSSION
Evading host defense mechanisms is a critical element in bacterial pathogenesis. Previous studies have indicated that one aspect of F. nucleatum virulence is its induction of apoptosis in host lymphocytes (Jewett et al., 2000). In this study, we identified homologs to the apoptosis-inducing protein Fap2 in the transformable F. nucleatum strain ATCC 23726, created an isogenic mutant for the aim1 gene, and demonstrated that the mutation impaired the ability of F. nucleatum to induce apoptosis. These findings document the first characterized mutation in F. nucleatum, and confirm the first apoptosis-inducing gene in this species. Together, these data provide a critical initial step in defining a genetic basis for F. nucleatum pathogenesis.
The creation of site-directed mutants represents a major advance in F. nucleatum research. Currently, genomic DNA sequences are available for 3 strains of F. nucleatum. The ATCC 23726 strain has not undergone genomic sequence analysis, but is related to the sequenced subspecies nucleatum strain ATCC 25586 at the subspecies level. Using the ATCC 25586 genomic sequence, we found that all 8 of the Fap2 homologs identified in the ATCC 25586 genome were present in ATCC 23726, and showed a high degree of similarity to the ATCC 25586 homologs over the sequenced fragment. Analysis of Fap2 showed that it contained 3 domains. The C-terminal autotransporter domain is predicted to be responsible for pore formation and transport of the protein to the cell surface. The central domain, occupying a majority of the protein, contains tandem and periodic repeats similar to those found in filamentous hemagglutinins and adhesins (Clantin et al., 2004). The central and C-terminal domains of Fap2 are similar to those of Fap2 homologs identified in the ATCC 25586 genome, whereas the N-terminal domain remains distinct. The contribution of the N-terminal domain to inducing apoptosis is unknown, but truncation of this domain in the gene fragment recovered from the E. coli library indicates that it may have no role. The central domain sequences of the Fap2 homologs vary, but all contain tandem and periodic repeats, extending from their N-terminus to the C-terminal autotransporter domain. There is a high degree of similarity between the autotransporter domain of Fap2 and the homologs. Our findings clearly indicate that transcription of the 3′ region of aim1, encoding the autotransporter domain and the 3′ portion of the central domain, is blocked in aimless1. Thus, the truncated aim1 protein is probably not secreted. Additional investigation at a protein level should confirm these predictions.
Phenotypic analysis of aimless1 in comparison with the parental wild-type strain revealed a role for the aim1 protein in the induction of apoptosis of Jurkat cells. These findings validate the earlier results indicating that Fap2 confers the apoptosis-inducing phenotype in E. coli. Demonstration that the downstream gene Fn2057 is transcribed at wild-type levels in aimless1 rules out possible polar effects caused by the insertion (Fig. 1C). The ability to induce apoptosis in eukaryotic cells is not unique to F. nucleatum. Bacterial proteins that induce apoptosis have been identified in several pathogens, including the vacuolating toxin of Helicobacter pylori (Henderson and Nataro, 2001; Cover et al., 2003), yet the molecular mechanisms by which these proteins cause disease have not been well-established. The aim1 mutation resulted in a partial, but highly reproducible, inhibition of Jurkat cell apoptosis. This is important, since it is likely that other F. nucleatum factors contribute to apoptosis induction, consistent with findings in other bacterial pathogens (Henderson and Nataro, 2001). The 7 additional Fap2 homologs are putative apoptosis-inducing OMPs, and studies are under way examining their potential role in pathogenesis. Two possible mechanisms of apoptosis induction by F. nucleatum OMPs have been hypothesized: (1) F. nucleatum OMPs directly deliver death signals through contact with lymphocyte cell-surface death receptors; and (2) F. nucleatum-mediated aggregation of lymphocytes allows lymphocyte death receptors to be crosslinked with their associated ligands on adjacent cells (Jewett et al., 2000). The results presented in this study, demonstrating the role of aim1 in apoptosis, do not allow for a distinction between the two hypotheses. It is likely that multiple mechanisms lead to the induction of F. nucleatum-mediated apoptosis, indicating that further experiments are required to clarify the distinct mechanisms involved and how their synergistic effect contributes to apoptosis induction.
The generation of aimless1 was possible with the use of a new F. nucleatum integration vector. The site-specific chromosomal insertional mutagenesis demonstrated with pIP-aim1 is consistent with the accepted mechanism of homologous recombination. Additional vectors for molecular analysis in F. nucleatum include shuttle plasmids and an additional selectable marker conferring clindamycin resistance (Haake et al., 2000), facilitating the mutagenesis of multiple genes, or mutagenesis and complementation of a single gene. Together, these tools allow for the molecular analysis of the F. nucleatum genome, and will aid in confirming the predicted function of genes with known homologs, as well as in determining the function of novel genes.
The induction of apoptosis in host lymphocytes provides a strategy that bacterial pathogens use to facilitate their survival through subverting the host innate immune response. The identification of aim1 as an F. nucleatum gene involved in suppressing the immune system represents an important first step in defining the molecular basis of F. nucleatum pathogenesis. The presence of additional Fap2 homologs in the F. nucleatum genome indicates that further investigation of their function is warranted, and creation of additional gene inactivation mutants is under way.
Acknowledgments
The authors acknowledge Julian I. Rood for providing the pJIR750 plasmid. This work was supported by Delta Dental grant WDS78956 (to W.S.) and by NIH/NIDCR PHS Grants No. DE12639 and DE015348 (to S.K.H.).
References
- Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, Villeret V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci USA. 2004;101:6194–6199. doi: 10.1073/pnas.0400291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Krishna US, Israel DA, Peek RM., Jr Induction of gastric epithelial cell apoptosis by Helicobacter pylori vacuolating cytotoxin. Cancer Res. 2003;63:951–957. [PubMed] [Google Scholar]
- Edson RS, Rosenblatt JE, Washington JA, 2nd, Stewart JB. Gas-liquid chromatography of positive blood cultures for rapid presumptive diagnosis of anaerobic bacteremia. J Clin Microbiol. 1982;15:1059–1061. doi: 10.1128/jcm.15.6.1059-1061.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gay MA, Sanchez-Andrade A, Cereijo MJ, Pulpeiro JR, Armesto V. Pyomyositis and septic arthritis from Fusobacterium nucleatum in a nonimmunocompromised adult. J Rheumatol. 1993;20:518–520. [PubMed] [Google Scholar]
- Haake SK, Yoder SC, Attarian G, Podkaminer K. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J Bacteriol. 2000;182:1176–1180. doi: 10.1128/jb.182.4.1176-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann JG, Lang CJ, Hartl H, Tomandl B. Multiple brain abscesses caused by Fusobacterium nucleatum treated conservatively. Can J Neurol Sci. 2003;30:266–268. doi: 10.1017/s0317167100002717. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect Immun. 2001;69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett A, Hume WR, Le H, Huynh TN, Han YW, Cheng G, et al. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68:1893–1898. doi: 10.1128/iai.68.4.1893-1898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousimies-Somer H, Savolainen S, Makitie A, Ylikoski J. Bacteriologic findings in peritonsillar abscesses in young adults. Clin Infect Dis. 1993;16(Suppl 4):S292–S298. doi: 10.1093/clinids/16.supplement_4.s292. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V, Ivanova N, Anderson I, Reznik G, Bhattacharyya A, Gardner WL, et al. Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Res. 2003;13:1180–1189. doi: 10.1101/gr.566003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F, Shi W. A unique nine-gene comY operon in Streptococcus mutans. Microbiology. 2005;151:157–166. doi: 10.1099/mic.0.27554-0. [DOI] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Good IJ, Burmeister JA, Palcanis KG, et al. Bacteriology of experimental gingivitis in young adult humans. Infect Immun. 1982a;38:651–667. doi: 10.1128/iai.38.2.651-667.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982b;38:1137–1148. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Cato EP, Burmeister JA, Palcanis KG, et al. Bacteriology of experimental gingivitis in children. Infect Immun. 1984;46:1–6. doi: 10.1128/iai.46.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Palcanis KG, et al. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507–519. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot S, Gal K, Goldblat MV, Eslami HH. The role of anaerobic bacteria in the pathogenesis of urinary tract infections. J Urol. 1981;126:852–853. doi: 10.1016/s0022-5347(17)54781-3. [DOI] [PubMed] [Google Scholar]
- Sabiston CB, Jr, Gold WA. Anaerobic bacteria in oral infections. Oral Surg Oral Med Oral Pathol. 1974;38:187–192. doi: 10.1016/0030-4220(74)90054-1. [DOI] [PubMed] [Google Scholar]
- Sundqvist GK, Eckerbom MI, Larsson AP, Sjögren UT. Capacity of anaerobic bacteria from necrotic dental pulps to induce purulent infections. Infect Immun. 1979;25:685–693. doi: 10.1128/iai.25.2.685-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talan DA, Citron DM, Abrahamian FM, Moran GJ, Goldstein EJC The Emergency Medicine Animal Bite Infection Study Group. Bacteriologic analysis of infected dog and cat bites. N Engl J Med. 1999;340:85–92. doi: 10.1056/NEJM199901143400202. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Truant AL, Menge S, Milliorn K, Lairscey R, Kelly MT. Fusobacterium nucleatum pericarditis. J Clin Microbiol. 1983;17:349–351. doi: 10.1128/jcm.17.2.349-351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]


