Abstract
The envelope (Env) glycoprotein of human immunodeficiency virus (HIV) contains 24 N-glycosylation sites covering much of the protein surface. It has been proposed that one role of these carbohydrates is to form a shield that protects the virus from immune recognition. Strong evidence for such a role for glycosylation has been reported for simian immunodeficiency virus (SIV) mutants lacking glycans in the V1 region of Env (J. N. Reitter, R. E. Means, and R. C. Desrosiers, Nat. Med. 4:679-684, 1998). Here we used recombinant vesicular stomatitis viruses (VSVs) expressing HIV Env glycosylation mutants to determine if removal of carbohydrates in the V1 and V2 domains affected protein function and the generation of neutralizing antibodies in mice. Mutations that eliminated one to six of the sites for N-linked glycosylation in the V1 and V2 loops were introduced into a gene encoding the HIV type 1 primary isolate 89.6 envelope glycoprotein with its cytoplasmic domain replaced by that of the VSV G glycoprotein. The membrane fusion activities of the mutant proteins were studied in a syncytium induction assay. The transport and processing of the mutant proteins were studied with recombinant VSVs expressing mutant Env G proteins. We found that HIV Env V1 and V2 glycosylation mutants were no better than wild-type envelope at inducing antibodies neutralizing wild-type Env, although an Env mutant lacking glycans appeared somewhat more sensitive to neutralization by antibodies raised to mutant or wild-type Env. These results indicate significant differences between SIV and HIV with regard to the roles of glycans in the V1 and V2 domains.
The human immunodeficiency virus (HIV) envelope protein (Env) is the target of virus-neutralizing antibodies, but it does not normally elicit a strong neutralizing antibody response in infected individuals. The ability of HIV to evade the immune system has been associated in part with both the rapid variability of the HIV Env protein sequence and the masking of epitopes by glycosylation (reviewed in reference 43). The HIV Env glycoprotein precursor, gp160, is a highly glycosylated protein of approximately 850 amino acids. During intracellular transport, the gp160 polyprotein is cleaved into two subunits that remain associated: gp41, which contains ecto-, transmembrane, and cytoplasmic domains, and gp120, which is noncovalently linked to the ectodomain of gp41 (29). All 24 potential N-linked sites are glycosylated on gp120 from the HIV IIIB strain expressed in Chinese hamster ovary (CHO) cells, including 13 that contain complex type oligosaccharides and 11 that contain a high-mannose type and/or hybrid type oligosaccharide structure (36). Several studies have shown that the presence of carbohydrates is especially critical during early steps of Env protein folding and cleavage (16, 37, 45, 64), but once Env achieves its final conformation, glycosylation is less critical (16, 40). The X-ray crystal structure of the gp120 core in ternary complex with CD4 and an antibody predicts that carbohydrates are exposed on the outer surface of gp120, probably providing protection from antibody recognition of the peptide backbone (50, 66, 68). The role of these carbohydrates in protein function and immune recognition has not yet been completely examined, and most studies have been performed with laboratory-adapted HIV strains. Primary isolates are often more difficult to neutralize than T-cell-line-adapted (TCLA) strains (41), although a range of neutralization sensitivities exists in both (8).
In order to determine which specific N-linked glycans are critical for Env protein function or immune escape, several recent studies have been directed to individual or multiple mutations of glycosylation sites. Effects of glycosylation on viral replication, gp160 cleavage, CD4 binding activity, and coreceptor usage have been documented (34, 42). Specific Env glycosylation sites also appear to have an important role in modulating the antibody response. For example, removal of an N-linked glycan in the HIV-1BRU Env V1 region can make the virus more resistant to neutralization by anti-V3 antibodies (22). HIV IIIB env clones lacking an N-glycan in the V3 loop of Env protein can become more sensitive to virus neutralization (2). By masking an immunodominant epitope in the V3 loop with additional N-linked carbohydrates, the antibody response can be shifted from the V3 epitope to the V1 epitope in an HIV HXB2 strain (19).
One of the most dramatic effects of carbohydrate removal from an envelope glycoprotein has been reported from studies with simian immunodeficiency virus (SIV) (48). Rhesus monkeys infected with SIVmac 239 mutants lacking glycosylation sites in the V1 region of gp120 produced high titers of neutralizing antibody against the mutant virus. Most importantly, the mutant viruses induced much higher titers of antibody to the wild-type (wt) virus than were induced by the wt itself. Related but less dramatic effects of glycosylation have been observed in the V3 domain of TCLA HIV type 1 (HIV-1) (2, 57). In addition, experiments in guinea pigs with HIVBRU Env containing mutated glycosylation sites in the V4 and V5 domains showed that immunizations with mutant viruses generated antibodies that neutralized mutant viruses twofold better than they neutralized wt virus. Similarly, immunizations with wt viruses generated antibodies that neutralized wt virus twofold better than they neutralized mutant viruses (3).
Based on the results with SIV showing that carbohydrate removal can significantly enhance the neutralizing antibody response to Env (48) and considering the need for more studies involving HIV primary isolates, we studied the functional roles of Env N-linked glycans from the HIV primary isolate 89.6. Single glycosylation mutations and combinations of glycosylation mutations were introduced across the entire V1 and V2 regions and were studied in a transient expression system. Recombinant vesicular stomatitis viruses (VSVs) expressing HIV Env glycosylation mutants were recovered, and these Env proteins were shown to be incorporated into the surfaces of VSV virions. The effects of Env carbohydrate removal on syncytium formation, CD4 binding activity, and cleavage were examined.
We also studied the antibody response to Env glycosylation mutants expressed in VSV vectors. Previous studies have shown that VSV vectors can induce neutralizing antibodies to HIV-1 89.6 both in mice and in rhesus macaques (53, 54). Mice were immunized with recombinant VSVs expressing HIV Env with four or six sequential glycosylation sites mutated in the V1 and V2 regions. Serum antibodies were tested for binding to oligomeric parental Env, binding to nonglycosylated peptides covering the mutated sites, and neutralization of parental or mutated Env.
MATERIALS AND METHODS
Plasmid construction.
A previously described construct, VSV-89.6gp160G (26), encodes a hybrid Env protein that includes the extracellular domain, the transmembrane domain, and 4 amino acids from the cytoplasmic tail of the HIV-1 strain 89.6 envelope protein, fused to 26 amino acids of the cytoplasmic tail of VSV G. This construct was amplified from pVSV-89.6gp160G (26) by PCR using Vent polymerase (New England Biolabs). The forward primer was 5′AACCTTGGAACCCGGGCTCGAGACAATGAGAGTGAAGGAGATCAGand contained an XhoI site (underlined). The reverse primer was 5′ GATCGGATCCGCGGCCGCGCTAGCGGTATCACAAGTTGATTTGG and contained NheI and NotI sites (underlined and boldfaced, respectively). The PCR product was digested with XhoI and NotI restriction enzymes (New England Biolabs) and cloned into BS-SKII (Stratagene), which had been previously digested with XhoI and NotI. The resulting plasmid was designated pBS-89.6G.
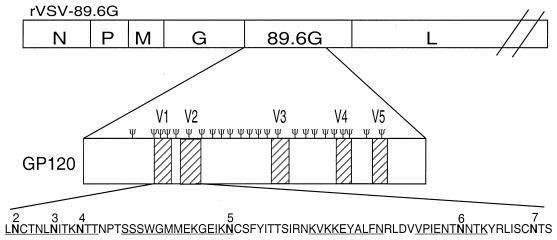
Mutations eliminating glycosylation sites were introduced into pBS-89.6G by using the Quick Change site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). Mutants were generated with reverse and forward primers containing the desired mutation. All mutations substituted the AAT parental sequence encoding asparagine with a CAA sequence encoding glutamine. After Maxi-Prep (Qiagen) purification of mutated plasmids, they were sequenced to confirm the presence of mutations. A total of six glycosylation sites in pBS-89.6G were mutated (Fig. 1); these were sites 2 to 7, numbered starting from the site closest to the N terminus of gp160. Table 1 summarizes the parental plasmids and primers used to generate each glycosylation mutant. For example, by using pBS-89.6G as a parental plasmid, the first glycosylation mutant generated was pBS-89.6G(4), which eliminated the fourth glycosylation site of gp120. The forward primer was 5′ GAATATCACTAAGCAAACTACTCAACCCACTAGTAGCAGC 3′, and the reverse primer was 3′ TTATAGTGATTCGTTTGATGAGTTGGGTGATCATCGTCGACCCCTTAC 5′ (Table 1). (Sequence changes that eliminated sites are boldfaced.) All glycosylation mutants were designated by the number of sites eliminated. We also mutated the asparagine codon at position 421 in the gp120 sequence (shown underlined), although glycosylation was not expected or observed at this position because of a proline residue between the asparagine and the threonine (20).
FIG. 1.
Diagram of a recombinant VSV encoding the 89.6G protein. The Ψ symbol shows the positions of all of the predicted glycans on the expanded gp120 diagram. The amino acid sequences of the V1 and V2 regions are expanded at the bottom of the figure, with individual glycosylation sites numbered. Note that the NPT sequence following site 4 is not a glycosylation site because of the proline. Glycosylation mutations were introduced at the numbered sites in the V1 and V2 regions of 89.6G gp120, replacing asparagine (boldfaced) with glutamine. Peptide sequences used in ELISAs are underlined.
TABLE 1.
Primers used to generate EnvG glycosylation mutants
| Primer set | Template DNA | Final plasmid |
|---|---|---|
| 5′ GAATATCACTAAGCAAACTACTCAACCCACTAGTAGCAGC 3′ | pBS-89.6G | pBS-89.6G(4) |
| 3′ TTATAGTGATTCGTTTGATGAGTTGGGTGATCATCGTCGACCCCTTAC 5′ | ||
| 5′ GTGTTACTTTACAATGCACTAATTTGCAAATCACTAAG 3′ | pBS-89.6G | pBS-89.6G(2,3) |
| 3′ CACAATGAAATGTTACGTGATTAAACGTTTAGTGATTCTTATGATGATTAGGGTG 5′ | ||
| 5′ GAAAATACTCAAAATACTAAGTATAGGTTAATAAGTTGTCAAACCTCAGTC 3′ | pBS-89.6G | pBS-89.6G(6,7) |
| 3′ CTTTTATGAGTTTTATGATTCATATCCAATTATTCAACAGTTTGGAGTCAG 5′ | ||
| 5′ GGAGAAAGGAGAAATAAAACAATGCTCTTTCTATATCACCACAAGC 3′ | pBS-89.6G(4) | pBS-89.6G(4,5) |
| 3′ CCTCTTTCCTCTTTATTTTGTTACGAGAAAGATATAGTGGTGTTCG 5′ | ||
| 5′ CTCTGTGTTACTTTACAATGCACTAATTTGCAAATCACTAAGCAAAC 3′ | pBS-89.6G(4) | pBS-89.6G(2-4) |
| 3′ GAGACACAATGAAATGTTACGTGATTAAACGTTTAGTGATTCGTTTG 5′ | ||
| 5′ GAAAATACTCAAAATACTAAGTATAGGTTAATAAGTTGTCAAACCTCAAGTC 3′ | pBS-89.6G(4) | pBS-89.6G (4,6,7) |
| 3′ CTTTTATGAGTTTTATGATTCATATCCAATTATTCAACAGTTTGGAGTTCAG 5′ | ||
| 5′ GGAGAAAGGAGAAATAAAACAATGCTCTTTCTATATCACCACAAGC 3′ | pBS-89.6G(2,3) | pBS-89.6G(2,3,5) |
| 3′ CCTCTTTCCTCTTTATTTTGTTACGAGAAAGATATAGTGGTGTTCG 5′ | ||
| 5′ GGAGAAAGGAGAAATAAAACAATGCTCTTTCTATATCACCACAAGC 3′ | pBS-89.6G(4,6,7) | pBS-89.6G(4-7) |
| 3′ CCTCTTTCCTCTTTATTTTGTTACGAGAAAGATATAGTGGTGTTCG 5′ | ||
| 5′ CTCTGTGTTACTTTAAATTGCACTAATTTGCAAATCACTAAGCAAAC 3′ | pBS-89.6G(4-7) | pBS-89.6G(3-7) |
| 3′ GAGACACAATGAAATTTAACGTGATTAAACGTTTAGTGATTCGTTTG 5′ | ||
| 5′ CTCTGTGTTACTTTACAATGCACTAATTTGCAAATCACTAAGCAAAC 3′ | pBS-89.6G(4-7) | pBS-89.6G(2-7) |
| 3′ GAGACACAATGAAATGTTACGTGATTAAACGTTTAGTGATTCGTTTG 5′ |
To generate plasmids for recovery of VSV recombinants expressing Env glycosylation mutants, pBS-89.6G, pBS-89.6G(4), pBS-89.6G(4-7), and pBS-89.6G(2-7) were digested with XhoI and NheI restriction enzymes (New England Biolabs) and cloned into XhoI and NheI sites between the VSV G and L genes of pVSV-XN2, which encodes the entire VSV antigenome (55). The resulting plasmids were designated pVSV-89.6G, pVSV-89.6G(4), pVSV-89.6G(4-7), and pVSV-89.6G(2-7).
To generate plasmids for recovery of VSVΔG recombinants expressing Env glycosylation mutants, the sequences of DNA encoding constructs 89.6G(4-7) and 89.6G(3-7) were amplified by PCR using Vent polymerase (New England Biolabs). The forward primer, 5′ CCGGGCCCCCCCACGCGTACAATGAGAGTG AAGGAGATCAGG 3′, contained an MluI site (boldfaced). The reverse primer, 5′ GATCGGATCCGCGGCCGCGCTAGCGGTATCACAAGTTGATTTGG 3′, contained an NheI site (underlined). The PCR product was digested with MluI and NheI restriction enzymes (New England Biolabs) and ligated to pVSVΔG-JRFLG-GFP (4), which had previously been digested with MluI and NheI to remove the JRFLG insert. The resulting plasmids were designated pVSVΔG-89.6G(4-7)-GFP and pVSVΔG-89.6G(3-7)-GFP.
Protein expression using the vaccinia virus-T7 system.
Approximately 2.5 × 105 baby hamster kidney (BHK) cells were plated onto 35-mm-diameter dishes and incubated for 18 h at 37°C. Cells were then washed twice with Dulbecco's modified Eagle's medium (DMEM) and infected for 0.5 h at a multiplicity of infection (MOI) of 10 with vTF7-3, a recombinant vaccinia virus that encodes T7 RNA polymerase (18). Cells were then transfected with 5 μg of either pBS-SKII, pBS-89.6G, or pBS-89.6G glycosylation mutants by using a cationic liposome reagent (52). The medium was removed after 3 h and replaced with DMEM supplemented with 10% fetal bovine serum (FBS). BHK cells were then washed twice with methionine-free medium and labeled for 1 h in 0.5 ml of methionine-free medium containing 100 μCi of [35S]methionine. Cells were washed with phosphate-buffered saline (PBS) and lysed with 0.5 ml of detergent solution (1% Nonidet P-40 [NP-40], 0.4% deoxycholate, 50 mM Tris-HCl [pH 8], 62.5 mM EDTA) with 5 U of aprotinin. Lysates were transferred to 1.5-ml Eppendorf tubes and centrifuged in an Eppendorf microcentrifuge at 10,000 rpm for 2 min to remove nuclei. Supernatants were transferred to new tubes, and sodium dodecyl sulfate (SDS) was added to a final concentration of 0.2%.
Immunoprecipitations.
Immunoprecipitations were performed by adding 2 μl of sheep polyclonal anti-gp120 serum (National Institutes of Health [NIH] AIDS Research and Reference Reagent Program) to the cell lysates and incubating for 0.5 h at 37°C. Samples were incubated with 40 μl of protein A-agarose for 15 min, pelleted, washed three times with ice-cold radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 1% deoxycholate, 1% NP-40, and 0.15 M NaCl in 10 mM Tris [pH 7.4]), and resuspended in 40 μl of sample buffer.
Recovery of VSV recombinants.
VSV recombinants expressing Env mutants were recovered as previously described (55). BHK cells were infected at an MOI of 10 with vTF7-3, and after 1 h they were transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, and 10 μg of either pVSV-89.6G, pVSV-89.6G(4), pVSV-89.6G(4-7), or pVSV-89.6G(2-7) by using a cationic liposome reagent. After incubation for 48 h at 37°C, cell supernatants were removed, filtered through a 0.2-μm-pore-size filter, and transferred to fresh BHK cells for 48 h. VSV recoveries were confirmed initially by observation of cytopathic effect in BHK cells and immunofluorescence microscopy. Virus stocks were prepared by using plaque-purified virus to infect BHK cells.
VSVΔG viruses expressing green fluorescent protein (GFP) and 89.6G glycosylation mutants were recovered as previously described (4). BHK cells (about 75% confluent) were plated onto 10-cm-diameter petri dishes. Cells were infected at an MOI of 10 with vTF7-3 (18). After 1 h of infection, cells were transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, 4 μg of pBS-G, and 10 μg of full-length VSV plasmid. At 48 h posttransfection, supernatants were filtered and passaged onto BHK-G cells (56), which had been induced 24 h earlier to express G. Cells were incubated for 48 h at 37°C, and viral supernatants were then filtered through a 0.2-μm-pore-size filter to remove vaccinia virus. Viral stocks were prepared by expansion of recovered virus in 107 BHK-G cells for 32 h and subsequent transfer of the entire supernatant to 107 BHK cells. After infection for 3.5 h at 37°C, the viral inoculum was removed, cells were washed three times with DMEM, and 10 ml of DMEM supplemented with 5% FBS was added. After an additional 11 h, supernatants were harvested, clarified by centrifugation at 1,250 × g for 5 min, and stored at −80°C.
Western blotting.
Purification of virions and Western blotting were performed as previously described (23). Approximately 106 BHK cells on 10-cm-diameter dishes were infected at an MOI of 0.01 for 0.5 h in serum-free (SF) DMEM, after which the medium was removed and replaced with DMEM supplemented with 5% FBS. Cells were incubated for 18 h, and supernatants were collected and centrifuged at 1,250 × g for 10 min at 21°C. Clarified supernatants were layered on top of a solution of 20% sucrose in TE buffer (10 mM Tris-Cl [pH 7.5]-1 mM EDTA) and centrifuged for 1 h at 4°C in a Beckman SW41 Ti rotor at 38,000 rpm. Viral pellets were then resuspended in 200 μl of TE buffer.
Samples containing equal amounts of protein (normalized by the Pierce bicinchoninic acid protein assay) were fractionated by electrophoresis on an SDS-8% polyacrylamide gel and transferred to a nitrocellulose filter at 4°C, for 18 h at 40 mA. The nitrocellulose filter was blocked in 15 ml of TTBS (0.02% Tween 20, 0.9% NaCl, 100 mM Tris-HCl [pH 7.5]) supplemented with 1% bovine serum albumin (BSA) (American Bioanalytical) at 21°C. After 2 h, 7.5 μl of sheep anti-gp120 (1:2,000 dilution) was added to the blocking solution. After 0.5 h, the nitrocellulose filter was washed three times for 5 min each time with TTBS. The nitrocellulose filter was then incubated with 15 ml of TTBS-1% BSA including 1.5 μl of a biotin-conjugated rabbit anti-sheep secondary antibody (Vector Laboratories) (1:10,000 dilution). After 0.5 h, the nitrocellulose filter was washed again and incubated in 15 ml of TTBS-1% BSA including 1.5 μl of horseradish peroxidase-conjugated streptavidin (Vector Laboratories) (1:10,000 dilution) for 0.5 h. Finally, the nitrocellulose filter was washed again and developed by chemiluminescence (ECL reagents; Amersham).
Pulse-chase metabolic labeling of VSV-infected cells.
Approximately 5 × 105 BHK cells on 35-mm-diameter plates were infected in 0.5 ml of DMEM for 0.5 h with wt or recombinant VSVs at an MOI of 100. DMEM supplemented with 10% FBS (1.5 ml) was added to the plates, and cells were incubated for 3.5 h at 37°C. Cells were then washed with methionine-free medium and pulse-labeled for 0.5 h with 100 μCi of [35S]methionine. The labeling medium was removed from the cells and replaced with DMEM supplemented with 2 mM methionine. Immediately after labeling, or at 1 and 4 h after the chase, cell supernatants and cell lysates were collected. Cell lysates were prepared as described above. Cell supernatants were collected in 1.5-ml Eppendorf tubes and centrifuged in an Eppendorf microcentrifuge for 2 min at 10,000 rpm. Supernatants were transferred to new tubes and adjusted to a solution containing 1% NP-40, 0.4% deoxycholate, 50 mM Tris-HCl (pH 8), 62.5 mM EDTA, and 0.2% SDS. Cell lysates and supernatants were precleared by addition of 2 μl of a polyclonal rabbit anti-VSV antibody for 0.5 h at 37°C, followed by addition of 40 μl of protein A-agarose and incubation for 10 min at 37°C. Samples were pelleted, and supernatants were transferred to new Eppendorf tubes. Sheep anti-gp120 (1.5 μl) was added to the samples for 0.5 h at 37°C, followed by incubation with 40 μl of protein A-agarose for 0.5 h at 37°C. Samples were pelleted, washed three times with RIPA buffer, and resuspended in 40 μl of sample buffer. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (8% acrylamide), followed by exposure to a PhosphorImager screen (Molecular Dynamics).
Protein expression in cells infected with VSVΔG recombinants.
Approximately 6 × 105 BHK cells on 6-cm-diameter plates were infected with recombinant VSVs at an MOI of 20 for 1 h in 3 ml of DMEM. The medium was then replaced with 3 ml of DMEM supplemented with 5% FBS for 8 h at 37°C. Cells were washed twice with a methionine-free medium and labeled in 3 ml of methionine-free medium containing 200 μCi of [35S]methionine. After 1 h, cells were washed again with PBS and lysed with 1.5 ml of detergent solution (1% NP-40, 0.4% deoxycholate, 50 mM Tris-HCl [pH 8], 62.5 mM EDTA) containing 5 U of aprotinin. Lysates were transferred to 1.5-ml Eppendorf tubes and centrifuged at 10,000 rpm for 2 min in an Eppendorf microcentrifuge, after which supernatants were transferred to new tubes. Samples were analyzed by SDS-PAGE (8% acrylamide), followed by exposure to a PhosphorImager screen (Molecular Dynamics).
Preparation of viral stocks for boosting.
Approximately 106 BHK cells on 10-cm-diameter dishes were infected at an MOI of 0.01 with either recombinant wild-type (rwt) VSV, VZV expressing influenza virus hemagglutinin (VSV-HA) (51), VSV-89.6G, VSV-89.6G(4-7), or VSV-89.6G(2-7) for 0.5 h in SF DMEM. The medium was removed and replaced with DMEM supplemented with 5% FBS. Cells were incubated for 18 h, and supernatants were collected and centrifuged at 1,250 × g for 10 min at 21°C. Clarified supernatants were layered on top of a solution of 20% sucrose in TE buffer and centrifuged for 1 h at 4°C in a Beckman SW41 Ti rotor at 38,000 rpm. Viral pellets were then resuspended in 200 μl of PBS. Viral titers, as well as viral protein concentrations, were determined.
Inoculation of mice.
Inoculations were performed as previously described (51, 54). Six-week-old female BALB/c mice (Charles River Laboratories) were inoculated intranasally with 25 μl of virus inoculum (105 PFU) in DMEM (day 0). Each group (four to five mice per group) was inoculated with one of five different recombinants: rwt VSV, VSV-HA (51), VSV-89.6G, VSV-89.6G(4-7), or VSV-89.6G(2-7). Mice were weighed daily in a plastic beaker on a Sartorius balance (model 1409). At day 31, mice were boosted intraperitoneally (i.p.) with 50 μg of the same VSV recombinant administered in the first inoculation. An identical i.p. boost was performed at day 84. At day 98, mice were boosted again intranasally with VSV recombinants.
At day 113, blood samples from mice in each group were pooled and allowed to clot at 21°C. Clots were removed, and samples were centrifuged in a TOMY MTX-150 centrifuge (TMA-11 fixed-angle rotor) at 4°C for 10 min at 5,500 rpm. Clarified sera were transferred to sterile Eppendorf tubes and heat inactivated at 56°C for 1 h.
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were performed as previously described (54). Ninety-six-well plates (Costar 9018) were first coated with 0.1 mg of concanavalin A (ConA; Sigma)/ml in 20 mM Tris-HCl-1 M NaCl (pH 8.5) at 21°C. After 2 h, 30 μl of gp140 supernatant (derived from vBD1 infections) diluted in PBS was added to the wells for 18 h at 4°C. vBD1 is a recombinant vaccinia virus vector that expresses an oligomeric 89.6 gp140 protein containing the extracellular domains of gp120 and gp41 but lacking the transmembrane and cytoplasmic domains of gp41 (49). After gp140 binding, blocking was performed by addition of PBS-10% calf serum for 30 min. Serial twofold dilutions of serum (from 1:100 to 1:800) were added to the wells for 2 h at 21°C. A horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce) diluted 1:20,000 was then added to the wells for 1 h at 21°C. For colorimetric analysis, 2,2′-azinobis(3-ethylbenzthiazoline-6 sulfonic acid) tablets (Immunopure ABTS; Pierce) were used. After each incubation step mentioned above, wells were washed three times with 200 μl of PBS-0.05% Tween 20, and immediately after the last incubation, an extra wash with 2× PBS-0.05% Tween was performed. Optical densities were determined at a wavelength of 405 nm.
ELISAs for peptides covering the V1 and V2 regions (Fig. 1) did not use ConA for the first coating step. Approximately 1 μg of peptide was added to each well in 150 μl of 100 mM NaHCO3 (pH 9.6). Coating was performed for 18 h at 4°C. The remaining steps were performed as described above.
VSV neutralization assay.
Serial twofold dilutions of sera in PBS were added to 96-well plates in a total volume of 50 μl. rwt VSVs (50 μl, containing 100 PFU) resuspended in SF DMEM were added to the wells and incubated at 37°C for 1 h. Approximately 1,500 BHK cells in 100 μl of DMEM-10% FBS were then added to each well, and plates were incubated for 2 to 3 days at 37°C. The neutralization titer was defined as the highest dilution which corresponded to complete inhibition of VSV cytopathic effect. All assays were performed in duplicate, and all results agreed within 1 dilution.
HIV-1 envelope neutralization.
Neutralization assays were performed as previously described (4). A day before the assay, approximately 20,000 HeLa-CD4 cells were plated in individual wells on 96-well plates. After 18 h, VSVΔG-89.6G-GFP or VSVΔG-89.6G(4-7)-GFP virus stocks were incubated with monoclonal antibody (MAb) I1 (1 μl/ml) against VSV G (35) in order to neutralize any infectivity due to residual G protein. After 10 min at 21°C, 25 μl of DMEM-10% FBS containing 100 infectious units (i.u.) of VSVΔG-89.6G-GFP or VSVΔG-89.6G(4-7)-GFP was mixed with serial dilutions (typically 1:10 to 1:160) in 25 μl of DMEM-10% FBS. After incubation for 30 min at 37°C, 50 μl of the virus-serum mixture was added to the HeLa-CD4 cells in duplicate wells containing 150 μl of DMEM-10% FBS. After 18 h of incubation, GFP-positive cells and syncytia were counted by fluorescence microscopy using an Olympus CK40 microscope with a 4× objective.
RESULTS
Generation of glycosylation mutations in HIV envelope V1 and V2 regions.
A plasmid (pVSV-89.6Ggp160G) encoding the HIV 89.6 envelope protein (11) with its cytoplasmic domain replaced by the VSV G protein cytoplasmic domain has been described previously (26). The region of this plasmid encoding the 89.6G protein was amplified by PCR and cloned into the pBS-SKII vector in order to generate glycosylation mutants and to further test protein expression and function. Glycosylation mutations were introduced into the V1 and V2 regions (Fig. 1) of gp120. All mutations replaced asparagine with the structurally related amino acid glutamine. A total of six glycosylation sites (sites 2 to 7) were mutated, and 10 different combinations of mutations were generated, ranging from single sites [pBS-89.6G(4)] to complete elimination of all sites [pBS-89.6G(2-7)].
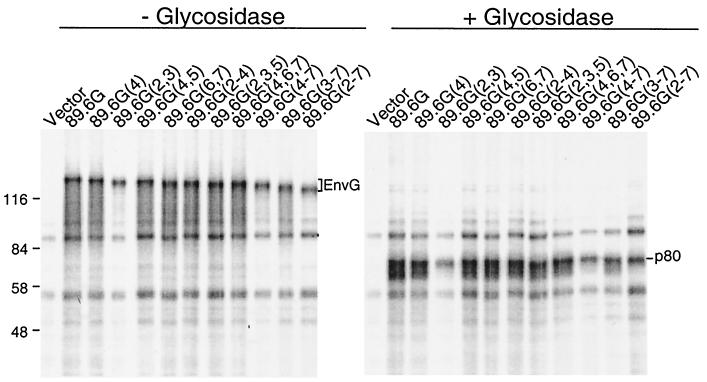
To confirm that mutated glycosylation sites were actually used in the parental protein, the mutant proteins were expressed in BHK cells by using the vaccinia virus-T7 system (18) and were labeled with [35S]methionine. Cell lysates were immunoprecipitated with sheep anti-gp120 and analyzed by SDS-PAGE (Fig. 2). An increase in protein mobility was observed for all 89.6G glycosylation mutants, and the increase was proportional to the number of sites mutated. To ensure that the differences in size between 89.6G envelope glycosylation mutants were due to differences in N glycosylation, all samples were treated with N-glycosidase F (Fig. 2), which removes all N-linked glycans (38). The predicted molecular size of the 89.6G protein without glycans is 80 kDa. All samples treated had the same mobility between the 58- and 84-kDa markers, indicating that glycosylation was responsible for mobility differences before treatment.
FIG. 2.
Immunoprecipitation of Env G glycosylation mutants from transfected cells and treatment with N-glycosidase F. BHK cells were infected with vTF7-3 and then transfected with pBS-SKII expressing either 89.6G or 89.6G glycosylation mutants for 6 h, and proteins were labeled with [35S]methionine for 1 h. Cell lysates were immunoprecipitated with a polyclonal sheep anti-gp120 serum and protein A-agarose. For N-glycosidase treatment (30), samples eluted from protein A were divided in two, peptide N-glycosidase F buffer G7 was added, and half of each sample was treated with peptide N-glycosidase F (New England Biolabs). Incubations of all samples were performed at 37°C for 1 h. Samples were analyzed by SDS-PAGE (8% acrylamide), followed by exposure to a PhosphorImager screen (Molecular Dynamics). Positions of Env G, p80, and molecular weight markers are indicated.
Cell surface expression and membrane fusion by Env glycosylation mutants.
To determine if the mutant proteins were expressed on the cell surface, indirect immunofluorescence was performed on BHK cells expressing each protein. All mutated proteins were clearly detectable on the cell surface and appeared to be expressed at similar levels (data not shown). The HIV 89.6 envelope gene is derived from a dualtropic R5X4 virus, and its expression results in syncytium formation in cells expressing CD4 and either the CXCR4 or the CCR5 coreceptor (14). To determine if the 89.6G glycosylation mutants retained membrane fusion activity resulting in syncytium formation, the parental and mutated proteins were expressed in HeLa-CD4 cells by using the vaccinia virus-T7 system. Formation of multinucleated cells (syncytia) was readily visible by phase-contrast microscopy (Fig. 3). A summary of the results is given in Table 2. All glycosylation mutants retained the ability to form syncytia, except for mutant 89.6G(2-7), which lacked all six sites. The 89.6G(4-7) and 89.6G(2,3,5) constructs formed fewer syncytia than the other mutants. The 89.6G(3-7) mutant, lacking five sites, formed only 1 or 2 syncytia per 2 × 105 cells. These results indicate that mutations affecting multiple glycosylation sites can alter protein function.
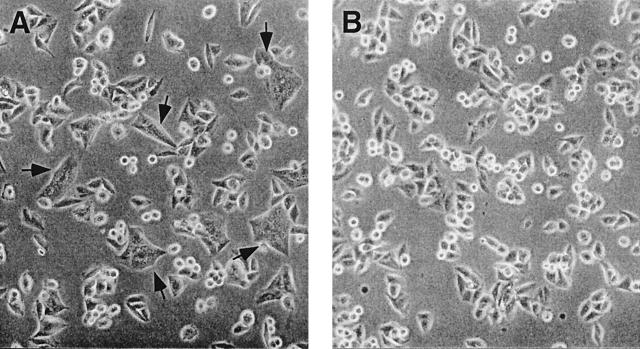
FIG. 3.
Examples of syncytium formation by 89.6G glycosylation mutants. HeLa-CD4 cells (2.5 × 105) were plated onto 35-mm-diameter dishes and incubated for 18 h at 37°C. Cells were infected with vTF7-3 and transfected with either pBS-89.6G(2-3) (A) or vector alone (B). At 8 h posttransfection, the numbers of syncytia in 10 fields per dish were counted by using a Nikon Diaphot phase microscope with a 20× objective. Arrows indicate syncytia.
TABLE 2.
Syncytium formation from all plasmid transfections in HeLa-CD4 cells
| Plasmid expressed | Syncytium formationa |
|---|---|
| Vector alone | − |
| 89.6G | +++ |
| 89.6G(4) | +++ |
| 89.6G(2,3) | +++ |
| 89.6G(4,5) | +++ |
| 89.6G(6,7) | +++ |
| 89.6G(2-4) | +++ |
| 89.6G(2,3,5) | ++ |
| 89.6G(4,6,7) | +++ |
| 89.6G(4-7) | ++ |
| 89.6G(3-7) | + |
| 89.6G(2-7) | − |
+++, 50 to 100 syncytia/10 fields; ++, 20 to 40 syncytia/10 fields; +, 1 to 2 syncytia/10 fields; −, no syncytia in 10 fields.
A mutant lacking fusion function binds CD4.
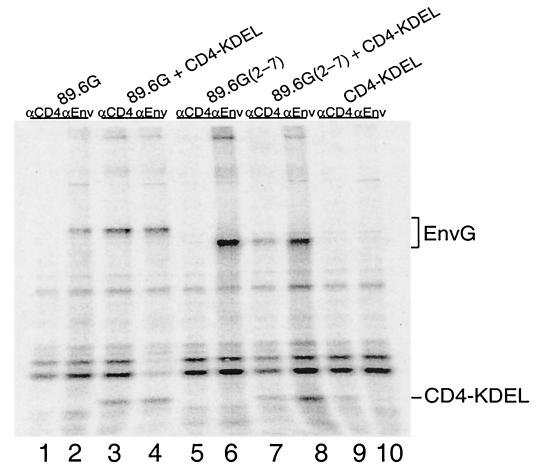
Because of the complete lack of membrane fusion function in the 89.6G(2-7) mutant, we wanted to determine if the protein could still bind to CD4. To do this we used an assay that measures binding of HIV Env to a soluble CD4 retained in the endoplasmic reticulum (ER), by a KDEL signal (6). DNA encoding 89.6G(2-7) was transfected into BHK cells in the presence or absence of DNA encoding CD4-KDEL. Proteins were labeled with [35S]methionine, and cell lysates were immunoprecipitated with either the anti-CD4 MAb OKT4 or sheep anti-gp120. Samples were analyzed by SDS-PAGE. If 89.6G(2-7) interacts with CD4-KDEL, it should be retained in the ER and both proteins should coimmunoprecipitate with anti-CD4 and anti-gp120. As shown in Fig. 4, when pBS-89.6G or pBS-89.6G(2-7) was transfected alone, the Env G protein was immunoprecipitated with anti-gp120 (lanes 2 and 6) but not with anti-CD4 (lanes 1 and 5). The CD4-KDEL protein was detected when cells were transfected with pBS-CD4-KDEL alone and immunoprecipitated with anti-CD4 but not with anti-gp120 (Fig. 4, lanes 9 and 10). When pBS-89.6G or pBS-89.6G(2-7) was transfected along with pBS-CD4-KDEL, both Env G and CD4 were coimmunoprecipitated with either anti-CD4 (Fig. 4, lanes 3 and 7) or anti-gp120 (lanes 4 and 8). Therefore, although 89.6G(2-7) protein does not induce syncytium formation, it retains its ability to bind CD4, and the lack of fusion activity must result from some other functional defect.
FIG. 4.
CD4 binding by Env glycoslation mutants. BHK cells were infected with vTF7-3 and transfected with either 10 μg of pBS-sCD4-KDEL (lanes 9 and 10), which encodes a soluble form of CD4 that is retained in the ER (6), 5 μg of pBS-89.6G (lanes 1 and 2) or pBS-89.6G(2-7) (lanes 5 and 6), or a combination of 10 μg of pBS-sCD4-KDEL and 5 μg of pBS-89.6G (lanes 3 and 4) or pBS-89.6G(2-7) (lanes 7 and 8). Cells were labeled with [35S]methionine for 1 h. Immunoprecipitations were performed with 2 μl of sheep polyclonal anti-gp120 serum (αEnv) or 2 μl of MAb OKT4 (αCD4) (58). Samples were analyzed by SDS-PAGE (8% acrylamide), followed by exposure to a PhosphorImager screen (Molecular Dynamics). The positions of Env G and sCD4-KDEL are indicated.
Recovery of VSVs expressing 89.6G glycosylation mutants.
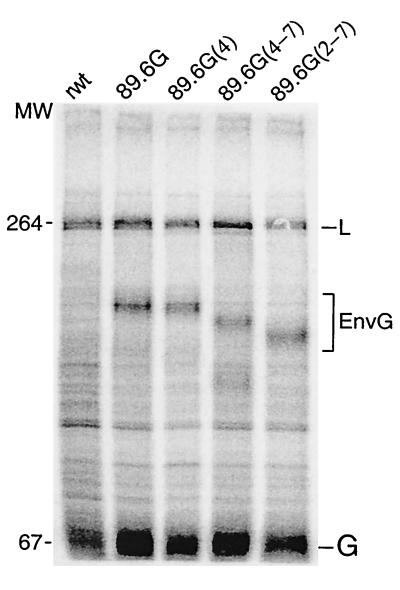
VSV recombinants expressing Env G hybrid protein are useful for studying protein transport to the cell surface and incorporation into VSV virions (26). To study the mutant Env protein with this system, we recovered a VSV recombinant expressing Env G with a single glycosylation mutation [VSV-89.6G(4)], one with four glycosylation sites mutated that still forms syncytia [VSV-89.6G(4-7)], and one with six glycosylation mutations that does not induce syncytium formation [VSV-89.6G(2-7)]. Surface expression of VSV G and Env G by these recombinants was confirmed by infection of BHK cells and immunofluorescence with sheep anti-gp120 and the mouse anti-VSV G MAbs I1 and I14 (data not shown). Expression of Env G proteins of the appropriate mobilities was tested by infecting BHK cells with VSV recombinants. Proteins synthesized in infected cells were labeled for 1 h with [35S]methionine, and cell lysates were analyzed by SDS-PAGE (Fig. 5). The positions of the VSV L and G proteins are indicated and are seen in each lysate. The Env G proteins encoded by the recombinants had mobilities consistent with their glycosylation statuses. The VSV N, P, and M proteins were intentionally run off the gel in order to display differences in mobilities of the mutated Env proteins.
FIG. 5.
Expression of 89.6G glycosylation mutants from VSV recombinants. Approximately 5 × 105 BHK cells on 35-mm-diameter plates were infected with wt or recombinant VSVs at an MOI of 10 for 1 h in 0.5 ml of DMEM. The medium was then replaced with 2 ml of DMEM supplemented with 5% FBS, and cells were incubated for an additional 7 h at 37°C. Cells were washed twice with methionine-free medium and labeled in 0.5 ml of methionine-free medium containing 100 μCi of [35S]methionine. After 1 h, cells were washed with PBS and lysed with 0.5 ml of detergent solution. Lysates were transferred to 1.5-ml Eppendorf tubes and centrifuged at 10,000 rpm in an Eppendorf microcentrifuge for 2 min, and supernatants were transferred to new tubes. About 10 μl of each sample was analyzed by SDS-8% PAGE, followed by exposure to a PhosphorImager screen (Molecular Dynamics). The positions of two VSV proteins (L and G) as well as HIV Env G are indicated. MW, molecular weight (in thousands).
It has been reported previously that Env G proteins are incorporated into VSV virions (26). To determine if 89.6G glycosylation mutants were also incorporated into VSV virions, BHK cells were infected with VSV recombinants for 18 h, and virions were purified from the culture medium. Samples of viruses were analyzed by SDS-PAGE and Western blotting using sheep anti-gp120. As shown in Fig. 6, VSV-89.6G, VSV-89.6G(4), and VSV-89.6G(4-7) incorporated cleaved Env G and only traces of uncleaved Env G into virions. However, VSV-89.6G(2-7) incorporated almost equal amounts of cleaved and uncleaved Env G, indicating a partial defect in cleavage. The defect in cleavage could be due to conformational changes caused by extensive removal of glycosylation sites.
FIG. 6.
Incorporation of 89.6G Env glycosylation mutants into VSV virions. BHK cells were infected with the indicated VSV recombinants at an MOI of 0.01 for 18 h. Cell debris was removed by centrifugation, and clarified supernatants were layered on top of a solution of 20% sucrose in TE buffer and centrifuged for 1 h at 4°C in a Beckman SW41 Ti rotor at 38,000 rpm. Western blotting to detect gp120 was performed by using sheep polyclonal anti-gp120 serum. Details of the Western blotting procedure are given in Materials and Methods. Molecular weight markers, as well as positions of cleaved and uncleaved Env G proteins, are indicated.
Processing of Env G glycosylation mutants.
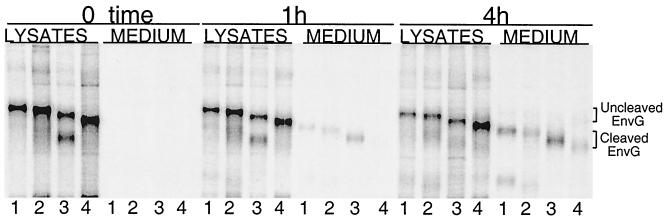
To further analyze the processing of Env G glycosylation mutants encoded by VSV recombinants, a pulse-chase experiment was performed (Fig. 7). BHK cells were infected with VSV recombinants for 4 h, labeled with [35S]methionine for 0.5 h, and then incubated with excess unlabeled methionine. At 0, 1, and 4 h after labeling, cell lysates and supernatants were immunoprecipitated with sheep anti-gp120. Samples were then analyzed by SDS-PAGE. As shown in Fig. 7, immediately after labeling (0 h of chase), all Env G proteins were present in cell lysates in uncleaved form. However, the 89.6G(4-7) protein showed early cleavage of a fraction of the molecules. After the 1-h chase period, nonmutated Env G was present in both cell lysates and supernatants. Some Env G was expected in supernatants due to shedding of mature gp120 protein from fully processed gp120 and gp41 or from budding of virions containing gp120 (26). The cleaved form of 89.6G(4-7) was present in greater amounts in the supernatants, consistent with faster cleavage seen immediately after the pulse, while mutant 89.6G(2-7) was not detectable in the supernatant. At 4 h after the chase, all proteins were in the supernatants, although there was clearly more 89.6G(2-7) retained in the cells. Both uncleaved Env G and cleaved Env G were observed in supernatants from VSV-89.6G(2-7)-infected cells, consistent with incorporation of both cleaved and uncleaved proteins into virions. Our results show that all glycosylation mutants can be expressed on the cell surface and incorporated into VSV virions, as seen earlier. However, mutations at multiple glycosylation sites can alter the rate and extent of Env G cleavage.
FIG. 7.
Immunoprecipitation of Env G from infected cell lysates and supernatants. BHK cells were infected with VSV-89.6G (lanes 1), VSV-89.6G(4) (lanes 2), VSV-89.6G(4-7) (lanes 3), or VSV-89.6G(2-7) (lanes 4) for 4 h and labeled with [35S]methionine for 30 min. Immediately after the labeling (time zero) or at 1 or 4 h after the chase, cell lysates and medium were immunoprecipitated with sheep polyclonal anti-gp120 serum. Samples were analyzed by SDS-PAGE (8% acrylamide), followed by exposure to a PhosphorImager screen (Molecular Dynamics). Positions of uncleaved and cleaved Env G are indicated.
Construction of VSVΔG-89.6G-GFP glycosylation mutants.
Previously, our group described ΔG VSV Env G-GFP viruses that propagate as live viruses on HeLa-CD4 cells and can be used to quantitate neutralizing antibodies directed to the HIV Env protein (4). Because we wanted to test the effects of these mutations on protein function in infection and on induction of neutralizing antibodies to both glycosylated and nonglycosylated Env, we constructed VSV mutants lacking the VSV glycoprotein gene (ΔG) but expressing 89.6 Env G proteins lacking glycosylation sites in the V1 and V2 regions, and a GFP marker protein (Fig. 8A). To generate these viruses expressing Env G glycosylation mutants, we cloned the 89.6G(4-7) and 89.6G(3-7) constructs into the pVSVΔG-GFP vector and recovered VSV recombinants from these constructs.
FIG. 8.
Diagram of recombinant VSV, VSV-89.6G, and VSVΔG-89.6G-GFP genomes, and expression of 89.6G glycosylation mutants in VSVΔG-GFP-infected cells. rwt VSV, VSV-89.6G, and VSV-89.6G glycosylation mutants were used for immunizations of mice. The 89.6G glycosylation mutants were also cloned into the MluI and NheI sites of VSVΔG-GFP cDNA between the M and GFP genes. These recombinant viruses were used for Env neutralization assays (A). BHK-G cells were infected with recombinant VSVs for 8 h and labeled with [35S]methionine for 1 h, and cell lysates were analyzed by SDS-PAGE. The positions of four VSV proteins (L, G, N, and P) as well as HIV Env G are indicated (B).
Analysis of ΔG VSV-Env G-GFP glycosylation mutants.
To examine the proteins encoded by the VSV ΔG recombinants, BHK-G cells were infected at an MOI of 20 and labeled with [35S]methionine. Crude cell lysates were then analyzed by SDS-PAGE (Fig. 8B). As controls, previously described VSV-GFP (4), VSV-89.6G (26), and VSV-89.6G(4-7) were used. VSV N, P, and L proteins were visible in lysates of cells infected with all recombinant viruses. As expected, none of the ΔG viruses expressed VSV G protein, while both VSV-GFP and VSV-89.6G did. Because expression of cellular genes is shut off by VSV infection, the VSV G encoded by the cells is not seen. The mobilities of the two Env glycosylation mutants were consistent with the loss of four or five N-linked glycans. Note that the GFP and VSV M proteins were electrophoresed out of the gel to allow observation of the small differences in Env G protein mobility.
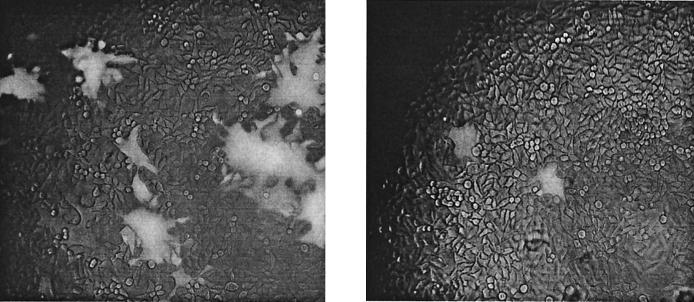
When HeLa-CD4 cells were infected with the VSVΔG-89.6G-GFP viruses, green fluorescent syncytia were observed (Fig. 9). Syncytia formed by VSVΔG-89.6G(4-7)-GFP looked similar to those formed by VSVΔG-89.6G-GFP in terms of size and number (Fig. 9A). However, many fewer infected cells were detected after VSVΔG-89.6G(3-7)-GFP infection, and these were mostly single cells with an occasional smaller syncytium (Fig. 9B). Titers of recovered ΔG virus determined by counting green fluorescent cells or syncytia on HeLa-CD4 cells were 3 × 103 i.u./ml for VSV-89.6G, 1 × 103 i.u./ml for VSVΔG-89.6G(4-7)-GFP, and only 50 i.u./ml for VSVΔG-89.6G(3-7)-GFP. The low titers for VSVΔG-89.6G(3-7)-GFP were probably a result of the reduced fusion activity of this Env G construct, which was noted above. Because of the very low titers, we were unable to use VSVΔG-89.6G(3-7)-GFP for neutralization assays.
FIG. 9.
Infection of HeLa-CD4 cells with VSVΔG recombinants. HeLa-CD4 cells were infected for 18 h with VSVΔG-89.6G(4-7)-GFP (A) or VSVΔG-89.6G(3-7)-GFP (B). Syncytium formation and green fluorescence were observed with a fluorescence microscope. A low level of bright-field light was included to detect background cells.
Immunization of mice with VSV vectors expressing Env glycosylation mutants.
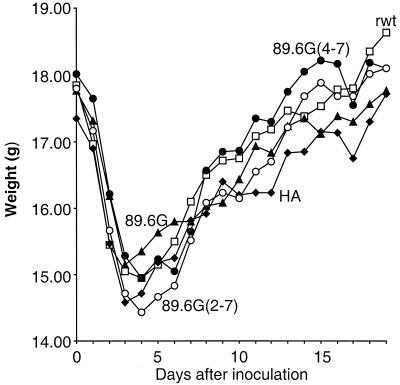
The antibody response to HIV Env was studied in sera from immunized BALB/c mice. We chose to immunize mice with VSV-89.6G(4-7), lacking sites 4 to 7 in V1 and V2, or VSV-89.6G(2-7), lacking all sites for glycosylation in V1 and V2. The latter mutant also lost fusion function but retained CD4 binding. rwt VSV, VSV-HA (51), and VSV-89.6G were used as immunization controls. Previous studies have shown that infection of BALB/c mice with VSV recombinants results in weight loss for 5 to 7 days (51). As shown in Fig. 10, mice lost 15 to 19% of their body weight (average of five mice per group) as a result of VSV infection and then recovered, as observed previously. These results also suggested that all VSV recombinants were replicating to similar extents.
FIG. 10.
Vector-associated pathogenesis in BALB/c mice. Five groups of four to five mice were inoculated intranasally (day 0) with 105 PFU of rwt VSV (□), VSV-HA (⧫), VSV-89.6G (▴), VSV-89.6G(4-7) (•), or VSV-89.6G(2-7) (○). Average-daily weights for mice in each group are shown.
As shown in previous studies, intranasal immunization does not generate neutralizing antibodies against 89.6 Env unless boosts are performed with alternative vectors that are not neutralized by antibodies to VSV G (Indiana) (54). However, we found here that neutralizing antibodies could be generated against 89.6G Env G if we boosted i.p. with purified, concentrated virus containing Env G protein. This boost is presumably effective as a protein boost, since Env G is present in recombinant virions and may not require vector replication.
Analysis of antibodies to glycosylated, oligomeric gp140 by use of ELISA.
Total serum antibody binding to a soluble, oligomeric form of 89.6 Env designated 89.6-gp140 was measured by a published ELISA (49, 54). The lectin ConA was used to bind gp140 to the ELISA plates so that gp140 was bound in multiple orientations and exposed as many epitopes as possible. After incubation with serial dilutions of mouse antisera, the plates were incubated with a horseradish peroxidase-coupled anti-mouse antibody. The amount of antibody bound was then determined by a colorimetric assay.
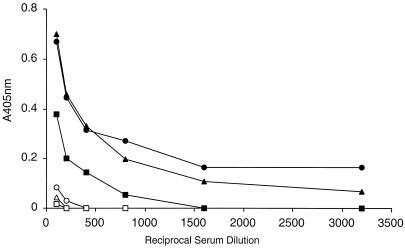
The results of this ELISA are shown in Fig. 11. At 1 month after the initial immunization, only low levels of anti-gp140 antibodies were detected. We did not detect neutralizing antibodies at this time (data not shown). Following two i.p. protein boosts with purified virus at days 31 and 84, and one more intranasal protein boost at day 98, anti-gp140 antibodies were readily detected by ELISA at day 113. Mice immunized with VSV-89.6G(4-7) and mice immunized with VSV-89.6G had similar titers of antibody to gp140, while mice immunized with VSV-89.6G(2-7) had approximately fourfold-lower titers. It is possible that the lower titers generated by 89.6G(2-7) reflect a shift in the immune response to epitopes not represented in the gp140 molecule used for the ELISA.
FIG. 11.
ELISA for oligomeric 89.6-gp140. The ELISA was performed as described in Materials and Methods by using serial dilutions of mouse sera. Backgrounds of control mouse sera run in parallel were subtracted. Triangles, sera from VSV-89.6G immunizations; circles, sera from VSV-89.6G(4-7) immunizations; squares, sera from VSV-89.6G(2-7) immunizations. Open and solid symbols represent sera obtained at day 31 or 113 after the initial immunization, respectively.
ELISA for nonglycosylated peptides in V1 and V2.
In the previous studies of glycosylation mutants of SIV, antibodies to the nonglycosylated mutants were detected by ELISAs using nonglycosylated peptides spanning the mutant regions (48). We also performed a similar ELISA using nonglycosylated peptides from the V1 and V2 regions of 89.6 Env protein. Figure 1 shows the sequences of the peptides (underlined) that were used. However, we did not detect any significant reactivity of sera from immunizations with VSV expressing wt Env or mutated Env using any of these peptides. These results indicate that the deglycosylated V1 and V2 domains present in the 89.6G Env construct are not good inducers of antibodies to linear epitopes in these domains.
Titers of neutralizing antibodies to VSV.
The VSV G protein is the target of antibodies neutralizing VSV (27), and immunization of mice with VSV recombinants typically elicits neutralizing titers in the range of 1:5,000 to 1:10,000. Because all of the vectors used for immunizations contained VSV G, we measured the anti-VSV neutralizing antibody titers for all mouse sera, using twofold serial dilutions and determination of the end point corresponding to complete neutralization. These values are given in Table 3. Titers generated by all recombinants were 1:10,240, except for VSV-89.6G, for which the titer was 1:5,120. These results indicate that all VSV recombinants replicated to similar levels in the mice, consistent with similar pathogenesis in all animals.
TABLE 3.
Neutralization titers to VSV and EnvG proteins
| Immunization or antibodya | Neutralization titerb against:
|
||
|---|---|---|---|
| VSV | VSVΔG-89.6G-GFP | VSVΔG-89.6G(4-7)-GFP | |
| rwt VSV | 1:10,240 | 0 | 0 |
| VSV-HA | 1:10,240 | 0 | 0 |
| VSV-89.6G | 1:5,120 | 1:40-1:80 | 1:160 |
| VSV-89.6G(4-7) | 1:10,240 | 1:20 | 1:160 |
| VSV-89.6G(2-7) | 1:10,240 | 0 | 1:20-1:40 |
| HIV Ig | 0 | 1:800 | 1:800 |
Mice were immunized with the indicated viruses. Pooled immunoglobulin from HIV-infected patients (HIV Ig) was used as a control in the last line.
Titers of neutralizing antibodies in sera of immunized mice.
Titers of neutralizing antibodies against HIV Env G proteins.
Neutralization of 89.6G and 89.6G(4-7) by mouse sera was detected by reductions in the numbers of green fluorescent cells and syncytia in HeLa-CD4 cells infected with VSVΔG-89.6G-GFP or VSVΔG-89.6G(4-7)-GFP. At day 113 after the initial inoculation, sera from mice immunized with control viruses failed to neutralize either ΔG-89.6G or ΔG-89.6G(4-7) (Table 3). Sera from mice immunized with VSV-89.6G(4-7) or VSV-89.6G(2-7) had titers of neutralizing antibodies against VSVΔG-89.6G(4-7) that were higher than those against fully glycosylated VSVΔG-89.6G, suggesting that the mutants might expose new epitopes that elicited an increased neutralizing antibody response to mutant Env. However, immunization with VSV-89.6G also generated a higher titer of neutralizing antibodies to the mutant VSVΔG-89.6G(4-7) than to VSVΔG-89.6G, suggesting that, instead, the mutant was simply more sensitive to neutralizing antibodies, including those elicited by wt Env. A control immunization with pooled immunoglobulin from HIV-infected patients gave identical neutralization titers (1:800) against VSVΔG-89.6G and VSVΔG-89.6G(4-7). The major conclusion from these data is that immunization with VSV recombinants expressing mutant Env proteins lacking glycans in V1 and V2 elicited lower neutralizing titers to wt Env than immunization with wt Env itself and therefore does not appear to be a useful approach for enhancing the neutralizing antibody response to the primary HIV isolate 89.6.
DISCUSSION
Here we have characterized Env protein mutants of an HIV primary isolate in which the glycosylation sites in the V1 and V2 regions were eliminated. We chose to study the V1 and V2 regions because SIV mutants lacking glycans in these regions are able to induce high titers of neutralizing antibodies to both mutant and wild-type SIV Env proteins (48). Because sequences in the V1 and V2 loops of other HIV strains have been associated with HIV tropism, infectivity, and fusion activity (1, 5, 7, 59, 65), our initial studies were directed toward determining how mutation of a single or multiple glycosylation sites affected 89.6 Env expression and function. All glycosylation sites (a total of six) in the V1 and V2 regions could be mutated without affecting expression of Env protein on the cell surface. However, not all constructs retained fusion competence. As many as three sequential glycosylation sites could be mutated without disrupting syncytium formation. However, mutations of four [89.6G(4-7)] or five [89.6G(3-7)] glycosylation sites reduced fusion activity, and mutation of all six glycosylation sites [89.6G(2-7)] eliminated all fusion activity. Because individual or double glycosylation mutants covering the whole V1 and V2 region showed normal fusion activity, the loss of function appears to be related to a cumulative effect of loss of multiple sites.
Previous studies have described the effects of multiple Env glycosylation mutations on viral replication. Different combinations of triple glycosylation mutations generated in SIV mac239 have resulted in replication-incompetent virus (44, 47). In simian-human immunodeficiency virus (SHIV) 89.6 containing the 89.6 envelope, passage in monkeys resulted in a more pathogenic SHIV-89.6P, which lacks a glycosylation site in V2 (site 6), resulting in decreased fusion activity (15). Also, mutation of a glycosylation site (equivalent to site 6 in 89.6 Env) in the V2 loop of Env in HIV-1 strain HXB2 resulted in nonviable virus (17). Triple or individual glycosylation mutations in the HXB2 Env V1 and V2 regions resulted in impaired virus infectivity (65). However, in our studies, single or double mutations at equivalent positions of the 89.6 Env protein had no effect on fusion activity. In another HIV strain (IIIB), the V1 and V2 loops have been deleted and the virus is still replication competent (7). These results, taken together, suggest that mutation of glycosylation sites in the V1 and V2 loops may impair folding of the Env protein or prevent conformational changes required for membrane fusion.
To test these mutants for immunogenic potential with VSV-based vectors, we constructed VSV recombinants expressing HIV Env with a single or multiple glycosylation sites mutated. Having the mutant Env proteins expressed in VSV allowed us to perform kinetic experiments on Env transport, cleavage, and incorporation into virions. This was not possible with the vaccinia virus-T7 system, in which HIV Env transport is much less efficient. Using the recombinant VSV system, we found that Env lacking all V1 and V2 glycosylation sites [pBS-89.6G(2-7)] was cleaved inefficiently, transported slowly, and incorporated into VSV virions as both cleaved and uncleaved forms. In contrast, the Env mutant lacking sites 4 to 7 showed faster cleavage than wt Env and more-rapid release of cleaved gp120 into the medium. The effects of glycan removal on multiple aspects of Env function all suggest indirect effects on protein folding, although no clear conformational changes were detected with a limited number of MAbs that recognized 89.6 HIV Env.
One possible explanation for the increased antigenicity of SIV Env glycosylation mutants is exposure of epitopes such as chemokine receptor-binding sites that are normally hidden. Chemokine receptor binding is thought to require CD4 binding and movement of the V1 and V2 loops to expose the chemokine receptor-binding site (30, 50, 60, 66, 67). A CD4-independent Env mutant derived from HIV IIIB has been identified recently, and its coreceptor binding site is exposed (25, 31). Sequence analysis of this mutant revealed 18 mutations not present in the original HIV IIIB Env. Determinants for CD4 independence mapped outside the regions that determine coreceptor specificity (the V1, V2, and V3 regions) but included loss of five glycosylation sites (one in the V2 region). In addition, the HIV-1 primary isolate Env (ADA) that acquired CD4 independence also lost a glycosylation site in V2 among other mutations (28).
We wanted to investigate whether mutation of glycosylation sites in the V1 and V2 regions had any effect in exposing the chemokine receptor-binding site. However, we could not detect consistent changes in recognition with MAbs 17B and 48D, which bind CD4-induced epitopes that overlap the chemokine receptor-binding site and that have been shown to react with CD4-independent Env (61, 62, 63). We also looked for CD4-independent entry into HOS cells expressing CXCR4 or CCR5 (12, 24, 33) by using VSVΔG recombinants expressing Env mutants 89.6G(2-4) and 89.6G(4-7) and were unable to detect it.
To determine the effect of carbohydrate removal in the HIV Env V1 and V2 domains on the antibody response against HIV, we inoculated mice with VSV recombinants expressing mutant Env proteins. Inoculations were performed with VSV vectors encoding Env protein of the primary HIV isolate 89.6 with mutations in four glycosylation sites in V1 or with mutations in all six sites in V1 and V2. Neutralizing antibodies directed against parental or mutated Env were measured by using VSVΔG viruses that express Env glycosylation mutants and the GFP marker protein. All constructs induced antibodies that recognized oligomeric 89.6 Env gp140 in an ELISA, but Env glycosylation mutants with mutations in V1 and V2 were no better than wt Env at inducing antibodies neutralizing wt Env. However, the mutant Env proteins did appear to be somewhat more sensitive to neutralizing antibodies raised against either wt or mutant Env, suggesting conformational changes in the mutant Env proteins.
The major goal of our study was to compare the neutralizing antibody response to HIV Env glycosylation mutants with mutations in the V1 and V2 domains to the results obtained with SIV (48). Infection of rhesus macaques with SIV carrying glycosylation mutations in or near the Env V1 domain can increase the neutralizing antibody response to SIV as much as 25-fold over that obtained with infection with wt SIV. Titers of neutralizing antibodies to the homologous SIV glycosylation mutant were even greater than titers of antibodies neutralizing SIV. The greater neutralization of wt SIV by sera from animals infected with the mutant viruses suggested that glycans were masking neutralizing epitopes that do not induce neutralizing antibodies when glycans are present. Our results with Env from an HIV primary isolate are very different in that the mutants were less effective than wt Env at inducing neutralizing antibodies against wt Env.
The SIV results are puzzling because it is difficult to visualize how antibodies generated to a nonglycosylated protein sequence would be able to recognize the same sequence in glycosylated form better than it is recognized by antibodies generated to the glycosylated sequence itself. Our results with HIV are more consistent with what might be expected and suggest that the SIV results may require an explanation other than direct epitope masking by glycans. It is possible, for example, that removal of glycans in the V1 region of SIV Env has effects on protein conformation, perhaps exposing epitopes elsewhere in SIV Env that do not normally elicit neutralizing antibodies. The SIV Env glycosylation mutants were subsequently reported to be CD4 independent (13), suggesting that the high neutralizing titers were due to the exposure of usually hidden intermediate epitopes such as the chemokine receptor-binding site. If these epitopes were only transiently exposed, for example, during virus entry, they might not normally elicit an antibody response. Antibodies raised to the nonglycosylated SIV Env protein exposing these hypothetical epitopes might, however, be able to neutralize wt virus during entry. A similar explanation was invoked to explain the broadly neutralizing “fusion-competent” antibodies described by Nunberg and colleagues (32). Structural differences between HIV Env and SIV Env could explain differences in conformational changes in HIV Env versus SIV Env caused by glycan removal in V1 and V2.
Although no detailed analysis of the effect of glycans in the V1 and V2 regions of HIV-1 has been reported, neutralizing epitopes in the V1 and V2 domains of HIV primary isolates have been identified (46). An earlier study has shown that acquisition of a glycan in the V1 region can mask a major neutralizing epitope of SIV Env (9). Similarly, addition of a glycan in the V3 domain of a TCLA HIV-1 strain has been shown to block neutralization by antibodies recognizing the V3 domain (21, 39). In fact, mutation of N-linked glycans in the HIV-1 V1 domain has been associated with resistance to neutralization. Removal of an N-linked glycan in the V1 domain of HIV BRU-LAV resulted in a strain more resistant to neutralization by MAbs to the V3 loop and to soluble CD4 (22). Also, in vivo adaptation of a SHIV clone expressing TCLA SF33 (SHIVSF33A) resulted in a neutralization-resistant variant by removal and addition of a glycosylation site in the V1 and V3 regions, respectively (10, 11). Effects of glycan removal in the V4 and V5 domains of TCLA HIV have been reported. Here the results showed that an antibody to mutant envelope neutralized the mutant envelope better than an antibody to the wt, but the antibody to mutant envelope did not exhibit greater neutralization of wt Env (3).
Other possible explanations for the differences between our results with HIV and those obtained for SIV are the use of mice instead of primates to generate neutralizing antibodies. However, the magnitude of the neutralizing antibody response to HIV Env generated by VSV vectors in mice is very similar to that seen in rhesus macaques (unpublished data). It thus seems very likely that there are significant differences in the effects of carbohydrate removal in the V1 region between HIV and SIV in terms of induction of a greater neutralization response to fully glycosylated Env proteins in the latter. Given the extensive sequence divergence among HIV isolates, it is also likely that there will be significant differences among HIV strains with regard to the role of glycans in the immune response.
Acknowledgments
This work was supported by NIH grant AI40357. M.I.Q.-K. acknowledges support from an NIH MARC predoctoral fellowship.
We thank Anjeanette Roberts and Karl Haglund for instruction in inoculation of mice, and we are grateful to Nina Rose for development of the ELISA and the neutralization assay and for instruction in their use. We are grateful to Tracey Ferguson and other members of the Yale Animal Resource Center, BCMM, for care and assistance with our mice. We thank Robert Doms and James Robinson for providing mouse and human anti-Env MAbs. We also thank the Rose laboratory members for helpful comments and suggestions during the preparation of the manuscript, and we thank JoAnn Falato for administrative assistance.
REFERENCES
- 1.Andeweg, A. C., P. Leeflang, A. D. Osterhaus, and M. L. Bosch. 1993. Both the V2 and V3 regions of the human immunodeficiency virus type 1 surface glycoprotein functionally interact with other envelope regions in syncytium formation. J. Virol. 67:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 3.Bolmstedt, A., S. Sjolander, J. E. Hansen, L. Akerblom, A. Hemming, S. L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 4.Boritz, E., J. Gerlach, J. E. Johnson, and J. K. Rose. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, M. T., G. R. Simpson, A. J. Cann, M. A. Johnson, and R. A. Weiss. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buonocore, L., and J. K. Rose. 1990. Prevention of HIV-1 glycoprotein transport by soluble CD4 retained in the endoplasmic reticulum. Nature 345:625-628. [DOI] [PubMed] [Google Scholar]
- 7.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers, R. 2001. Avoidance of antibody recognition by SIV-and HIV-encoded envelope proteins. In 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill.
- 14.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 15.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, M. Fernandes, K. Liou, R. Gomila, J. Lee, and J. Sodroski. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fennie, C., and L. A. Lasky. 1989. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J. Virol. 63:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox, D. G., P. Balfe, C. P. Palmer, J. C. May, C. Arnold, and J. A. McKeating. 1997. Length polymorphism within the second variable region of the human immunodeficiency virus type 1 envelope glycoprotein affects accessibility of the receptor binding site. J. Virol. 71:759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W. P. Tsai, G. Lin, J. J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 20.Geetha-Habib, M., H. R. Park, and W. J. Lennarz. 1990. In vivo N-glycosylation and fate of Asn-X-Ser/Thr tripeptides. J. Biol. Chem. 265:13655-13660. [PubMed] [Google Scholar]
- 21.Goudsmit, J., C. Debouck, R. H. Meloen, L. Smit, M. Bakker, D. M. Asher, A. V. Wolff, C. J. Gibbs, Jr., and D. C. Gajdusek. 1988. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc. Natl. Acad. Sci. USA 85:4478-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gram, G. J., A. Hemming, A. Bolmstedt, B. Jansson, S. Olofsson, L. Akerblom, J. O. Nielsen, and J. E. Hansen. 1994. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch. Virol. 139:253-261. [DOI] [PubMed] [Google Scholar]
- 23.Haglund, K., J. Forman, H. G. Krausslich, and J. K. Rose. 2000. Expression of human immunodeficiency virus type 1 Gag protein precursor and envelope proteins from a vesicular stomatitis virus recombinant: high-level production of virus-like particles containing HIV envelope. Virology 268:112-121. [DOI] [PubMed] [Google Scholar]
- 24.He, J., and N. R. Landau. 1995. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J. Virol. 69:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, J. E., M. J. Schnell, L. Buonocore, and J. K. Rose. 1997. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J. Virol. 71:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley, J. M., S. U. Emerson, and R. R. Wagner. 1972. The glycoprotein of vesicular stomatitis virus is the antigen that gives rise to and reacts with neutralizing antibody. J. Virol. 10:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 30.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaCasse, R. A., K. E. Follis, M. Trahey, J. D. Scarborough, D. R. Littman, and J. H. Nunberg. 1999. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science 283:357-362. [DOI] [PubMed] [Google Scholar]
- 33.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 66:5110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, W. R., W. J. Syu, B. Du, M. Matsuda, S. Tan, A. Wolf, M. Essex, and T. H. Lee. 1992. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:2213-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121:168-174. [DOI] [PubMed] [Google Scholar]
- 36.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 37.Li, Y., L. Luo, N. Rasool, and C. Y. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maley, F., R. B. Trimble, A. L. Tarentino, and T. H. Plummer, Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195-204. [DOI] [PubMed] [Google Scholar]
- 39.Matsushita, S., M. Robert-Guroff, J. Rusche, A. Koito, T. Hattori, H. Hoshino, K. Javaherian, K. Takatsuki, and S. Putney. 1988. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J. Virol. 62:2107-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montefiori, D. C., W. E. Robinson, Jr., and W. M. Mitchell. 1988. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 85:9248-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama, E. E., T. Shioda, M. Tatsumi, X. Xin, D. Yu, S. Ohgimoto, A. Kato, Y. Sakai, Y. Ohnishi, and Y. Nagai. 1998. Importance of the N-glycan in the V3 loop of HIV-1 envelope protein for CXCR-4- but not CCR-5-dependent fusion. FEBS Lett. 426:367-372. [DOI] [PubMed] [Google Scholar]
- 43.Nara, P. L., R. R. Garrity, and J. Goudsmit. 1991. Neutralization of HIV-1: a paradox of humoral proportions. FASEB J. 5:2437-2455. [DOI] [PubMed] [Google Scholar]
- 44.Ohgimoto, S., T. Shioda, K. Mori, E. E. Nakayama, H. Hu, and Y. Nagai. 1998. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J. Virol. 72:8365-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal, R., G. M. Hoke, and M. G. Sarngadharan. 1989. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:3384-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinter, A., W. J. Honnen, S. C. Kayman, O. Trochev, and Z. Wu. 1998. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine 16:1803-1811. [DOI] [PubMed] [Google Scholar]
- 47.Reitter, J. N., and R. C. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 49.Richardson, T. M., Jr., B. L. Stryjewski, C. C. Broder, J. A. Hoxie, J. R. Mascola, P. L. Earl, and R. W. Doms. 1996. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J. Virol. 70:753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, A., E. Kretzschmar, A. S. Perkins, J. Forman, R. Price, L. Buonocore, Y. Kawaoka, and J. K. Rose. 1998. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 72:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 53.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 54.Rose, N. F., A. Roberts, L. Buonocore, and J. K. Rose. 2000. Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J. Virol. 74:10903-10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schnell, M. J., J. E. Johnson, L. Buonocore, and J. K. Rose. 1997. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell 90:849-857. [DOI] [PubMed] [Google Scholar]
- 57.Schonning, K., B. Jansson, S. Olofsson, J. O. Nielsen, and J. S. Hansen. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 58.Shaw, A. S., K. E. Amrein, C. Hammond, D. F. Stern, B. M. Sefton, and J. K. Rose. 1989. The lck tyrosine protein kinase interacts with the cytoplasmic tail of the CD4 glycoprotein through its unique amino-terminal domain. Cell 59:627-636. [DOI] [PubMed] [Google Scholar]
- 59.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker, B. D., M. Kowalski, W. C. Goh, K. Kozarsky, M. Krieger, C. Rosen, L. Rohrschneider, W. A. Haseltine, and J. Sodroski. 1987. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc. Natl. Acad. Sci. USA 84:8120-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, W. K., M. Essex, and T. H. Lee. 1996. Single amino acid substitution in constant region 1 or 4 of gp120 causes the phenotype of a human immunodeficiency virus type 1 variant with mutations in hypervariable regions 1 and 2 to revert. J. Virol. 70:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 67.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 68.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]