Abstract
Retrotransposition of the Ty1 element of Saccharomyces cerevisiae is temperature sensitive. Transposition activity of Ty1 is abolished at temperatures above 34°C. In this report, we show that the major block to transposition at high temperature is the inhibition of processing of the Gag-Pol-p199 polyprotein and the concomitant reduction of reverse transcriptase (RT) activity. Expression of a Ty1 protease construct in Escherichia coli shows that protease enzymatic activity is inherently temperature sensitive. In yeast, Gag processing is only partially inhibited at high temperature, while cleavage of the Gag-Pol polyprotein is completely inhibited. Sites of proteolytic processing are differentially susceptible to cleavage during growth at high temperature. Overall levels of the Gag-Pol polyprotein are reduced at high temperature, although the efficiency of the requisite +1 frameshifting event appears to be increased. RT activity is inherently relatively temperature resistant, yet no cDNA is made at high temperature and the amount of RT activity is greatly reduced in virus-like particles formed at high temperature. Taken together, these results suggest that alterations in Ty1 proteins that occur at high temperature affect both protease activity and RT activity, such that Ty1 transposition is abolished.
The Ty1 element of the yeast Saccharomyces cerevisiae is a 6-kb retrotransposon with long terminal repeats (LTRs) at each end (4). Transcription of the element is directed by the 5′ LTR and produces an RNA molecule from which the element-encoded proteins Gag and Gag-Pol are translated. Host protein Spt3p has been shown to be required for Ty1 element transcription (30). These proteins are subsequently assembled into structures termed virus-like particles (VLPs) and processed by the element-encoded protease (PR). VLPs are essential replication intermediates in which reverse transcription occurs (12, 14).
The 49-kDa Gag protein is the most abundant translational product and is the major structural determinant of VLPs (25). A +1 frameshift signal 15 nucleotides (nt) 5′ to the stop codon for Gag results in the production a smaller amount of a 199-kDa Gag-Pol fusion protein (2, 7). This fusion protein contains the Gag structural protein, as well as the enzymes PR, integrase (IN), and reverse transcriptase (RT)/RNase H (1, 12, 24, 32).
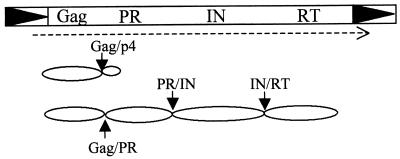
PR processes both the Gag and the Gag-Pol translation products (Fig. 1). Cleavage of the C-terminal 40 amino acids of Gag yields a processed Gag-p45 (CA) protein. The Gag-Pol polyprotein undergoes a semiordered cleavage by PR, releasing each of the protein components (Fig. 1). The Gag-PR cleavage site portion is hydrolyzed first, at the same location at which the smaller Gag-p49 protein is processed. This step is essential, and blocking this cleavage inhibits all further processing (21, 22). The remaining p160-Pol polyprotein is cleaved to release PR, IN (90 kDa), and RT/RNase H (60 kDa). During the assembly process, the Ty1 RNA is packaged within the VLPs and subsequently reverse transcribed into a full-length cDNA. In the final step of transposition, the cDNA is integrated into a new site in the host genome, and the cycle can begin anew with transcription of the newly transposed element. Although they employ a virus-like replication process, Ty1 VLPs are noninfectious and do not leave the host cell. Previous studies indicated that proteolytic processing was regulated in cells and that overproducing Ty1 mRNA overcomes a defect in processing by Ty1 (9).
FIG. 1.
Ty1 processing sites. Translation of the Ty1 element yields Gag-p49 and Gag-Pol-p199 protein products, which are subsequently processed by PR. The Gag/PR processing site is present in both translational products. This site is cleaved first in the Gag-Pol-p199 polyprotein, followed by cleavage at the two downstream sites (21).
Growth at high temperature has a multitude of cellular consequences, and it has been known for some time that Ty1 transposition is inherently temperature sensitive (5, 26). Transposition frequency is maximal at 22°C and is still readily detectable at 30°C, the standard growth temperature for S. cerevisiae. At higher temperatures, the level of transposition drops rapidly, and transposition is abolished above 34°C. In this report, we show that the major block to transposition at high temperature is the inhibition of processing of the Gag-Pol-p199 polyprotein and a concomitant reduction of RT activity. Autoprocessing of a Gag-PR fusion protein expressed in Escherichia coli is also inhibited at high temperature. Correspondingly, in yeast, no processing of the Gag-Pol-p199 is detected at high temperature. Cleavage of the Gag-p49 protein is variable and is slightly reduced at high temperature in some strains, indicating variability in the temperature sensitivity of the various processing sites. No cleavage of the Gag-p49 protein is seen in a PR− mutant grown at high temperature. Previous studies of PR cleavage site mutants have shown that the processing of the Gag/PR site is a prerequisite to further processing (21). At high temperature, the Gag-Pol polyprotein yield is reduced and the processing of this polyprotein phenocopies a PR− active-site mutant. Ty1 cDNA synthesis is undetectable at high temperature, which is further evidence for a virtually complete Gag-Pol processing defect. Although exogenous RT activity is not innately temperature sensitive, the RT activity in VLPs formed at high temperature is greatly reduced. Thus, we propose that alterations in Ty1 proteins that occur at high temperature affect both PR activity and RT activity, such that Ty1 transposition is completely blocked.
MATERIALS AND METHODS
Yeast strains.
The yeast strains and Ty1 plasmids used for transposition assays and biochemical analysis of VLPs are given in Table 1.
TABLE 1.
Yeast strains and Ty1 plasmids
| Strain | Genotype | Ty1 plasmida (reference) | Ty1 marker | Strain background |
|---|---|---|---|---|
| YH8 | MATα ura3-167 his3Δ200 leu2Δ1 trp1Δ1 | GRF167 | ||
| YH50 | MATaura3-167 his3Δ200 leu2Δ1 trp1Δ1 spt3-202 | S288C | ||
| YH51 | MATahis4-539 lys2-801 ura3-52 spt3-202 | pJEF1105 (6) | neo | S288C |
| YH82 | MATahis4-539 lys2-801 ura3-52 trp1Δ63 leu2Δ1 | pJEF724 (3) | None | S288C |
| JKc1015 | MATα ura3-167 his3Δ200 leu2Δ1 | pGTy1H3m his3AI (10) | his3AI | GRF167 |
| JKc1031 | MATahis4-539 lys2-801 ura3-52 spt3-202 | pJEF1105 (6) | neo | S288C |
| JKc1032 | MATahis4-539 lys2-801 ura3-52 spt3-202 | pJEF1105PR- (6) | neo | S288C |
| JKc1054 | MATα ura3-167 his3Δ200 leu2Δ1 | pJEF1105 (6) | neo | GRF167 |
| JKc1055 | MATα ura3-167 his3Δ200 leu2Δ1 | pJEF1105PR- (6) | neo | GRF167 |
| W3031-B | MATα ade2-1 can1-100 his3-11,15 leu2-3 trp1-1 ura3-1 | pGTy1H3m his3AI (10) | his3AI | W303 |
| Hansen BY4741 | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | pGTy1H3m his3AI (10) | his3AI | S288C |
| YPH652 | MATα ura3-52 his3Δ200 leu2Δ1 | pGTy1H3m his3AI (10) | his3AI | YPH (S288C) |
All Ty1 plasmids are GAL-Ty1-H3 URA3 2μm.
Transposition assays.
For quantitation, strains were initially grown as ∼12- by 12-mm patches on SC-Ura medium to maintain plasmid selection. Cells were then replica plated to galactose medium and incubated at the appropriate temperature for 44 to 48 h to induce transposition. Following galactose induction patches were printed to SC-His medium. The cells remaining on the galactose plates were transferred to 10 ml of sterile water (dilution 1). Fifty microliters of this dilution was transferred to 5 ml of sterile water (dilution 2). One hundred microliters of dilution 1 was plated to SC-His medium, except at temperatures above 32°C, in which case cells in dilution 1 were pelleted, resuspended in ∼200 μl of water, and plated to SC-His. Fifty microliters of dilution 2 was plated to yeast extract-peptone-dextrose (YPD). Following incubation at 30°C (YPD, 2 days; SC-His, 4 days) the resulting colonies were counted. Transposition frequency was calculated by dividing the number of colonies on SC-His by the total number of cells plated on SC-His, as determined by the colony number on YPD, factoring in the dilution and original volume of dilution 1 plated. If no colonies appeared on SC-His, transposition frequency was taken to be less than the frequency calculated from a single colony.
Cell homogenates.
The cell growth procedure was based on a protocol previously described (22). Specifically, cultures were grown at either 22 or 37°C. The starting density was an A600 of ∼0.2, and cells were collected when the density reached an A600 of ∼2 (about 12 h at 37°C and about 36 h at 22°C). Aliquots (0.5 ml) of the culture were collected by centrifugation, resuspended in 40 μl of buffer B (10 mM HEPES-KOH [pH 8.0], 15 mM KCl, 5 mM EDTA), and frozen at −75°C. Aliquots were thawed on ice, and the total volume was brought to 200 ml with buffer B. Cold glass beads were added to the meniscus. Forty microliters of 100% trichloroacetic acid (TCA) was added, and the samples were vortexed at top speed for 4 min. Samples were placed immediately on ice, and 1 ml of ice-cold 5% TCA was added. Samples were spun for 20 min at 14,000 × g in the cold. The liquid was aspirated, and the pellet was resuspended in 1 ml of cold water. Samples were spun for 10 min as before, and the supernatant was aspirated. Proteins were extracted by resuspending the pellet in 150 μl of sample buffer (6% sodium dodecyl sulfate [SDS], 0.5 M Tris base) and incubating the suspension at 50°C for 10 min. Samples were spun for 1 min (14,000 × g), and the supernatant was removed to a fresh tube. The extraction process was repeated, and the supernatants were pooled. One-third volume of a solution of 0.25 M dithiothreitol, 50% glycerol, and 0.2% bromphenol blue was added, and the samples were spun (14,000 × g) for 3 min. The supernatant was transferred to a fresh tube.
VLP preparation.
Cells were grown and lysed as described previously (12). Extract (7.5 ml) was loaded onto a 70/30/20 (5-/5-/15-ml) step gradient and centrifuged for 180 min at 28,000 rpm in a Sorvall AH629 swinging-bucket rotor. The remaining extract was saved for use as whole-cell extract. Fractions were collected by puncturing the bottom of the tube and collecting 1.2-ml fractions. To pellet VLPs, peak fractions 4, 5, and 6 were pooled, diluted to 11 ml with buffer B, and pelleted for 1 h at 35,000 rpm in a Sorvall A1256 fixed-angle rotor. The pellet was resuspended in 150 μl of buffer B.
Immunoblotting.
Whole-cell extracts and purified VLPs were mixed with an equal volume of 2× sample loading buffer (20% [vol/vol] glycerol, 0.125 M Tris-Cl [pH 6.8], 5% [wt/vol] SDS, 10% [vol/vol] 14 M β-mercaptoethanol, 0.2% [wt/vol] bromphenol blue) and boiled (3 min) prior to loading on 10 (Gag blots) or 7.5% (Pol blots) SDS gel. Gels were transferred to nitrocellulose (for Gag blots) or a polyvinylidene difluoride (PVDF) membrane (for Pol blots) in Tris-glycine buffer containing 10% methanol at 24 V for 1 h. Membranes were blocked in phosphate-buffered saline containing 5% nonfat dried milk. Membranes were then probed with antibodies as described previously (21). Antibody binding was detected with the appropriate secondary antibody followed by ECL (nitrocellulose) or ECL-Plus (PVDF) reagent and exposure to X-ray film. Anti-Gag (anti-VLP polyclonal serum R2-F) and anti-IN (monoclonal antibody 8B-11) are described elsewhere (12, 23).
Processing in E. coli.
A Ty1 Gag-PR construct (PPR) was prepared as described previously (19). Briefly, this construct contains a methionine codon preceding codon 348 of Ty1 Gag and a TAG codon after codon 182 of PR. The frameshift signal was removed via a single nucleotide deletion to permit expression in E. coli. Cells were lysed in 5 volumes of 1.2× Laemmli buffer. The lysate was centrifuged at 14,000 × g at room temperature, and the supernatant was transferred to a fresh tube. The samples were resolved by SDS-14% polyacrylamide gel electrophoresis and transferred to a PVDF membrane at 300 mA for 2 h. Membranes were probed with rabbit polyclonal antibody JH695 as described previously (19).
Biochemical assays.
Frameshift assays were done as described previously (27). Plasmids pACTTy and pACTy were a kind gift from M. Cassan. Ty1 cDNA synthesis assays were performed exactly as described previously (21). RT activity assays were done as described previously (12).
Glucose chase experiments.
Galactose induction/glucose chase experiments were done using strain YH82. For high-temperature induction, cells were inoculated at a density of ∼8 × 106 to 10 × 106 cells/ml into selective medium (to maintain the Ty1 plasmid) containing 1% raffinose and incubated at 22°C for 3 h. Cultures were then shifted to 37°C for 2 h prior to induction. Galactose was added to 2% final concentration, and cultures were incubated overnight. Glucose was then added to 2% final concentration to stop induction. The culture was divided in half and incubated at 37 or 30°C, with 5-ml aliquots being removed at each time point. Cells from each aliquot were pelleted and frozen prior to protein isolation as described previously (22).
RESULTS
Transposition is temperature sensitive.
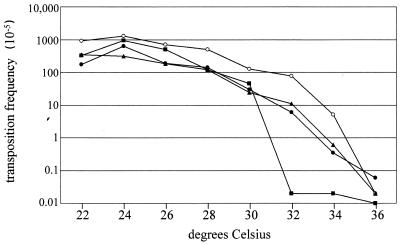
Ty1 transposition is temperature sensitive. Transposition of endogenous Ty1 elements is sharply reduced at 30°C relative to that at 22°C and is undetectable at 37°C (5, 26). We quantitated Gal-induced transposition of a Ty1 element, marked with the his3AI reporter gene, in 2°C increments from 22 to 36°C in several strain backgrounds. Transposition frequency is highest in most strains at ∼24°C (Fig. 2). Similar to what was found for genomic elements, the frequency of galactose-induced transposition decreases steadily as temperature increases, dropping sharply above 32°C. Similar results are observed when a Ty1 element containing an uninterrupted reporter gene is used (data not shown). All strains show greater-than-1,000-fold reduction in transposition frequency from 22 to 36°C. Transposition at 36°C is undetectable in most strains.
FIG. 2.
Transposition is temperature sensitive. Several wild-type yeast strains containing a galactose-inducible Ty1 element on plasmid pGTy1m his3AI were quantitated for transposition following galactose induction at the indicated temperatures (as described in Materials and Methods). Strains are as follows: JKc1015 (open circles), W3031-B (solid squares), Hansen BY4741 (solid circles), and PH652 (solid triangles). Full strain genotypes are given in Table 1.
Ty1 PR is temperature sensitive.
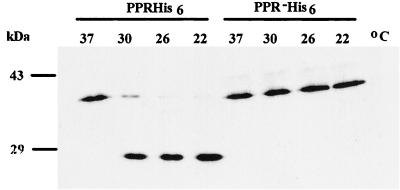
During studies of the Ty1 PR protein, Ty1 PR was cloned and heterologously expressed in E. coli (19). The Ty1 PR expression construct used contains an ATG codon immediately upstream of the codon for amino acid 348 of the Ty1 Gag protein, followed in frame by the coding sequence for Ty1 PR. The frameshift signal was “erased” by a single base pair deletion that does not alter protein coding with respect to Gag or PR. The coding sequence for a carboxy-terminal hexahistidine tag was affixed, followed by a termination codon. Autoprocessing of the fusion protein yields an ∼28-kDa PR protein. Immunoblot analysis of the purified protein probed with an anti-Gag polyclonal antibody shows that autoprocessing occurs efficiently at 22 and 26°C and to a lesser extent at 30°C and is abolished at 37°C (Fig. 3). The fusion protein remains soluble irrespective of the temperature at which the cells were grown. As a control, a construct bearing an inactivating mutation in the PR active site was also expressed. This construct is not processed at any temperature examined, indicating that the processing observed in the wild-type construct is not the result of an adventitious E. coli protease. The Ty1 PR is therefore inactive with respect to the Gag-PR cleavage site at high temperature when expressed in E. coli as a Gag-PR fusion. Although the context of the Gag cleavage site in this construct is likely quite different than that for the native Ty1 Gag or Gag-Pol protein, the effect of temperature on PR cleavage is severe. This result led us to further investigate Ty1 PR activity at high temperatures in yeast.
FIG. 3.
Ty1 PR expressed in E. coli is temperature sensitive. A fusion protein (PPRHis6) containing the Gag/PR cleavage site followed by the Ty1 PR and six histidines was expressed in E. coli. Immunoblots of culture extracts were probed with polyclonal antibody JH695 to Gag. The fusion protein undergoes autoprocessing in cells grown at 22, 26, or 30°C but not in cells grown at 37°C. In the PPR−His6 construct, the PR active site is mutated, abolishing autoprocessing.
Processing of Pol, but not Gag, is defective at high temperature.
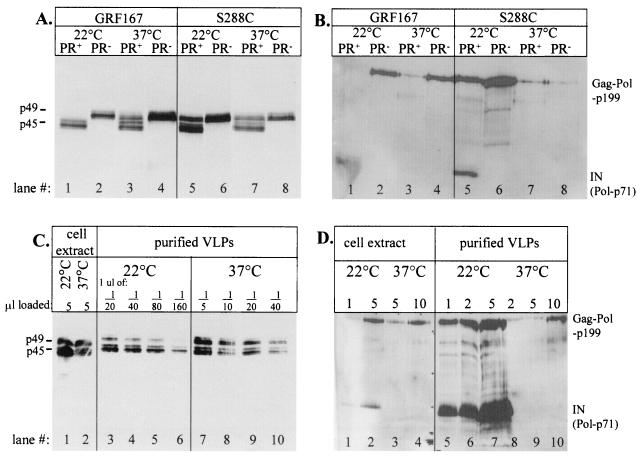
Since PR activity is apparently innately temperature sensitive, we examined the processing of Ty1 proteins at various temperatures in yeast strains containing a Ty1 element transcriptionally regulated by the GAL1 promoter. A PR− mutant version of this Ty1 element was used in parallel to assess nonspecific Ty1 protein degradation at high temperature. Cells were induced to express Ty1 by adding galactose to actively growing cells at 22 or 37°C. A polyclonal antiserum (R2-F) that detects both precursor Gag-p49 and processed Gag-p45 was used (21) to probe homogenates prepared from cells induced at either 22 or 37°C (Fig. 4A). The levels of Gag-p49 processing at 22°C vary by strain origin (Fig. 4A, lane 1 versus 5). At 37°C, processing is slightly inhibited in a GRF167 strain. However, in an S288C strain, although the total protein yield is less at 37°C, processing is not affected, as the ratio of p45/p49 does not change (Fig. 4A, lane 5 versus 7). This is in stark contrast to the processing of the Gag-PR construct in E. coli, where no processing was detected at 37°C. The processing of Gag-p49 in yeast at high temperature is not due to nonspecific activation of cellular proteases, as no processing of Gag-p49 is seen using the PR− construct.
FIG. 4.
Processing of Ty Pol proteins is temperature sensitive. (A) Immunoblots of cellular homogenates from yeast cultures induced by galactose at 22 (10 μl/lane) or 37°C (20 μl/lane) and probed with anti-Gag. The Gag-p49 and processed-Gag-p45 bands each appear as doublets. Two yeast strains of differing origins, each harboring a galactose-inducible element with either a functional (PR+) or a mutant (PR−) PR, were used. (B) Same as panel A, except that immunoblots were probed with anti-IN. Gag-Pol-p199 and IN (Pol-p71) are indicated. Homogenate from cells induced at 22 (20 μl/lane) or 37°C (40 μl/lane) was loaded onto the gel. (C) Immunoblot of whole-cell extracts and purified VLPs from a GRF167 strain (JKc1015) induced at 22 or 37°C and probed with anti-Gag. The numbers above the lanes indicate the volumes of original sample loaded. A fraction above a lane indicates the dilution of VLPs loaded. (D) Immunoblot of whole-cell extracts and purified VLPs from a GRF167 strain (JKc1015) induced at 22 or 37°C and probed with anti-IN. The numbers above the lanes indicate the volume (in microliters) of original sample loaded.
Anti-IN monoclonal antibody 8B-11 detects processed IN (Pol-p71; apparent molecular mass, ∼90 kDa) as well as the full-length Gag-Pol-p199 polyprotein and any processing intermediates containing the IN epitope. 8B-11 was used to assess efficiency of Pol processing. Immunoblots of whole-cell homogenates prepared from cells induced at either 22 or 37°C were probed with an anti-IN antibody (Fig. 4B). While processed IN is readily detected in cells grown at 22°C, it is undetectable in cells grown at 37°C. Gag-Pol-p199 polyprotein yield is markedly reduced, but detectable, at 37°C. Thus, total cellular Gag-Pol-p199 levels are reduced and proteolysis of the Ty1 Pol polyprotein appears to be defective at high temperature. As seen with Gag-p49, the processing efficiency of Gag-Pol-p199 at 22°C varies by yeast strain (Fig. 4B, lane 1 versus 5). This result suggests that there is a “context sensitivity” for the processing of the Gag-PR cleavage site. Since Gag-PR cleavage of the Gag-Pol-p199 polyprotein is essential for further processing, we propose that, at high temperature, Gag-Pol is not processed at the Gag-PR site but that Gag-p49 is processed to some extent.
The lack of visible processed IN at 37°C (Fig. 4B, lanes 3 and 7) could alternatively be explained by the marked reduction in Gag-Pol polyprotein in these whole-cell extracts. Thus, we further investigated production and processing of Ty1 proteins by immunoblotting purified VLPs. Cell cultures of GRF167 origin were induced by galactose at 22 and 37°C, and cell extracts were subsequently fractionated on a sucrose gradient. Peak fractions were pelleted to concentrate and purify VLPs. VLPs are concentrated by this fractionation, indicating that stable VLPs are formed at high temperature, albeit at reduced yield. Immunoblot analysis of purified VLPs shows an approximately threefold reduction in the overall level of Ty1 Gag protein produced at 37°C (Fig. 4C). The reduction of the Pol protein product is greater, as shown by immunoblots of purified VLPs probed with an anti-IN antibody (Fig. 4D). There is, however, a sufficient Gag-Pol-p199 protein signal present in both cell extracts and VLPs induced at 37°C to detect processed IN, if it had been present (Fig. 4D, lane 2 versus 4 and lane 5 versus 10). The reduction in protein levels at high temperature is significant but far less than the concomitant 1,000-fold reduction in transposition frequency.
Transcriptional and translational effects are not significant contributors to the temperature sensitivity of transposition.
The levels of steady-state Ty1 mRNA for cells grown in galactose at 22 and 37°C were found to be the same (data not shown). As Ty1 mRNA is very abundant in both conditions, it is unlikely to be a limiting component. The total amount of Gag protein detected in VLPs when cells are grown at 37°C is reduced by approximately threefold compared to that when cells are grown at 22°C. However, the difference in Pol protein production appears to be much greater. Thus, although translation initiation efficiency and stability of Ty1 proteins at high temperature are unlikely to contribute significantly to reduced transposition, the difference in the relative amounts of Gag and Gag-Pol could, in principle, result from an effect of temperature on frameshifting. A +1 frameshift signal near the coding sequence for the C terminus of Gag regulates the relative amounts of Gag and Gag-Pol proteins. An AGG codon near the 3′ end of the Gag coding sequence requires decoding by a rare tRNA, frequently resulting in a ribosomal stall (2, 7, 31). A +1 frameshift occurs ∼10% of the time while the stalled ribosome awaits a tRNA to decode the AGG codon, producing the Gag-Pol fusion protein. A temperature-sensitive effect on frameshifting could significantly affect the ratios of Ty1 protein products. Retrotransposition is blocked when the Gag-to-Gag-Pol ratio is disturbed by (i) alteration of expression levels of a single-copy tRNA gene that inhibits frameshifting or (ii) expression, in trans, of a protein known to inhibit +1 ribosomal frameshifting (11, 15, 28, 31).
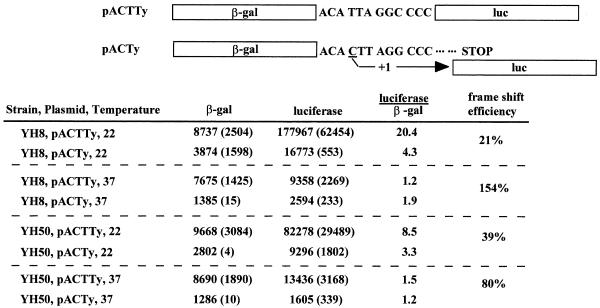
Frameshifting efficiency was investigated by using a construct containing the β-galactosidase open reading frame (ORF), followed by the Ty1 frameshift signal and the luciferase ORF (27). A control construct encodes luciferase as an in-frame fusion to β-galactosidase (Fig. 5). A successful +1 frameshift in the first construct thus results in translation of the luciferase reporter protein. Levels of luciferase activity were assayed and standardized to β-galactosidase activity. The results (Fig. 5) show that frameshifting in the context of the pACTy construct at 22°C is strain dependent and is, at best, fourfold more efficient than the 10% frameshifting seen in the context of a Ty1 element mRNA. The relative change in the luciferase-to-β-galactosidase activity ratios at 37°C implies an increase in frameshifting efficiency that approaches or exceeds 100% at high temperature. However, it is notable that β-galactosidase activity in the context of pACTy is reduced by twofold at high temperature, thereby artifactually increasing the apparent frameshift efficiency. Additionally, we cannot rule out effects of differences in enzyme stability at high temperatures, as luciferase activity is notably reduced in both constructs at 37°C. However, an increase in frameshifting efficiency would be expected to result in increased readthrough translation and a greater amount of Gag-Pol protein than Gag produced at 37°C. Our immunoblots of VLPs show a reduction in Gag-Pol protein synthesis compared to that of Gag at 37°C. Thus, the modest increase in frameshifting efficiency is presumably insignificant with respect to Ty1 transposition.
FIG. 5.
Temperature effects on frameshifting. The diagram shows the constructs used to measure frameshifting. pACTTy contains the luciferase reporter gene in frame with the β-galactosidase reporter gene. In the pACTy construct, the Ty1 frameshift sequence has been inserted between the two ORFs. Expression of the luciferase reporter indicates the efficiency of the Ty frameshift site. The reporter gene activity from the constructs was measured in triplicate in two yeast strains grown at 22 and 37°C. The β-galactosidase (β-gal) and luciferase activities are the averages of the three measurements, and the standard deviations are in parentheses. Luciferase activity, normalized to β-galactosidase activity, is used to calculate the percent frameshift efficiency for each strain at both temperatures. Frameshift efficiency is calculated as 100 times the ratio of luciferase activity/β-galactosidase activity for the pACTy plasmid divided by the corresponding ratio for the pACTTy plasmid.
cDNA synthesis is defective.
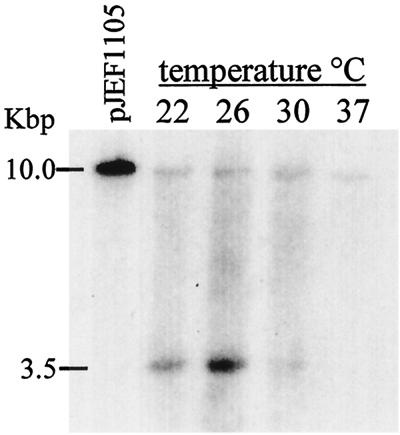
Since the cleavage of the Gag-Pol-p199 protein was inhibited, we examined how this affected further steps in the Ty1 life cycle. Cleavage of the Gag-Pol-p199 sites is a semiordered process in that Gag/PR cleavage must occur first, but the subsequent cleavages do not appear to be ordered. Mutational analysis of the cleavage sites has shown that cleavage of PR/IN and IN/RT sites is not required for cDNA synthesis but that cleavage of the Gag/PR site is required. We therefore examined Ty1 cDNA synthesis by Southern blotting total DNA isolated from cell cultures grown at different temperatures (Fig. 6). No Ty1 cDNA is detectable in Southern blots of DNA isolated at 36°C. Similarly, VLPs isolated from cells grown at 36°C do not contain any cDNA. The amount of cDNA detected at 30°C is ∼20% of the amount seen at 22°C, consistent with the levels of transposition observed (Fig. 2). The outcome of the reverse transcription process at high temperature phenocopies that of a PR− element at low and normal growth temperatures.
FIG. 6.
Ty1 cDNA synthesis as a function of temperature. Cells (YH51; Table 1) containing a galactose-inducible Ty1 element were grown at the indicated temperatures. Southern blot analysis of extracted DNA indicates that production of the 3.5-kbp cDNA product is markedly reduced at 30°C and is undetectable at 37°C. Nucleic acids were extracted, digested with EcoRI, treated with RNase, electrophoretically separated (1% agarose gel), and transferred. The membrane was probed with a [32P]-labeled neo cDNA probe. The larger ∼10.0-kbp band corresponds to the Ty1 plasmid.
RT activity is not inherently temperature sensitive but is reduced in VLPs formed at high temperature.
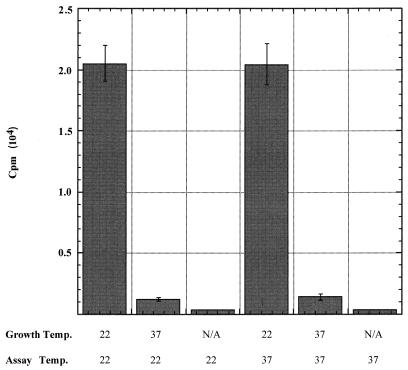
Since no cDNA is detectable, we investigated RT activity in vitro at high temperature. VLPs prepared from galactose-induced cultures at 22 and 37°C were assayed for in vitro RT activity at both 22 and 37°C. Since VLP yield is reduced at high temperature, the amounts of VLPs added to the reaction mixtures were normalized to levels of Gag protein in the VLPs (Fig. 4C). As seen in Fig. 7, RT activity is not inherently temperature sensitive; VLPs formed at 22°C have the same levels of RT activity at 22 and 37°C. However, VLPs formed at 37°C have ∼15-fold reduced activity at either temperature. In vitro RT activity can readily be detected in the context of the full-length polyprotein, as both PR− active-site mutants and Gag*PR cleavage site mutants have RT activity on an exogenous primer template (21, 32). Despite the reduction in Gag-Pol polyprotein levels at 37°C, there is a significant amount of RT activity above background in these VLPs, suggesting that what little RT is present still possesses activity in the in vitro assay.
FIG. 7.
RT activity is not temperature sensitive. VLPs were purified from cells (JKc1015; Table 1) induced with galactose at 22 or 37°C. RT activity using exogenously added primers and templates was measured at 22 and 37°C for both VLP preparations. Assay samples were normalized to the Gag protein (Fig. 4C) by adding a 3.5-fold-greater volume of the 37°C VLP preparation. N/A (none added), background level of incorporated radioactivity when buffer B is added in place of VLPs.
The PR defect is not reversible.
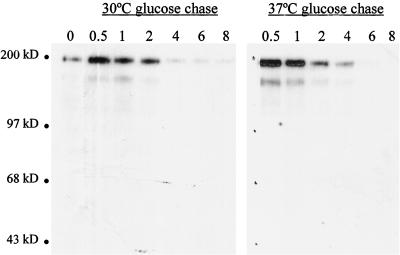
To determine whether the processing defect is reversible, yeast cultures were induced by galactose at 37°C overnight, after which glucose was added to halt Ty1 gene expression. Cell aliquots were further incubated at 37°C or shifted to 30°C at various intervals. Immunoblots of total cell protein from aliquots were probed with an anti-IN antibody. As shown in Fig. 8, IN was not processed from the polyprotein upon subsequent incubation at 30°C. Additionally, the half-lives of the polyprotein at 30 and 37°C are the same, as shown by the similar degradation rates of the Gag-Pol-p199 polyprotein. Thus, a shift down to the permissive temperature after galactose induction does not recover processing.
FIG. 8.
The PR defect is not reversible. Cells (YH82; Table 1) were induced with galactose overnight at 37°C. Following addition of glucose, the cultures were shifted down to 30°C or maintained at 37°C. Numbers indicate the time points, in hours, at which aliquots were removed. Total cell protein was analyzed by immunoblotting using an antibody to IN. No processed IN was detected.
DISCUSSION
In this paper we demonstrate that a major block to transposition at high temperature is the inhibition of PR activity, exacerbated by a decrease in overall Ty1 protein levels. The decrease in overall Gag-Pol protein level observed at 37°C could be a consequence of the decreased PR activity or it could be due to some unrelated change in cellular physiology. When expressed in E. coli, recombinant Ty1 PR is nonfunctional at high temperature, suggesting an intrinsic temperature sensitivity. In yeast, cleavage of the Gag-Pol-p199 polyprotein is completely blocked at high temperature, and cleavage of the Gag protein is slightly reduced in some strains. In VLPs formed at high temperature, exogenous RT activity is significantly reduced but detectable, presumably reflecting a decrease in total RT protein, as this exogenous RT activity is not inherently temperature sensitive in vitro. Endogenous RT activity, determined by measuring Ty1 cDNA production, decreases with increasing growth temperature, consistent with reduced RT amounts and activity, and is undetectable at 37°C. Thus, the exogenous RT activity detected in purified VLPs is insufficient to produce detectable cDNA in vivo. This conclusion agrees with earlier studies in which PR-deficient mutants failed to synthesize cDNA (21, 32). We hypothesize that a temperature-induced conformational change in the Ty1 Gag-Pol polyprotein reduces the activity of both PR and RT, resulting in nearly undetectable levels of transposition. It is likely that the reduction in Ty1 protein levels at 37°C also contributes to the severe transposition defect, as Ty1 transposition does not necessarily correlate with the amount of detected protein. Curcio and Garfinkel previously showed that expression of a galactose-inducible Ty1 element increases transposition efficiency at the posttranslational level, and their studies suggested that the activity of endogenous Ty1 elements may be limited by the levels of PR activity (9). Correspondingly, we have shown that a major defect in high-temperature transposition is posttranslational. It is possible that a limiting factor for transposition falls below a threshold level at temperatures above 32°C, contributing to the rapid drop-off in transposition.
A recently published study determined the necessity of each of the Ty1 cleavage sites for Ty1 transposition (21). Proteolytic processing of the three Ty1 cleavage sites occurs in a regulated and semiordered manner. Gag/PR must be cleaved first, after which cleavage of the other two sites occurs without required order. Blockage of either of the cleavage sites in Pol (PR/IN and IN/RT), independently or in combination, did not affect cDNA production in these mutants, and the affected site(s) shows a cleavage defect. However, blocking the Gag*PR cleavage site abolished cleavage at all three sites and also blocked cDNA production. A PR− active-site mutant, in which no processing occurs, was also defective in cDNA synthesis. We found that, in VLPs formed at high temperature, a different phenotype was observed. Processing at the two Pol sites (PR/IN and IN/RT) was completely abolished. However, unlike what was found for PR− and Gag*PR processing site mutants, processing at high temperature in yeast was not completely blocked, as indicated by the presence of processed Gag-p45 protein from the Gag-p49 primary translation product. This processing of p49 required active Ty1 PR. Taken together, these results point to a context-dependent decrease in PR activity wherein Gag-Pol is not processed at the Gag-PR junction but Gag-p49 is processed. Curiously, both of these processing events occur at the same scissile bond located between histidine 401 and asparagine 402 (Fig. 1).
Since we observe processing of Gag, it appears that VLPs formed at high temperature harbor an additional defect, other than processing, that blocks cDNA production. Production of cDNA is gradually reduced as temperature increases. A mild increase in temperature (to 30°C) reduces the amount of cDNA produced, although Gag-Pol-p199 processing occurs readily at this temperature. Thus, as temperature increases, VLPs become less efficient at producing cDNA. The observed reduction of RT activity was still expected to produce a small but detectable amount of residual cDNA synthesis at 37°C. However, we failed to detect any cDNA. The cDNA synthesis defect might therefore be exacerbated by the slight decrease in Gag processing. In summary, it is likely that the temperature sensitivity of Ty1 retrotransposition reflects the combined effects of deficiencies in both proteolytic processing and in vivo cDNA synthesis.
Previous studies have shown that PR− mutants are defective in cDNA production, not because RT is inactive but because of a defect in accessing the endogenous primer or template or a defect in forming dimeric RNA or both (13, 21, 32). The initial step to cDNA synthesis in Ty1 is the annealing of the host-encoded tRNAiMet primer to the primer-binding site (PBS) on the Ty RNA. Data from studies of PBS region mutants suggest that formation of the primer/template complex is a temperature-sensitive step in transposition (18). A G:U mismatch introduced into the primer/template complex reduces transposition activity to approximately one-third wild-type levels at 27°C. Conversely, extension of the PBS complementarity region from 10 to 12 nt conferred a slight temperature resistance to transposition (17). At 34°C, transposition levels of an element containing a 12-nt PBS are nearly 100-fold greater than that of a wild-type element. However, transposition in these mutants is still substantially less than that seen at 22°C. Therefore, the lack of cDNA at high temperature may be attributed only in part to reduced efficiency of primer/template formation. It is also possible that temperature-induced conformational changes in the template/primer complexes that form at high temperature further reduce the efficiency of RT initiation.
Transposition is very sensitive to the conformation of the primer/template complex. Introduction of a G·U mismatch, while not expected to significantly affect primer/template annealing, does result in a slightly temperature-sensitive phenotype. Additionally, a single mismatch in the primer/template complex reduces transposition dramatically (16). Thus, temperature may have an effect on priming at two steps: high temperature may reduce the efficiency of primer/template annealing and may affect the conformational structure of complex that forms. Recent studies suggest that the amino terminus of the Ty1 PR plays an important role in the initiation of reverse transcription in vivo and that Gag is involved in primer-template annealing (8, 20).
Primer/template formation, the initial step in cDNA formation, is followed by RT-mediated DNA synthesis. The formation of cDNA in VLPs is a measure of the endogenous activity of RT. In this study, we also measured the ability of RT to synthesize DNA from an exogenous primer/template complex. Formation of this exogenous oligo(dG)/poly(C) primer/template complex is not temperature dependent. Our data show that RT activity is not inherently temperature sensitive. RT activities at 22 and 37°C in VLPs formed at 22°C are comparable. However, RT activity is greatly reduced in VLPs formed at high temperature, regardless of whether the in vitro assay is performed at 22 or 37°C. PR− mutant VLPs prepared at 22°C have readily detectable RT activity in these assays, indicating that processing is not required for activity and that RT is functionally active as part of the Gag-Pol-p199 polyprotein (21). These results argue that high temperature affects the conformation of the RT protein attained during synthesis, rendering it inactive. That the RT activity is not inherently temperature sensitive suggests that RT folding occurs during particle formation as part of the polyprotein and that RT, once folded, remains stable. Thus, particle formation at high temperature could result in an inactive conformation. Folding is likely to be a complex process, affected by interactions with other Ty1 or host factors (29). An alternative hypothesis is that exogenous RT activity is reduced simply because there is markedly less RT protein in VLPs formed at high temperature. We normalized to Gag proteins in the exogenous RT assays, and activity was still reduced 15-fold. Therefore, the inactivation of RT is significant.
The Gag-Pol-p199 protein is readily detectable at 37°C, but the overall levels of Pol protein products are reduced relative to levels seen at 22°C. Our immunoblot analyses suggest that the reduction of total Pol protein products at 37°C is greater than the reduction in Gag protein levels. We tested the effect of temperature on frameshifting and found that frameshifting is modestly affected, being approximately twofold more efficient at 37°C than at 22°C. A more efficient frameshifting mechanism would be expected to yield relatively more Pol protein product, not less. However, our findings resemble the results of a previous study on the effects of increased frameshifting efficiency. Frameshifting is mediated by a ribosomal stalling event caused by a rare tRNAArg codon at the +1 frameshifting site. Deletion of the gene for this tRNA increases frameshifting 3- to 17-fold (15). Interestingly, this mutation did not result in an accumulation of Pol products; rather the particles displayed a processing defect in that processed IN is undetectable in the mutant strain. As for VLPs made at high temperature, the processing of Gag in this mutant was not affected. Thus, an increase in frameshifting results in a processing defect, perhaps due to reduced PR activity in the context of particles forming in the presence of skewed Gag/Gag-Pol protein ratios. It therefore remains possible that a slight increase in frameshifting efficiency at high temperature could contribute to the observed Pol processing defect.
We have shown that the temperature sensitivity of transposition is due to a reduction in both PR and RT activity, resulting in a profound processing defect and a lack of any detectable cDNA in VLPs formed at 37°C. We hypothesize that aberrant folding at high temperature adversely affects the activities of both of these enzymes. Additionally, reduced protein levels, primer/template formation, and frameshifting effects may contribute to the phenotype of high-temperature VLPs. It is unknown whether the activity of IN is also affected by high temperature in vivo. Although reduced PR cleavage and RT function are major blocks to transposition at high temperature, the complete inhibition may well be due to the additive effects of defects in other steps as well. Such a complex control system allows adjustment to multiple environmental signals. Host mutants that partially restore transposition and processing at high temperature have been isolated (J. B. Keeney, unpublished data). Further characterization of these mutants will identify additional host cell-mediated events in the complex control of transposition.
Acknowledgments
This work was supported in part by NIH grants GM36481 to J.D.B. and GM54291-02 to J.B.K. and grants from The William J. von Liebig Foundation and the Howard Hughes Medical Institute (no. 52002813) to Juniata College.
REFERENCES
- 1.Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49:111-119. [DOI] [PubMed] [Google Scholar]
- 2.Belcourt, M. F., and P. J. Farabaugh. 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62:339-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., and S. B. Sandmeyer. 1991. Yeast transposable elements, p. 193-261. In J. Broach, E. Jones, and J. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- 5.Boeke, J. D., C. A. Styles, and G. R. Fink. 1986. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol. Cell. Biol. 6:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke, J. D., H. Xu, and G. R. Fink. 1988. A general method for the chromosomal amplification of genes in yeast. Science 239:280-282. [DOI] [PubMed] [Google Scholar]
- 7.Clare, J. J., M. Belcourt, and P. J. Farabaugh. 1988. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc. Natl. Acad. Sci. USA 85:6816-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristofari, G., D. Ficheux, and J.-L. Darlix. 2000. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 275:19210-19217. [DOI] [PubMed] [Google Scholar]
- 9.Curcio, M. J., and D. J. Garfinkel. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12:2813-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichinger, D. J., and J. D. Boeke. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955-966. [DOI] [PubMed] [Google Scholar]
- 13.Feng, Y. X., S. P. Moore, D. J. Garfinkel, and A. Rein. 2000. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 74:10819-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami, K., S. Pande, B. Faiola, D. P. Moore, J. D. Boeke, P. J. Farabaugh, J. N. Strathern, Y. Nakamura, and D. J. Garfinkel. 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeney, J. B., K. B. Chapman, V. Lauermann, D. F. Voytas, S. U. Astrom, U. von Pawel-Rammingen, A. Bystrom, and J. D. Boeke. 1995. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol. 15:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauermann, V., and J. D. Boeke. 1997. Plus-strand strong-stop DNA transfer in yeast Ty retrotransposons. EMBO J. 16:6603-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauermann, V., and J. D. Boeke. 1994. The primer tRNA sequence is not inherited during Ty1 retrotransposition. Proc. Natl. Acad. Sci. USA 91:9847-9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawler, J. F., Jr., G. V. Merkulov, and J. D. Boeke. 2001. Frameshift signal transplantation and the unambiguous analysis of mutations in the yeast retrotransposon Ty1 Gag-Pol overlap region. J. Virol. 75:6769-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawler, J. F., Jr., G. V. Merkulov, and J. D. Boeke. 2002. A nucleocapsid functionality contained within the amino-terminus of the Ty1 protein that is distinct and separable from proteolytic activity. J. Virol. 76:346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkulov, G. V., J. F. Lawler, Jr., Y. Eby, and J. D. Boeke. 2001. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J. Virol. 75:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkulov, G. V., K. M. Swiderek, C. B. Brachmann, and J. D. Boeke. 1996. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 70:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monokian, G. M., L. T. Braiterman, and J. D. Boeke. 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene. 139:9-18. [DOI] [PubMed] [Google Scholar]
- 24.Muller, F., K. H. Bruhl, K. Freidel, K. V. Kowallik, and M. Ciriacy. 1987. Processing of TY1 proteins and formation of Ty1 virus-like particles in Saccharomyces cerevisiae. Mol. Gen. Genet. 207:421-429. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, K. J., W. Tichelaar, N. Myers, N. R. Burns, S. J. Butcher, A. J. Kingsman, S. D. Fuller, and H. R. Saibil. 1997. Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J. Virol. 71:6863-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paquin, C. E., and V. M. Williamson. 1984. Temperature effects on the rate of Ty transposition. Science 226:53-55. [DOI] [PubMed] [Google Scholar]
- 27.Stahl, G., L. Bidou, J. P. Rousset, and M. Cassan. 1995. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23:1557-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tumer, N. E., B. A. Parikh, P. Li, and J. D. Dinman. 1998. The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae. J. Virol. 72:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm, M., M. Boutabout, and F. X. Wilhelm. 2000. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem. J. 348:337-342. [PMC free article] [PubMed] [Google Scholar]
- 30.Winston, F., K. J. Durbin, and G. R. Fink. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675-682. [DOI] [PubMed] [Google Scholar]
- 31.Xu, H., and J. D. Boeke. 1990. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc. Natl. Acad. Sci. USA 87:8360-8364. (Erratum, 88:2612, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youngren, S. D., J. D. Boeke, N. J. Sanders, and D. J. Garfinkel. 1988. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 8:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]