Abstract
bic is a novel gene identified at a common retroviral integration site in avian leukosis virus-induced lymphomas and has been implicated as a collaborator with c-myc in B lymphomagenesis. It lacks an extensive open reading frame and is believed to function as an untranslated RNA (W. Tam, Gene 274:157-167, 2001; W. Tam, D. Ben-Yehuda, and W. S. Hayward, Mol. Cell. Biol. 17:1490-1502, 1997). The oncogenic potential of bic, particularly its ability to cooperate with c-myc in oncogenesis, was tested directly by expressing c-myc and bic, either singly or in pairwise combination, in cultured chicken embryo fibroblasts (CEFs) and in chickens using replication-competent retrovirus vectors. Coexpression of c-myc and bic in CEFs caused growth enhancement of cells. Most importantly, chick oncogenicity assays demonstrated that bic can cooperate with c-myc in lymphomagenesis and erythroleukemogenesis. The present study provides direct evidence for the involvement of untranslated RNAs in oncogenesis and provides further support for the role of noncoding RNAs as riboregulators.
The proto-oncogene c-myc plays a critical role in the development of lymphomas in birds and mammals (11, 40). However, it is evident from work in both the murine and avian systems that activation of c-myc alone is not sufficient for full malignancy, and cooperation of c-myc with other proto-oncogenes is required to induce lymphomas (1, 2). Examples of myc collaborators in lymphomagenesis include the nuclear protein bmi-1 (22, 23, 29), ras G protein (3, 32, 50), the serine/threonine kinases PIM-1 (58-60) and Raf (32), the nonreceptor protein-tyrosine kinase Abl (24, 33), and the mitochondrial membrane protein bcl-2 (36, 51, 54).
In birds, avian leukosis virus (ALV)-induced lymphomas have provided a useful model for identification of proto-oncogenes which can cooperate with c-myc. The development of avian lymphoid leukosis is bursa dependent and believed to follow an orderly progression through distinct clinical stages: (i) transformed follicles; (ii) macroscopic bursal nodules; and (iii) metastasis (10). Insertional activation of c-myc appears to be an early event sufficient for the formation of transformed follicles (10, 57). Transformed follicle formation, however, is necessary but not sufficient for the development for avian leukosis, indicative of the requirement for multiple genetic events (5). cDNA microarray analysis for gene expression during myc-induced neoplastic transformation in the bursa of Fabricius shows that myc is more important in the early induction of lymphomas than in the maintenance of late-stage metastases (41).
To identify the genetic events necessary for inducing tumor progression in ALV-induced lymphomas, a double infection protocol involving sequential infection of chickens with ALVs of two different subgroups was used to generate multiple insertional mutations. This approach resulted in acceleration of lymphomagenesis and the identification of a common retroviral integration site (9). A putative proto-oncogene, bic, was isolated at this locus and characterized (56). bic is activated by promoter insertion and lacks an extensive open reading frame (ORF). The human and murine homologues of bic were recently cloned and show significant homology with the avian homolog over a short stretch of nucleotide sequence (55). However, similar to the avian homologue, the human and mouse bic genes do not have a long ORF. In addition, there is no homology among the multiple short ORFs present in these cDNAs. Moreover, the region of sequence homology is predicted by computer analysis to form a conserved imperfect RNA duplex. The lack of a conserved open reading frame and the evolutionary conservation of RNA secondary structures strongly suggest that bic functions as a noncoding RNA. Recent evidence has emerged that these noncoding RNAs can potentially function as riboregulators to control cell growth and differentiation, as well as transformation and oncogenesis (4, 13, 31).
Since integrations at the bic locus were usually found in conjunction with c-myc integrations in ALV-induced lymphomas, and the frequency of bic integrations was higher in metastatic tumors than in primary tumors, bic is implicated as a collaborator with c-myc in B lymphomagenesis and is presumably involved in late stages of tumor progression (9). bic gene rearrangements were also detected in several bursa-independent tumors (K. Parks and W. Hayward, unpublished data) induced by HB-1, which carries a recombinant c-myc/v-myc sequence (12). Therefore, it is likely that bic cooperates with myc in the pathogenesis of bursa-independent lymphomas. In view of the strong circumstantial evidence of the involvement of bic in tumorigenesis, we set out to demonstrate directly its oncogenic potential both in vitro and in vivo, particularly its collaborative interaction with c-myc in oncogenesis.
To characterize the oncogenic potential of bic, an efficient gene delivery system was needed. The replication-competent avian retroviral vector RCASBP provides a way of expressing the gene both in vitro and in vivo (11). This retrovirus vector may be used for studying oncogene cooperation in the avian system. Chicken cells can be made to express both the c-myc and bic genes by using two RCASBP viruses, each carrying an env gene of a different subgroup, which permits double infection to occur (15). Therefore, this approach facilitates studies in oncogene cooperation in the avian system in a way analogous to the crossing of transgenic mice carrying different oncogenes.
In an attempt to define directly the oncogenic potential of bic, in particular the possibility of cooperative interaction between c-myc and bic, we used these retroviruses to express bic either alone or in combination with c-myc in chicken embryo fibroblast (CEF) cultures and in chickens. The effects of overexpressing both c-myc and bic on cultured cell morphology and growth characteristics, as well as the incidence and latency of tumors in animals, were compared to the effects of overexpressing c-myc or bic alone. We demonstrated that overexpression of bic induced an enhancement of growth in CEFs overexpressing c-myc. Moreover, in vivo chicken oncogenicity assays showed that bic can collaborate with c-myc in the pathogenesis of lymphomas and erythroblastosis.
MATERIALS AND METHODS
Construction of RCASBP(A)-myc and RCASBP(B)-bic retroviruses.
The construction and characterization of the avian retrovirus vectors RCASBP(A) and RCASBP(B) have been described in detail elsewhere (15, 27, 45). RCASBP(A)-myc, previously designated RCASBP(A)c-myc (Δexon 1), was constructed as described and contains the chicken c-myc major open reading frame but lacks exon 1 sequences (44).
RCASBP(B)-bic was derived by insertion of a 505-bp ClaI insert from p Bic.exon2a.T3s into the ClaI site of the RCASBP(B) vector. p Bic.exon2a.T3s was constructed by ligating a 221-bp ClaI-BspHI fragment containing nucleotides 1 to 216 of bic exon 2 and a 304-bp BspHI-XhoI fragment of pXba-Ase into pBluescript SK plus (Stratagene) between the ClaI and XhoI sites. The ClaI-BspHI fragment was generated by PCR amplification of the cT465.R plasmid (56) using primer 1 as an upstream primer and primer 2 as a downstream primer under standard conditions, followed by digestion with ClaI and BspHI. pXba-Ase was derived by inserting a 433-bp insert fragment generated by digestion with AseI/Klenow and XbaI of pX-H0.5 between the XbaI and HindIII/Klenow sites of pBluescript SK plus (Stratagene). The ligation between the AseI/Klenow and HindIII/Klenow sites regenerated the HindIII sites. pX-H0.5 contains a 0.5-kb XbaI-HincII bic genomic fragment cloned into pBluescript SK plus (Stratagene). Primer 1 was 5′-CCATCGATTGTATCTCAAGGGGGAAAAAAACAG-3′, and primer 2 was 5′-TGTCACATGGAGGTCTTCTCAGCGTG-3′.
RNA analysis.
Total cellular RNA was isolated from confluent 10-cm plates of CEFs using RNAzol B (Biotecx Laboratories, Inc.) according to the manufacturer's instructions, which are modified from a single-step guanidinium isothiocyanate procedure for RNA isolation (8). Northern hybridization was performed as described before (56). DNA probes labeled to high specific activity (see below) were used in the Northern blots.
Protein analysis.
Total cell lysate was prepared from CEF cultures by homogenizing cultured cells in protein lysis buffer and pelleting the debris (30). Then 50 μg of total protein (as determined by the Pierce BCA assay) was subjected to standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) through a 10% gel, transferred to an Immobilon P membrane (Millipore), and blocked overnight at 4°C with 5% milk and 0.1% Tween 20. The membrane was then incubated with the primary antibody, which is a c-Myc monoclonal antibody (Santa-Cruz Biotechnology, Inc.), for 1 h at room temperature. The membrane was then rinsed for a total of 45 min and reacted with the secondary antibody for 1 h at room temperature. After rinsing for a total of 30 min, chemiluminescent detections were performed with the ECL detection kit (Amersham).
CEF transfection and virus production.
Primary CEF cultures (C/O, gs− chf−) prepared by standard techniques (20) were kindly provided by H. Hanafusa (The Rockefeller University). Cells were maintained at 37°C in F-10 medium containing 5% calf serum, 0.3% (wt/vol) tryptose phosphate broth, 0.19% sodium hydrogen carbonate, penicillin, and streptomycin.
CEF cultures were transfected with retrovirus vector DNA by the calcium phosphate precipitation method essentially as described before (52). Then 5 μg of highly purified DNA [RCASBP(A)-myc, RCASBP(B)-bic, RCASBP(A), or RCASBP(B)] was used in these transfections. Three passages after transfection, virus titer in overnight tissue culture supernatants was estimated by reverse transcriptase assays (60). Mass infection of the cells was routinely obtained after three passages, as judged from the virus titer in the tissue culture supernatant. These virus stocks were frozen at −80°C and used for infection of CEFs and chickens.
CEF infection.
For simultaneous double infection, CEFs were seeded on 60-mm plates at a density of 106 cells per plate and grown overnight in normal growth medium. After growing overnight in normal growth medium, RCASBP virus with A env [RCASBP(A)-myc and RCASBP(A)] was mixed with RCASBP virus with B env [RCASBP(B)-bic and RCASBP(B)] and added to cells at a multiplicity of infection of 5 to 10 each. The medium was then changed after 24 h. Mass infection was routinely obtained after 5 to 7 days, as demonstrated by Northern analysis and reverse transcriptase assays.
CEF cell growth assays.
For the cell growth assays, cells infected with different combinations of RCAS viruses were initially seeded on 60-mm plates at a density of 105 cells/plate and refed every 2 days. Cells were counted every 2 days with a hemacytometer. This experiment was done in duplicate.
Chicken oncogenicity assays.
Eighteen-day gs−chf− embryos (SPAFAS, Inc.) received 0.1 ml of a virus suspension containing 106 infectious units of RCAS virus of each envelope type per ml injected into a chorioallantoic vein. One-day-old chicks were transferred to a separate room of the Sloan-Kettering Institute animal facility for maintenance. Chickens inoculated with different viruses were maintained in separate cages and observed for disease. Chickens that appeared severely ill were euthanized in a CO2 chamber. All surviving birds were sacrificed at 16 weeks [for animals infected with RCASBP(A)-myc and RCASBP(B)-bic or RCASBP(A)-myc and RCASBP(B)] or at 20 weeks (for animals infected with RCASBP(A) and RCASBP(B)-bic or RCASBP(A) and RCASBP(B)]. Postmortem examinations were performed on all chickens. Tissues for histopathologic analyses were placed in 10% buffered Formalin and processed. Histopathological diagnoses were made with the assistance of H. Nguyen (Weill Medical College of Cornell University). Samples were taken from various diseased organs and frozen at −70°C for further analysis.
Isolation of high-molecular-weight tumor DNA.
Isolation of high-molecular-weight DNA was performed as described with modifications (12). From 0.8 to 1 g of tumor tissue stored frozen at −70°C was minced to frozen powder by using a pestle and homogenized in ice-cold TNE (0.15 M NaCl, 0.05 M Tris-HCl [pH 8.0], 5 mM EDTA) by hand in a Dounce homogenizer. Proteinase K (Boehringer Mannheim) was added to a final concentration of 500 μg/ml and SDS to a final concentration of 1%. Preparations were incubated for 3 h at 68°C and extracted twice with phenol, once with phenol-chloroform-isoamyl alcohol (25:24:1), and once with chloroform-isoamyl alcohol (24:1). The DNA was then ethanol precipitated and redissolved in 4 ml of TE (Tris-EDTA).
To remove residual RNA, DNase-free RNase (Boehringer Mannheim) was added to a final concentration of 1 μg/ml and SDS to a final concentration of 0.1%. After incubating at 37°C for 1 h, the DNA sample was extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform-isoamyl alcohol. Finally, the DNA was ethanol precipitated and redissolved in 1 ml of TE.
Restriction enzyme digestion and Southern blotting.
Restriction enzyme digestion of 15 μg of high-molecular-weight tumor DNA was carried out in a volume of 200 μl with a fivefold excess (80 U) of enzymes. After digestion was complete, the sample was extracted with phenol-chloroform and ethanol precipitated.
Digested DNAs were run on 0.7% agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer at 1 V/cm for 14 h and blotted to Zeta-probe GT membranes (Bio-Rad Laboratories) according to the procedure of Southern (53). Blots were rinsed briefly in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and baked in a vacuum oven at 80°C for 30 min. The filters were prehybridized, hybridized, and washed according to the manufacturer's recommendations (Bio-Rad).
DNA probes.
Radioactively labeled DNA probes were labeled to high specific activity (>5 × 108 cpm/μg of DNA) by incorporation of [α-32P]dCTP (3,000 Ci/mmol; NEN) into DNA by the random priming method (16) using the random primed DNA labeling kit (Boehringer Mannheim). The myc probe was a ClaI-EcoRI fragment isolated from pBN22.exon3, which was made by subcloning the ClaI-EcoRI fragment from pBN22 into pBluescript SK plus (Strategene). pBN22 was derived from pc myc-1 (39) by subcloning a SalI-EcoRI fragment into pBR322. The 0.96-kb ClaI-EcoRI fragment from pBN22.exon3 represents exon 3 of c-myc. The bic probe is a 0.5-kb ClaI fragment isolated from p Bic.exon2a (see above). The probe for chicken immunoglobulin light chain was a 2.7-kb BamHI-SalI fragment of the λ(J plus C) plasmid kindly provided by Jean-Claude Weill and Claude-Agnes Reynaud (49).
Statistical methods.
Comparisons of Kaplan-Meier survival curves were performed using the log-rank test. The incidence of tumors in different groups was compared using either the chi-square test or the Fisher exact test. The latency of tumors was compared using the Mann-Whitney U test. All statistical computations were performed by the Statview (version 4.5) statistical analysis program (Abacus Concepts, Inc.)
RESULTS
Construction of RCASBP(A)-myc and RCASBP(B)-bic.
As a vehicle for the expression of c-myc and bic cDNA sequences in vitro and in vivo, the replication-competent avian retrovirus vectors RCASBP(A) and RCASBP(B) were used, which contain the subgroup A env gene and subgroup B env gene, respectively (15, 27). The myc-expressing retrovirus, designated here RCASBP(A)-myc, contained a cDNA representing the coding exons (2 and 3) of chicken c-myc (for details, see Materials and Methods). Since a stop codon in frame with the ATG start codon of the gag gene (in-frame terminator) is present downstream of the splice acceptor in RCASBP(A)-myc (Fig. 1), the p55 myc protein is expressed from the normal c-myc initiation codon at the 5′ end of the c-myc cDNA (27, 44).
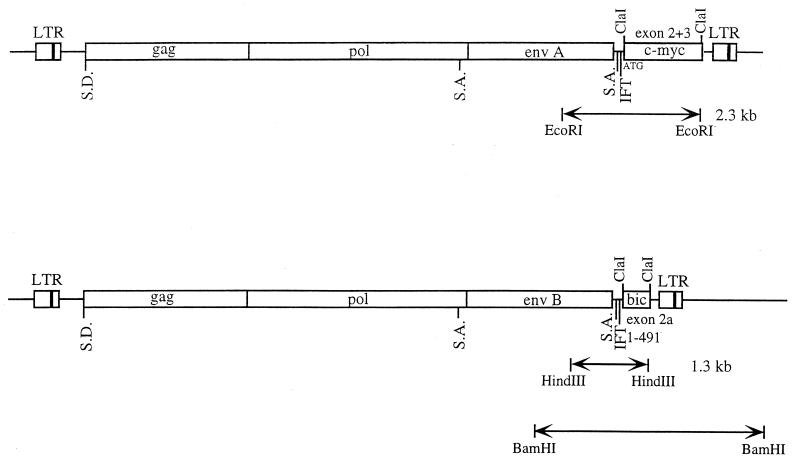
FIG. 1.
Structures of the RCASBP(A)-myc and RCASBP(B)-bic proviruses. c-myc and bic cDNA sequences were inserted into the ClaI sites of the replication-competent retrovirus vectors RCASBP(A) and RCASBP(B), respectively. The c-myc insert consists of chicken c-myc exons 2 and 3. The bic insert comprises nucleotides 1 to 491 of bic exon 2. The virus internal myc-specific (2.3-kb EcoRI) and bic-specific (1.3-kb HindIII) fragments are shown. The BamHI RCASBP(B)-bic virus-cell junction fragment is also depicted. S.D., splice donor; S.A., splice acceptor; IFT, in-frame terminator.
Based on the structures of the 2.6-kb and 1.0-kb tumor chimeric transcripts derived from alternative polyadenylation, the biological activity of avian bic is believed to reside in exon 2a, the small second exon (56). Therefore, the bic-expressing retrovirus was designed to generate subgenomic mRNAs that were similar in structure to the shorter chimeric bic transcripts seen in the tumors. An artificially constructed avian bic exon 2a was introduced into RCASBP(B), and the resulting vector was designated RCASBP(B)-bic (see Materials and Methods and also Fig. 1). The bic cDNA insert in RCASBP(B)-bic consisted of nucleotides 1 to 491 of bic exon 2. The introduction of bic exon 2a into an RCASBP vector with the subgroup B env segment facilitated the studies of collaboration between c-myc and bic, both in vitro and in vivo. Since the vectors RCASBP(A) and RCASBP(B) themselves are capable of inducing tumors in chickens by retroviral insertion, they were included in all controls in the double infection protocol.
Expression of c-myc and bic in CEFs using retrovirus vectors.
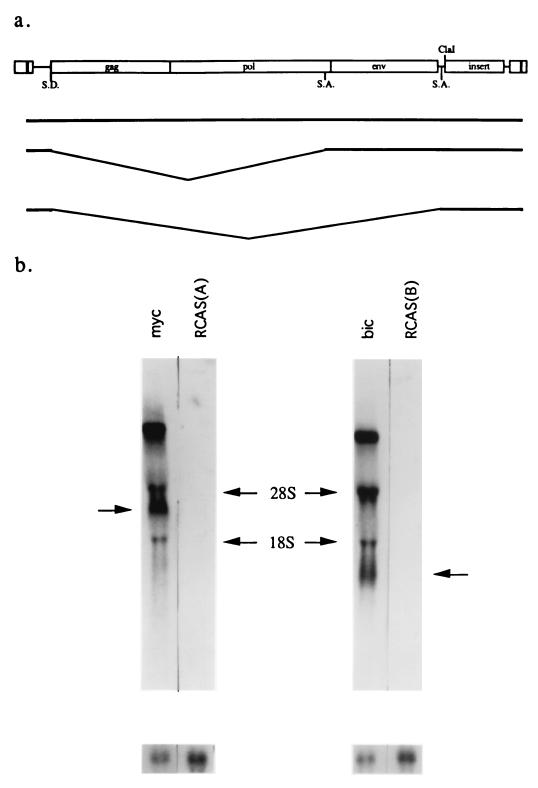
RCASBP(A)-myc and RCASBP(B)-bic vector DNAs were transfected into separate CEF cultures, and the transfected cells were passaged every 3 to 4 days. After three passages (about 10 to 12 days), mass infection was achieved, as indicated by the high titer of virus estimated by three successive reverse transcriptase assays. RNA expression from these two RCAS viruses in the virus-producing cultures was then assayed by Northern analysis. Three RNA species were expected from each of these constructs: full-length viral genome RNA; transcripts that are spliced from the gag splice donor to the splice acceptor 5′ of env; and transcripts that are spliced from the gag splice donor to the src splice acceptor immediately upstream of the ClaI site. c-myc and bic were expressed from the smallest spliced subgenomic RNAs (Fig. 2a). Northern hybridizations using exon 3 of c-myc or exon 2a of bic as a probe detected the expected transcripts of 8.9, 4.2, and 2.4 kb for RCASBP(A)-myc and 8.0, 3.3, and 1.5 kb for RCASBP(B)-bic (Fig. 2b). The two bands with mobilities similar to the 28S and 18S rRNAs most likely represent hybridization of probe to viral RNAs which comigrated nonspecifically with the rRNAs.
FIG. 2.
Expression of c-myc and bic in CEFs using RCASBP(A)-myc and RCASBP(B)-bic. (a) Schematic of expected RNAs generated from RCASBP(A)-myc and RCASBP(B)-bic constructs. These vectors express cDNA inserts as spliced subgenomic mRNA. SD, splice donor site; SA, splice acceptor site. (b) Northern blot analysis of c-myc and bic expression. Total RNA derived from CEFs mass-infected with RCASBP(A)-myc (lane myc), RCASBP(A) [lane RCAS(A)], RCASBP(B)-bic (lane bic), or RCASBP(B) [lane RCAS(B)] were probed with either a radiolabeled c-myc exon 3 cDNA probe (left) or a bic exon 2 probe (right). The subgenomic mRNA expressing c-myc or bic is indicated by an arrow. The blot was stripped and rehybridized to a chicken actin probe as a control for loading. The radioactive bands observed at the 28S and 18S regions are probably due to hybridization of probe sequence to viral RNAs that comigrated nonspecifically with the rRNAs.
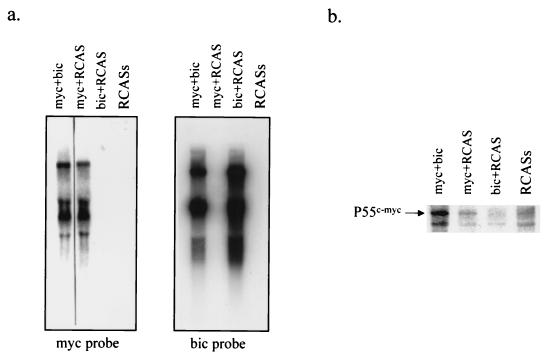
As an initial effort to characterize the biological potential of bic, we determined the effects of overexpression of c-myc and bic relative to c-myc alone in vitro, as well as the effects of overexpression of bic alone. CEF cultures infected with RCASBP(A)-myc plus RCASBP(B)-bic, RCASBP(A)-myc plus RCASBP(B), RCASBP(A) plus RCASBP(B)-bic, and RCASBP(A) plus RCASBP(B) were generated through a simultaneous double infection protocol (see Materials and Methods). These cultures were first examined for c-myc and bic mRNA expression by Northern blot analysis and for c-myc protein expression by Western blot analysis. RCASBP(A)-myc plus RCASBP(B)-bic-infected and RCASBP(A)-myc plus RCASBP(B)-infected cells expressed significantly higher levels of c-myc transcripts (Fig. 3a, left panel) and protein (Fig. 3b). In contrast, RCASBP(A) plus RCASBP(B)-bic-infected and RCASBP(A) plus RCASBP(B)-infected cells expressed only very low levels of c-myc (Fig. 3a, right panel, and 3b). High levels of bic transcripts were detected in RCASBP(A)-myc plus RCASBP(B)-bic-infected and RCASBP(A) plus RCASBP(B)-bic-infected cells, while the expression of endogenous bic was undetectable in RCASBP(A)-myc plus RCASBP(B)-infected and RCASBP(A) plus RCASBP(B)-infected cells. bic expression did not significantly alter expression of c-myc at either the mRNA (Fig. 3a) or protein (Fig. 3b) level, and similarly, c-myc expression did not affect bic expression (Fig. 3a).
FIG. 3.
Expression of c-myc and bic in doubly infected CEFs. (a) CEF cultures were simultaneously infected with two viruses as indicated, and total RNA from these cultures was subjected to Northern analysis as described in Materials and Methods. myc plus bic, RCASBP(A)-myc and RCASBP(B)-bic; myc plus RCAS, RCASBP(A)-myc and RCASBP(B); bic plus RCAS, RCASBP(A) and RCASBP(B)-bic; RCASs, RCASBP(A) and RCASBP(B). (b) Protein lysates prepared from doubly infected CEF cultures were separated by electrophoresis and subjected to Western blot analysis using monoclonal antibodies against c-Myc. The p55 myc protein expressed by RCASBP(A)-myc is indicated. The lanes are designated as in a. The faster-migrating bands represent cross-reactive proteins.
Enhanced growth of CEFs infected with RCASBP(A)-myc and RCASBP(B)-bic.
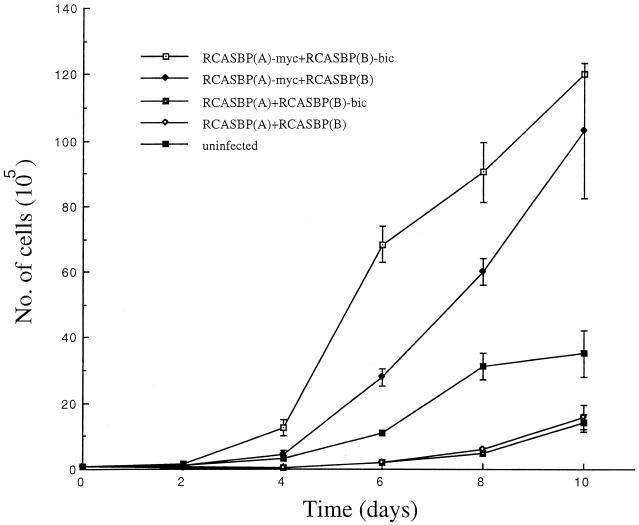
When the growth curves for CEFs mass infected with different combinations of RCASBP viruses were determined over a period of 10 days, the overall growth rate for the 10-day period for CEFs infected with RCASBP(A)-myc and RCASBP(B)-bic was not significantly different compared to that of CEFs infected with RCASBP(A)-myc and RCASBP(B). However, we found that the growth rate for the former group was significantly higher during the initial and middle phases of the growth curves (days 2 to 6) compared to the latter (Fig. 4). On the other hand, the CEFs infected with RCASBP(A) and RCASBP(B)-bic had a growth rate similar to that of CEFs infected with RCASBP(A) and RCASBP(B), implying that bic alone does not promote cell growth. In fact, both of these CEF cultures grew more slowly than uninfected fibroblasts, probably because of the cytotoxic effects caused by RCASBP(B) (Fig. 4). These results suggest that bic can synergize with c-myc to promote growth of CEFs, possibly in a cell density-dependent manner.
FIG. 4.
Enhanced growth of CEFs overexpressing c-myc and bic. CEFs doubly infected with the indicated viruses were plated at a starting density of 105 cells/plate. The cultures were trypsinized and counted at the indicated time points. The experiment was done in duplicate. The error bars represent the standard deviation at each time point.
Chickens infected with both RCASBP(A)-myc and RCASBP(B)-bic show poorer overall survival.
The oncogenic potential of bic was further characterized in vivo by infecting chickens with different combinations of RCAS viruses. Four groups of chickens were infected at day 18 of embryonation with one of the following combinations of viruses: (i) RCASBP(A)-myc plus RCASBP(B)-bic; (ii) RCASBP(A)-myc plus RCASBP(B); (iii) RCASBP(A) plus RCASBP(B)-bic; and (iv) RCASBP(A) plus RCASBP(B). Comparison of the incidence and latency of tumors in these groups of animals allows determination of whether myc and bic can collaborate in oncogenesis. Different groups of animals were reared in separate cages and monitored for the appearance of diseases. Chickens which died or were sacrificed because of overt signs of severe distress were necropsied, and pathological diagnoses were made microscopically.
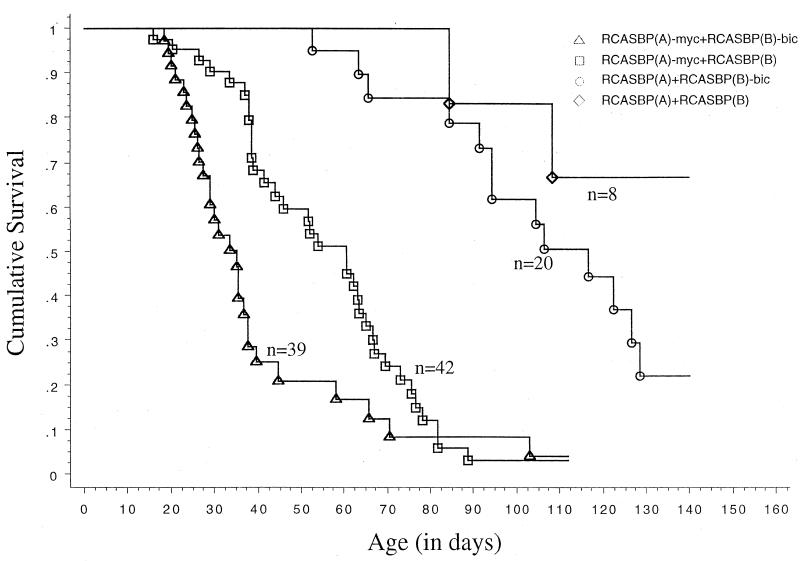
Approximately 78% of the animals died of neoplastic diseases. Survival curves for the four groups of animals which died of neoplastic diseases are shown in Fig. 5. Infection with RCASBP(A)-myc and RCASBP(B)-bic was associated with a significantly higher mortality from neoplastic disease compared to the other three groups [P < 0.01 versus RCASBP(A)-myc plus RCASBP(B) group and P < 0.0001 versus RCASBP(A) plus RCASBP(B)-bic and RCASBP(A) plus RCASBP(B) groups, respectively]. The median survival time for animals infected with RCASBP(A)-myc and RCASBP(B)-bic was 35.0 days, compared to 60.5 days for those infected with RCASBP(A)-myc and RCASBP(B) and 116.5 days for those infected with RCASBP(A) and RCASBP(B)-bic. Fewer than 50% of the animals died of neoplastic diseases in the RCASBP(A) plus RCASBP(B) group before the end of the study.
FIG. 5.
Kaplan-Meier survival curves (neoplastic diseases only) for animals infected with different retroviruses. Eighteen-day chick embryos were doubly infected with retrovirus vectors as indicated. Animals inoculated with different viruses were maintained in separate cages and observed for disease. Each death event (indicated by a vertical line) represents the death of an animal due to neoplastic disease.
Based on the above results, the poorer survival of animals infected with RCASBP(A)-myc and RCASBP(B)-bic relative to those infected with RCASBP(A)-myc and RCASBP(B) can be attributed to synergism between RCASBP(A)-myc and RCASBP(B)-bic, implying that bic may cooperate with c-myc in oncogenesis. Though not statistically significant, animals infected with RCASBP(A) and RCASBP(B)-bic tended to have a higher mortality from neoplastic disease than those infected with RCASBP(A) and RCASBP(B) (P = 0.13). This may reflect insertional activation by RCASBP(A) of bic-complementing proto-oncogenes, for instance, c-myc.
Retroviral infections induced various neoplasms in all four groups of animals.
Tumors were frequently induced in all four groups by retroviral infections. Various neoplastic diseases occurred in these animals, including lymphomas, erythroblastosis, myelocytomatosis, sarcomas, and adenocarcinomas.
Lymphomas in this experiment predominantly involved the liver. In many cases, particularly in histologically high-grade lymphomas, the liver was grossly enlarged, with homogeneous white areas. However, contrary to classic avian lymphoid leukosis (10), macroscopic bursal nodules were absent in all but one case, which was found in an animal infected with RCASBP(A) and RCASBP(B)-bic. In some of the bursas, transformed follicles may be seen microscopically. Histologically, lymphomatous foci in the liver consisted either of mainly medium and large lymphocytes or a mixture of small, medium, and large lymphocytes. These histological observations with the lymphomas appeared similar to HB-1-induced lymphomas (12). The lymphomas were classified as either low-grade or high-grade based on the degree of infiltration in the liver by lymphomatous tissues. In low-grade lymphomas, the liver contained multiple, relatively small lymphoid aggregates. High-grade lymphomas were characterized by more diffuse and extensive infiltration by neoplastic lymphoid cells. Two of the high-grade lymphomas observed were of the nodular type. When the extent of infiltration was not great enough to justify the diagnosis of lymphoma, the diagnosis of lymphoid hyperplasia was given.
These lymphomas were examined for the B-cell phenotype by testing for the presence of rearrangements in the immunoglobulin light-chain λ locus (49). Only one of the lymphomas (836L) showed obvious light-chain rearrangement (data not shown). A faint rearranged restriction fragment was observed in the Southern blot for another lymphoma (318L) which was heavily infiltrated by neoplastic lymphoid cells, implying that only a small portion of tumor cells in that lymphoma had the light-chain rearrangement (data not shown). The above results indicate that the vast majority of lymphomas observed in this experiment were of early B-cell or T-cell origin.
Erythroblastosis is a leukemia of cells of the erythroid lineage. The target cell for transformation is thought to be an intravascular hemacytoblast in the bone marrow (46). Microscopic examinations of the visceral organs, particularly the liver and kidneys, showed accumulation of erythroblasts in the capillaries, often resulting in dilation of the hepatic sinuses.
Myelocytomatosis usually develops as masses at the skull and sternal and pelvic bones (38) and is a neoplasm of cells in the myeloid lineage. However, in this experiment, the liver was the predominant organ involved. Grossly, the liver was enlarged and infiltrated with diffuse and nodular yellow-white growths. The tumor consisted of compact masses of neoplastic myelocytes, which destroy and replace the hepatic parenchyma.
Sarcomas observed in this experiment consisted of fibrosarcomas, myxosarcomas, and chondrosarcomas. These tumors principally involved the liver and kidneys. Adenocarcinomas observed in this experiment occurred most frequently in the liver. They were also rarely found in the pancreas and lungs.
bic collaborates with c-myc in the pathogenesis of lymphomas and erythroblastosis.
The incidence of different neoplasms among the four groups of animals is shown in Table 1. The median time to death of animals with these neoplasms is also indicated. Animals infected with RCASBP(A)-myc and RCASBP(B) had a relatively high incidence of mylelocytomatosis, sarcomas, and adenocarcinomas compared to those infected with RCASBP(A) and RCASBP(B), none of which developed any of the three tumors. Statistical significance was only demonstrated for myelocytomatosis (P < 0.05), however, presumably because of the small sample size of the RCASBP(A) plus RCASBP(B)-infected group. Moreover, lymphomas developed with a shorter latency in the RCASBP(A)-myc plus RCASBP(B)-infected group relative to the RCASBP vector-infected control group (P < 0.05). Our results are consistent with previous studies in both the avian and murine systems, which demonstrated that myc plays a principal role in the pathogenesis of a wide variety of tumors (12, 26, 35, 38).
TABLE 1.
Summary of pathology of infected animalsa
| Group | Lymphoma
|
Erythroblastosis
|
Myelocytomatosis
|
Sarcoma
|
Adenocarcinoma
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) | MTD (days) | No. (%) | MTD (days) | No. (%) | MTD (days) | No. (%) | MTD (days) | No. (%) | MTD (days) | |
| RCASBP(A)-myc + RCASBP(B)-bic (n = 39) | 15 (38.5) | 27.5b | 4 (10.3)c | 32.5d | 5 (12.8)e | 44.5 | 7 (18.0) | 58.0 | 6 (15.4) | 51.2 |
| RCASBP(A)-myc + RCASBP(B) (n = 42) | 13 (31.0) | 53.0 | 0 (0.0) | — | 18 (42.9)f | 51.8 | 10 (23.8) | 65.8 | 11 (26.2) | 62.0 |
| RCASBP(A) + RCASBP(B)-bic (n = 20) | 10 (50.0) | 99.5 | 3 (15.0) | 94.5 | 0 (0.0) | — | 1 (5.0) | 65.5 | 0 (0.0) | — |
| RCASBP(A) + RCASBP(B) (n = 8) | 2 (25.0) | 96.5 | 0 (0.0) | — | 0 (0.0) | — | 0 (0.0) | — | 0 (0.0) | — |
Data are number (percent) of animals. MTD, median time to death.
P < 0.05 versus other three groups.
P < 0.05 versus the RCASBP(A)-myc plus RCASBP(B) group.
P < 0.05 versus the RCASBP(A) plus RCASBP(B)-bic group.
P < 0.01 versus the RCASBP(A)-myc plus RCASBP(B) group.
P < 0.05 versus the RCASBP(A) plus RCASBP(B) group.
Comparison of the incidence and latency of tumors for animals infected with RCASBP(A)-myc and RCASBP(B)-bic with those for the animals infected with RCASBP(A)-myc and RCASBP(B) revealed three major differences. First, lymphomas developed with a shorter latency in the former group (P < 0.02), although the incidence of lymphomas was similar for the two groups (Table 1). This difference can mainly be attributed to the acceleration in development of high-grade lymphomas (P < 0.05), although there appears to be a tendency towards shorter latency (P < 0.10) in the development of low-grade lymphomas for the RCASBP(A)-myc plus RCASBP(B)-bic-infected group as well (Table 2). On the other hand, animals infected with RCASBP(A) and RCASBP(B)-bic developed high-grade lymphomas with a relatively long latency [P < 0.001 versus RCASBP(A)-myc plus RCASBP(B)-bic group]. Second, the incidence of erythroblastosis was significantly greater for animals infected with RCASBP(A)-myc and RCASBP(B)-bic (P < 0.05). While 3 of 20 of the animals infected with RCASBP(A) and RCASBP(B)-bic also developed erythroblastosis, its latency was significantly longer compared with that for animals infected with RCASBP(A)-myc and RCASBP(B)-bic (P < 0.05).
TABLE 2.
Incidence and latency of high-grade and low-grade lymphomasa
| Group | High-grade
|
Low-grade
|
||
|---|---|---|---|---|
| No. (%) | MTD (days) | No. (%) | MTD (days) | |
| RCASBP(A)-myc plus RCASBP(B)-bic (n = 39) | 8 (20.5) | 32.3b | 8 (20.5) | 25.5 |
| RCASBP(A)-myc plus RCASBP(B) (n = 42) | 7 (16.7) | 60.5 | 7 (16.7) | 38.0 |
| RCASBP(A) plus RCASBP(B)-bic (n = 20) | 10 (50.0) | 99.5 | 0 (0.0) | — |
| RCASBP(A) plus RCASBP(B) (n = 8) | 2 (25.0) | 96.5 | 0 (0.0) | — |
MTD, median time to death.
P < 0.05 versus other three groups.
In view of the above results, we can conclude that c-myc and bic cooperate in the pathogenesis of lymphomas and erythroblastosis. The collaboration between c-myc and bic is seemingly restricted to hematopoietic malignancies of the lymphoid and erythroid lineage, since there was no difference in latency of other tumors between the RCASBP(A)-myc plus RCASBP(B)-bic- and RCASBP(A)-myc plus RCASBP(B)-infected groups. Lastly, animals infected with RCASBP(A)-myc and RCASBP(B)-bic had a lower incidence of myelocytomatosis compared to those infected with RCASBP(A)-myc and RCASBP(B) (P < 0.01). This is most likely due to the loss of susceptible animals from high-grade lymphomas and erythroblastosis in the former group. Both of these tumors had a shorter latency than myelocytomatosis.
Of the 20 animals infected with RCASBP(A) and RCASBP(B)-bic, 10 developed lymphomas (all high grade), 3 had erythroblastosis, and 1 had sarcoma. No myelocytomatosis or adenocarcinomas were found. No statistically significant difference was detected in the incidence or latency of the tumors when animals infected with RCASBP(A) and RCASBP(B)-bic were compared with animals infected with RCASBP(A) and RCASBP(B) (Tables 1 and 2). Despite the lack of statistical significance, the incidence of both erythroblastosis and lymphomas in the RCASBP(A) plus RCASBP(B)-bic-infected group appeared to be somewhat higher than in the RCASBP vector-infected control group (3 of 20 versus 0 of 8 and 10 of 20 versus 2 of 8, respectively). It is conceivable that the former group of animals may indeed have a higher tendency to develop lymphomas and erythroblastosis than the latter, probably as a result of insertional activation by RCASBP(A) of proto-oncogenes which can cooperate with bic, for example, c-myc. This predisposition may not have been statistically demonstrated in this experiment because of the small sample sizes of the two groups.
Proviral integrations of RCASBP(A)-myc and RCASBP(B)-bic in lymphomas and erythroblastosis.
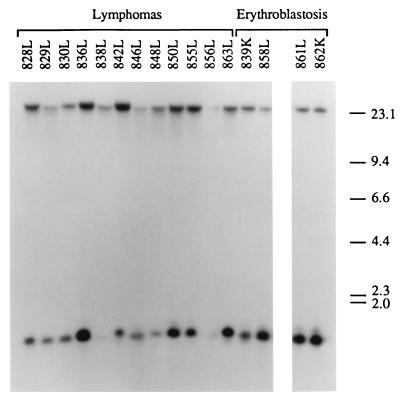
The presence of proviral integrations of RCASBP(A)-myc and RCASBP(B)-bic in lymphomas and erythroblastosis tumors from animals infected with RCASBP(A)-myc and RCASBP(B)-bic were determined by detecting myc-containing and bic-containing proviral fragments in the host genome. Restriction digestion of RCASBP(A)-myc and RCASBP(B)-bic proviruses with EcoRI and HindIII produces a 2.3-kb myc-positive fragment and a 1.3-kb bic-positive fragment, respectively (see Fig. 1). Hybridization of Southern blots of EcoRI-restricted tumor genomic DNA with a c-myc exon 3 probe identified the 2.3-kb myc-positive fragment, as well as the endogenous 15-kb c-myc EcoRI fragment (data not shown). Southern hybridization of HindIII-digested tumor genomic DNA with a bic exon 2a probe also detected the 1.3-kb bic-positive fragment and the endogenous 23-kb bic locus HindIII fragment in all the tumors examined (Fig. 6). The detection of myc- and bic-positive proviral fragments indicates that both RCASBP(A)-myc and RCASBP(B)-bic were present in these tumors.
FIG. 6.
Detection of proviral integrations of RCASBP(B)-bic in lymphomas and erythroblastosis tumors. High-molecular-weight DNA isolated from the tumor tissues (L, liver; K, kidney) of birds infected with RCASBP(A)-myc and RCASBP(B)-bic were digested with HindIII and subjected to Southern blot analysis using a radiolabeled bic exon 2a probe (see Fig. 1). The band of lower mobility represents the endogenous bic gene restriction fragment. Fragment sizes are indicated in kilobases.
No deletions or rearrangements of these two proviruses were detected in the tumors. The endogenous myc and bic loci appeared intact, as no novel c-myc- or bic-containing fragments were detected in addition to the expected endogenous and proviral fragments. The absence of proviral integrations in the vicinity of the myc and bic loci is expected, since these two oncogenes are carried by the proviruses.
Clonality of lymphomas and erythroblastoses containing both RCASBP(A)-myc and RCASBP(B)-bic proviruses.
The lymphomas and erythroblastosis tumors were then tested for clonality by examining the presence of junction fragments by Southern analysis. Junction fragments are restriction fragments consisting of viral sequences linked to adjacent cellular sequences. Junction fragments for independent proviral integrations differ in size, and they would not be detected by Southern analysis if proviruses have integrated at multiple different sites in the host genome in the cell population. However, Southern analysis would allow the detection of discrete viral junction fragments if the tumors are clonal.
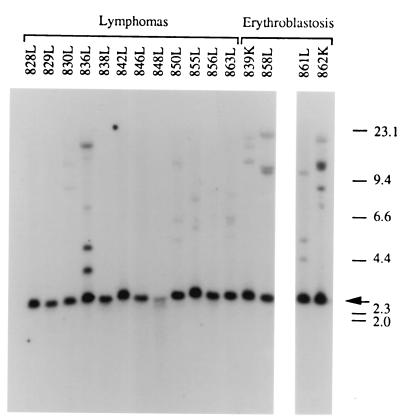
For detection of clonality with respect to bic, DNA isolated from tumors containing both RCASBP(A)-myc and RCASBP(B)-bic was digested with BamHI, which generates a virus junction fragment by cutting the RCASBP(B)-bic provirus in the env gene and the cellular genome 3′ of the provirus (see Fig. 1). Virus junction fragments were then detected using the bic exon 2a probe on Southern analysis. As shown in Fig. 7, the 2.5-kb germ line bic locus BamHI fragment was present in all the tumors examined. Virus junction fragments were detected in 5 of 12 lymphomas examined and 4 of 4 erythroblastosis tumors. At least two junction fragments were detected in each of these nine tumors. Each of these junction fragments may actually represent an independent clonal cell population. Alternatively, a clonal cell population within these tumors may harbor two or more RCASBP(B)-bic proviral integrations, resulting in the appearance of radioactive bands of approximately equal intensities in Southern blots. Therefore, the detection of multiple junction fragments in these tumors may indicate monoclonality or oligoclonality. However, tumors 836L and 862L are most likely to be oligoclonal, since multiple junction fragments of different intensities were observed in Southern analysis.
FIG. 7.
Detection of RCASBP(B)-bic virus junction fragments in lymphomas and erythroblastosis tumors. BamHI-digested tumor DNA was hybridized to a radiolabeled bic exon 2a probe in Southern blot analysis (see Fig. 1). The 2.5-kb endogenous BamHI fragment is indicated by an arrow. Bands of lower mobilities represent virus junction fragments. Sizes are shown in kilobases.
All five of the lymphomas with detectable virus junction fragments were high-grade lymphomas, and none of the five low-grade lymphomas examined showed virus junction fragments on Southern blots. These results suggest that while the low-grade lymphomas observed in this experiment are polyclonal, the high-grade lymphomas are monoclonal or oligoclonal. Based on these clonality analyses, it is likely that additional genetic alterations besides deregulated c-myc and bic are required for the development of high-grade lymphomas and erythroblastosis.
Rearrangements of c-myc and bic genes in lymphomas from animals infected with RCASBP(A) and RCASBP(B)-bic or RCASBP(A)-myc and RCASBP(B).
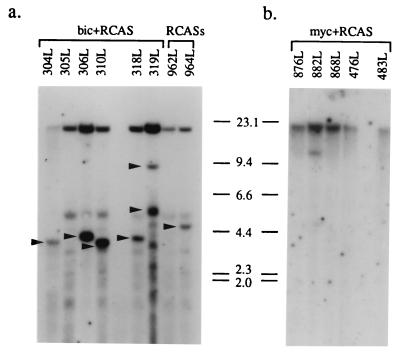
If myc can collaborate with bic in lymphomagenesis, it is likely that lymphomas from animals infected with RCASBP(A) plus RCASBP(B)-bic or RCASBP(A)-myc plus RCASBP(B) harbor c-myc or bic integrations, respectively. Indeed, c-myc rearrangements were detected in five of six tumors examined (304L, 306L, 310L, 318L, and 319L) from the RCASBP(A) plus RCASBP(B)-bic-infected group using a c-myc exon 3 probe (Fig. 8a). Examination of the two lymphomas (962L and 964L) from animals infected with RCASBP(A) and RCASBP(B) also revealed c-myc rearrangements in one of them (964L) (Fig. 8a).
FIG. 8.
Detection of c-myc and bic gene arrangements in lymphomas. (a) Lymphoma DNA from animals doubly infected with RCASBP(A) and RCASBP(B)-bic (bic plus RCAS) or RCASBP(A) plus RCASBP(B) (RCASs) was digested with EcoRI and hybridized to a radiolabeled c-myc exon 3 probe in Southern blot analysis. The 15-kb germ line EcoRI fragment is present in all samples. Bands which represent rearranged c-myc alleles are indicated by arrowheads. The multiple bands of similar sizes which are observed in all the lanes are likely due to nonspecific hybridization of probe sequences. (b) High-grade lymphoma DNA from animals infected with RCASBP(A)-myc and RCASBP(B) (myc plus RCAS) was digested with HindIII, and Southern hybridization was performed using a radiolabeled bic exon 2a probe. The 23-kb endogenous HindIII fragment is present in all samples. The additional fragment observed in lane 882L represents a rearranged bic allele.
To test if proviral integrations at bic were present in lymphomas from animals infected with RCASBP(A)-myc and RCASBP(B), HindIII-digested tumor DNAs from eight of these lymphomas were hybridized to a bic exon 2a probe in Southern blots. As shown in Fig. 8b, bic gene rearrangement was observed in one of the high-grade lymphomas (882L) examined. Interestingly, this lymphoma has a relatively short latency compared to the other high-grade lymphomas. These results are consistent with a cooperative interaction between c-myc and bic in lymphomagenesis.
DISCUSSION
Cooperation of c-myc and bic in lymphomagenesis and erythroleukemogenesis.
Coexpression of c-myc and bic does not fully transform cultured cells. Although cells infected with RCASBP(A)-myc and RCASBP(B)-bic showed enhanced growth at a relatively low cell density, these cells did not demonstrate additional morphological alterations compared to RCASBP(A)-myc plus RCAS(B)-infected cells. Moreover, CEFs expressing both c-myc and bic did not form colonies in soft agar despite repeated attempts (data not shown). This finding suggests that expression of c-myc and bic alone may not be sufficient for full transformation and that additional genetic alterations are probably required.
Two mechanisms may account for the enhanced growth observed in cells expressing c-myc and bic. First, bic may further increase the proliferation rate of c-myc-expressing cells. Alternatively, the apparent enhanced growth may be due to suppression by bic of myc-induced apoptosis. Besides being a positive regulator of cellular proliferation, c-myc has also been demonstrated to be a potent inducer of apoptosis when expressed in the absence of serum or growth factors (14). It is conceivable that bic may prevent apoptosis in these cells, resulting in an apparent increase in cell growth. However, in the present studies, CEF cultures were maintained in high concentrations of serum and hence were not expected to undergo apoptosis with c-myc overexpression, based on the results with Rat-1 fibroblasts.
Although a recent study showed that overexpression of c-myc was capable of inducing apoptosis in a small fraction of CEFs in non-growth-limiting conditions (44), the difference in growth between cells overexpressing c-myc and bic and those overexpressing c-myc alone appears too large to be accounted for only by the protection of a small fraction of cells from apoptosis (see Fig. 4). Therefore, it is likely that bic causes growth enhancement of c-myc-expressing cells by further promoting cell proliferation. However, we cannot completely exclude the possibility that suppression of apoptosis may also play a role. Further experiments are necessary to establish the basis of cooperation of c-myc and bic in CEF growth enhancement. In addition, it will also be interesting to determine if bic can cooperate with c-myc in other aspects of cell growth regulated by c-myc, for instance, in cell size regulation (28).
Most importantly, the present study demonstrates that c-myc and bic can act in synergy in the pathogenesis of lymphomas. The collaboration is primarily evident in high-grade lymphomas, although the possibility that bic also plays a role in the pathogenesis of low-grade lymphomas cannot be excluded. Apparently, overexpression of c-myc alone is sufficient to generate the polyclonal, low-grade lymphomas. The oligo- or monoclonality of the high-grade lymphomas in animals infected with RCASBP(A)-myc and RCASBP(B)-bic, as well as their longer latency compared to low-grade lymphomas, indicates that additional genetic alterations besides c-myc and bic activations are necessary for their pathogenesis and suggest that they represent a progression from low-grade lymphomas.
The lymphomas observed in these studies appear to be bursa independent. Transformed follicles were not often associated with these lymphomas, and macroscopic nodules were not seen except in one case. Furthermore, some lymphomas were unusual in that they consisted of small, medium, and large lymphocytes. These observations suggest that these lymphomas are different from the classic lymphoid leukosis and may resemble phenotypically the bursa-independent lymphomas induced by HB-1 (12). Consistent with this notion, most of the lymphomas examined to date did not exhibit λ light-chain rearrangements. It is likely that the neoplastic lymphoid cells consist of early B cells and/or T cells. The collaboration of c-myc and bic to generate this type of lymphoma is not unexpected, since bic rearrangements have also been observed in bursa-independent lymphomas induced by HB-1 (K. Parks and W. Hayward, unpublished data). It is unclear why lymphomas resembling classic avian lymphoid leukosis were not observed in this experiment. A possible explanation is that the target cells for this type of lymphoma are not very accessible to RCAS viral infection.
Although no statistically significant difference was detected in the incidence or latency of lymphomas for animals infected with RCASBP(A) and RCASBP(B)-bic and those infected with RCASBP(A) and RCASBP(B), the incidence of lymphomas in the former group was nevertheless higher, which is consistent with insertional activation by RCASBP(A) of c-myc or other proto-oncogenes capable of collaborating with an activated bic provided directly by RCASBP(B)-bic. The failure to detect a statistically significant difference in either incidence or latency of lymphomas between animals infected with RCASBP(A) and RCASBP(B)-bic and animals infected with RCASBP(A) and RCASBP(B) may be explained by the small sample sizes for the two groups, particularly for the RCASBP(A) plus RCASBP(B) group.
The mechanism(s) by which c-myc and bic cooperate in lymphomagenesis remains to be determined. bic may contribute to lymphomagenesis by suppressing myc-induced apoptosis in vivo. Interestingly, preneoplastic bursal stem cell populations induced by a v-myc oncogene were hypersensitive to induction of apoptosis by follicular dispersion and radiation (42). This observation is consistent with myc's being a positive regulator of both cellular proliferation and apoptosis (14). Therefore, apoptosis potentially serves as a protective mechanism to prevent tumorigenicity elicited by a deregulated myc. Abrogation of this potential safeguard mechanism will therefore contribute to myc-induced tumorigenesis. Indeed, suppression of apoptosis has been implicated in the neoplastic progression of avian bursal lymphomas. While normal and transformed follicle cells underwent apoptosis when the bursal follicles were irradiated or mechanically disrupted in vitro, a bursal lymphoma cell line was resistant to apoptosis induced by radiation (42).
Recently, it was found that c-myc overexpression promotes the formation of transformed follicles by blocking differentiation and by retention of lymphocytes while increasing proliferation only modestly (6). bic may promote the late stages, for example, metastasis, of lymphoma development by causing emigration of bursal lymphocytes, a hypothesis supported by the observation that retroviral integrations in bic are seen preferentially in metastatic lymphomas (9). Alternatively, bic may further increase the proliferative capacity of neoplastic lymphoid cells expressing c-myc.
In addition, some of the animals infected with RCASBP(A)-myc and RCASBP(B)-bic developed acute erythroblastosis. In contrast, no erythroblastosis was found in animals infected with RCASBP(A)-myc and RCASBP(B). It is possible that erythroblastosis induced in the latter group is of such long latency that all of the animals died from other neoplastic diseases before they had an opportunity to develop erythroblastosis. Interestingly, 15% (3 of 20) of the chickens infected with RCASBP(A) and RCASBP(B)-bic also developed erythroblastosis, albeit with a relatively long latency, while none of the animals infected with RCASBP(A) and RCASBP(B) had erythroblastosis. Although no statistical significance was shown, these results suggest that there may be an increased susceptibility for animals infected with RCASBP(A) and RCASBP(B)-bic to develop erythroblastosis because of the constitutive overexpression of bic in vivo. Increasing the sample size of the RCASBP(A) plus RCASBP(B) group may be useful to evaluate this possibility.
Erythroblastosis is the predominant neoplasm induced by the leukemia virus AEV-ES4, which contains both v-erbA and v-erbB, indicating synergism of these two oncogenes in the pathogenesis of this disease (19, 25). Our observations show that c-myc and bic can also cooperate in this neoplasm, whereas c-myc or bic alone appears to be weakly oncogenic, if at all, in erythroleukemogenesis. Collaboration of myc with another oncogene in avian erythroleukemia has not been directly demonstrated before this study. Apparently, activations of myc and bic are not sufficient to generate a full leukemic phenotype. Based on analyses of tumor viral junction fragments, the erythroblastosis induced in this experiment appears to be oligoclonal or monoclonal, implying that additional genetic events besides myc and bic activations are necessary. Given the propensity of involvement of c-erbB in erythroblastosis (17, 18, 34, 37, 43, 47), it will be interesting to see if proviral insertions in the c-erbB locus represent one of these additional genetic events.
bic may represent a novel class of myc collaborators.
To our knowledge, this is the first report of a noncoding RNA functioning as a collaborator with c-myc. Previously, it was demonstrated that H19, a gene which has no long open reading frame and is believed to function through its RNA (7), has tumor-suppressing potential (21). Moreover, the 3′ untranslated region of α-tropomyosin RNA can act as a tumor suppressor (48). Our present data provide direct evidence that bic can function as a riboregulator which plays a role in tumorigenesis. Furthermore, untranslated RNAs are likely to represent a novel class of myc collaborators. The molecular mechanism(s) underlying this intriguing cooperativity awaits further investigations.
Acknowledgments
We thank E. Lacy and A. Zelenetz for helpful comments and discussions. W.T. expresses gratitude to Y. Liu and B. Shen for support and encouragement.
This work was supported by NIH grants R37 CA32926 (to P.B.) and CA16599 (to W.S.H.). W.T. was a fellow in the Tri-Institutional M.D.-Ph.D. Program and is currently a research fellow of the Leukemia and Lymphoma Society. Research was sponsored in part by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Adams, J. M., and S. Cory. 1992. Oncogene cooperation in leukaemogenesis. Cancer Surv. 15:119-141. [PubMed] [Google Scholar]
- 2.Adams, J. M., A. W. Harris, A. Strasser, S. Ogilvy, and S. Cory. 1999. Transgenic models of lymphoid neoplasia and development of a pan-hematopoietic vector. Oncogene 18:5268-5277. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, W. S., O. Bernard, S. Cory, and J. M. Adams. 1989. Lymphomagenesis in E mu-myc transgenic mice can involve ras mutations. Oncogene 4:575-581. [PubMed] [Google Scholar]
- 4.Askew, D. S., and F. Xu. 1999. New insights into the function of noncoding RNA and its potential role in disease pathogenesis. Histol. Histopathol. 14:235-241. [DOI] [PubMed] [Google Scholar]
- 5.Baba, T. W., and E. H. Humphries. 1985. Formation of a transformed follicle is necessary but not sufficient for development of an avian leukosis virus-induced lymphoma. Proc. Natl. Acad. Sci. USA 82:213-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandvold, K. A., D. L. Ewert, S. C. Kent, P. Neiman, and A. Ruddell. 2001. Blocked B cell differentiation and emigration support the early growth of Myc-induced lymphomas. Oncogene 20:3226-3234. [DOI] [PubMed] [Google Scholar]
- 7.Brannan, C. I., E. C. Dees, R. S. Ingram, and S. M. Tilghman. 1990. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Clurman, B. E., and W. S. Hayward. 1989. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 9:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, M. D., L. N. Payne, P. B. Dent, B. R. Burmester, and R. A. Good. 1968. Pathogenesis of avian lymphoid leukosis. I. Histogenesis. J. Natl. Cancer Inst. 41:373-378. [PubMed] [Google Scholar]
- 11.Cory, S., and J. M. Adams. 1988. Transgenic mice and oncogenesis. Annu. Rev. Immunol. 6:25-48. [DOI] [PubMed] [Google Scholar]
- 12.Enrietto, P. J., L. N. Payne, and M. J. Hayman. 1983. A recovered avian myelocytomatosis virus that induces lymphomas in chickens: pathogenic properties and their molecular basis. Cell 35:369-379. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann, V. A., M. Z. Barciszewska, M. Szymanski, A. Hochberg, N. de Groot, and J. Barciszewski. 2001. The non-coding RNAs as riboregulators. Nucleic Acids Res 29:189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-Myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 15.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 16.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 17.Fung, Y. K., W. G. Lewis, L. B. Crittenden, and H. J. Kung. 1983. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell 33:357-368. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin, R. G., F. M. Rottman, T. Callaghan, H. J. Kung, P. A. Maroney, and T. W. Nilsen. 1986. c-erbB activation in avian leukosis virus-induced erythroblastosis: multiple epidermal growth factor receptor mRNAs are generated by alternative RNA processing. Mol. Cell. Biol. 6:3128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graf, T., and H. Beug. 1978. Avian leukemia viruses: interaction with their target cells in vivo and in vitro. Biochim. Biophys. Acta 516:269-299. [DOI] [PubMed] [Google Scholar]
- 20.Hanafusa, H. 1969. Rapid transformation of cells by Rous sarcoma virus. Proc. Natl. Acad. Sci. USA 63:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao, Y., T. Crenshaw, T. Moulton, E. Newcomb, and B. Tycko. 1993. Tumour-suppressor activity of H19 RNA. Nature 365:764-767. [DOI] [PubMed] [Google Scholar]
- 22.Haupt, Y., W. S. Alexander, G. Barri, S. P. Klinken, and J. M. Adams. 1991. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 65:753-763. [DOI] [PubMed] [Google Scholar]
- 23.Haupt, Y., M. L. Bath, A. W. Harris, and J. M. Adams. 1993. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene 8:3161-3164. [PubMed] [Google Scholar]
- 24.Haupt, Y., A. W. Harris, and J. M. Adams. 1993. Moloney virus induction of T-cell lymphomas in a plasmacytomagenic strain of E mu-v-abl transgenic mice. Int. J. Cancer 55:623-629. [DOI] [PubMed] [Google Scholar]
- 25.Hayman, M. J., and H. Beug. 1992. Avian erythroblastosis: a model system to study oncogene co-operation in leukemia. Cancer Surv. 15:53-68. [PubMed] [Google Scholar]
- 26.Hayward, W. S., B. G. Neel, and S. M. Astrin. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475-480. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iritani, B. M., and R. N. Eisenman. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs, J. J., B. Scheijen, J. W. Voncken, K. Kieboom, A. Berns, and M. van Lohuizen. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanter, M. R., R. E. Smith, and W. S. Hayward. 1988. Rapid induction of B-cell lymphomas: insertional activation of c-myb by avian leukosis virus. J. Virol. 62:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley, R. L., and M. I. Kuroda. 2000. Noncoding RNA genes in dosage compensation and imprinting. Cell 103:9-12. [DOI] [PubMed] [Google Scholar]
- 32.Langdon, W. Y., A. W. Harris, and S. Cory. 1989. Acceleration of B-lymphoid tumorigenesis in E mu-myc transgenic mice by v-H-ras and v-raf but not v-abl. Oncogene Res. 4:253-258. [PubMed] [Google Scholar]
- 33.Largaespada, D. A., D. A. Kaehler, H. Mishak, E. Weissinger, M. Potter, J. F. Mushinski, and R. Risser. 1992. A retrovirus that expresses v-abl and c-myc oncogenes rapidly induces plasmacytomas. Oncogene 7:811-819. [PubMed] [Google Scholar]
- 34.Lax, I., R. Kris, I. Sasson, A. Ullrich, M. J. Hayman, H. Beug, and J. Schlessinger. 1985. Activation of c-erbB in avian leukosis virus-induced erythroblastosis leads to the expression of a truncated EGF receptor kinase. EMBO J. 4:3179-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leder, A., P. K. Pattengale, A. Kuo, T. A. Stewart, and P. Leder. 1986. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell 45:485-495. [DOI] [PubMed] [Google Scholar]
- 36.McDonnell, T. J., and S. J. Korsmeyer. 1991. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18). Nature 349:254-256. [DOI] [PubMed] [Google Scholar]
- 37.Miles, B. D., and H. L. Robinson. 1985. High-frequency transduction of c-erbB in avian leukosis virus-induced erythroblastosis. J. Virol. 54:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mladenov, Z., U. Heine, D. Beard, and J. W. Beard. 1967. Strain MC29 avian leukosis virus. Myelocytoma, endothelioma, and renal growths: pathomorphological and ultrastructural aspects. J. Natl. Cancer Inst. 38:251-285. [PubMed] [Google Scholar]
- 39.Neel, B. G., G. P. Gasic, C. E. Rogler, A. M. Skalka, G. Ju, F. Hishinuma, T. Papas, S. M. Astrin, and W. S. Hayward. 1982. Molecular analysis of the c-myc locus in normal tissue and in avian leukosis virus-induced lymphomas. J. Virol. 44:158-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neiman, P. E. 1994. Retrovirus-induced B cell neoplasia in the bursa of Fabricius. Adv. Immunol. 56:467-484. [DOI] [PubMed] [Google Scholar]
- 41.Neiman, P. E., A. Ruddell, C. Jasoni, G. Loring, S. J. Thomas, K. A. Brandvold, R. Lee, J. Burnside, and J. Delrow. 2001. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proc. Natl. Acad. Sci. USA 98:6378-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neiman, P. E., S. J. Thomas, and G. Loring. 1991. Induction of apoptosis during normal and neoplastic B-cell development in the bursa of Fabricius. Proc. Natl. Acad. Sci. USA 88:5857-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsen, T. W., P. A. Maroney, R. G. Goodwin, F. M. Rottman, L. B. Crittenden, M. A. Raines, and H. J. Kung. 1985. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell 41:719-726. [DOI] [PubMed] [Google Scholar]
- 44.Petropoulos, C. J., I. Givol, and S. H. Hughes. 1996. Comparative analysis of the structure and function of the chicken c-myc and v-myc genes: v-myc is a more potent inducer of cell proliferation and apoptosis than c-myc. Oncogene 12:2611-2621. [PubMed] [Google Scholar]
- 45.Petropoulos, C. J., and S. H. Hughes. 1991. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J. Virol. 65:3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purchase, H. G., and L. N. Payne. 1984. Neoplastic diseases: leukosis/sarcoma group, p. 360-405. In M. S. Hofstad, H. J. Barnes, B. W. Calnek, W. M. Reid, and H. W. Yoder, Jr. (ed.), Diseases of poultry, 8th ed. Iowa State University Press, Ames.
- 47.Raines, M. A., W. G. Lewis, L. B. Crittenden, and H. J. Kung. 1985. c-erbB activation in avian leukosis virus-induced erythroblastosis: clustered integration sites and the arrangement of provirus in the c-erbB alleles. Proc. Natl. Acad. Sci. USA 82:2287-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rastinejad, F., M. J. Conboy, T. A. Rando, and H. M. Blau. 1993. Tumor suppression by RNA from the 3′ untranslated region of alpha-tropomyosin. Cell 75:1107-1117. [DOI] [PubMed] [Google Scholar]
- 49.Reynaud, C. A., V. Anquez, A. Dahan, and J. C. Weill. 1985. A single rearrangement event generates most of the chicken immunoglobulin light chain diversity. Cell 40:283-291. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz, R. C., L. W. Stanton, S. C. Riley, K. B. Marcu, and O. N. Witte. 1986. Synergism of v-myc and v-Ha-ras in the in vitro neoplastic progression of murine lymphoid cells. Mol. Cell. Biol. 6:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shinto, Y., M. Morimoto, M. Katsumata, A. Uchida, K. Aozasa, M. Okamoto, T. Kurosawa, T. Ochi, M. I. Greene, and Y. Tsujimoto. 1995. Moloney murine leukemia virus infection accelerates lymphomagenesis in E mu-bcl-2 transgenic mice. Oncogene 11:1729-1736. [PubMed] [Google Scholar]
- 52.Simon, M. C., W. S. Neckameyer, W. S. Hayward, and R. E. Smith. 1987. Genetic determinants of neoplastic diseases induced by a subgroup F avian leukosis virus. J. Virol. 61:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. Bio/Technology 24:122-139. [PubMed] [Google Scholar]
- 54.Strasser, A., A. W. Harris, M. L. Bath, and S. Cory. 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348:331-333. [DOI] [PubMed] [Google Scholar]
- 55.Tam, W. 2001. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274:157-167. [DOI] [PubMed] [Google Scholar]
- 56.Tam, W., D. Ben-Yehuda, and W. S. Hayward. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 17:1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, C. B., E. H. Humphries, L. M. Carlson, C. L. Chen, and P. E. Neiman. 1987. The effect of alterations in myc gene expression on B cell development in the bursa of Fabricius. Cell 51:371-381. [DOI] [PubMed] [Google Scholar]
- 58.van Lohuizen, M., S. Verbeek, P. Krimpenfort, J. Domen, C. Saris, T. Radaszkiewicz, and A. Berns. 1989. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56:673-682. [DOI] [PubMed] [Google Scholar]
- 59.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 60.Verbeek, S., M. van Lohuizen, M. van der Valk, J. Domen, G. Kraal, and A. Berns. 1991. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol. Cell. Biol. 11:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]