Abstract
The human subgroup C adenoviral E1B 55-kDa protein cooperates with the viral E4 Orf6 protein to induce selective export of viral, late mRNAs from the nucleus to the cytoplasm. Previous studies have suggested that such preferential transport of viral mRNA and the concomitant inhibition of export of cellular mRNAs are the result of viral colonization of specialized microenvironments within the nucleus. However, neither the molecular basis of this phenomenon nor the mechanism by which the E1B 55-kDa protein acts has been elucidated. We therefore examined viral late mRNA metabolism in HeLa cells infected with a series of mutant viruses that carry insertions at various positions in the E1B protein coding sequence (P. R. Yew, C. C. Kao, and A. J. Berk, Virology 179:795-805, 1990). All the mutations examined impaired cytoplasmic accumulation of viral L2 mRNAs and reduced L2 mRNA export efficiency. However, in most cases these defects could be ascribed to reduced E1B 55-kDa protein concentration or the unexpected failure of the altered E1B proteins to enter the nucleus efficiently. The latter property, the pleiotropic defects associated with all the mutations that impaired nuclear entry of the E1B protein, and consideration of its primary sequence suggest that these insertions result in misfolding of the protein. Insertion of four amino acids at residue 143 also inhibited viral mRNA export but resulted in increased rather than decreased accumulation of the E1B 55-kDa protein in the nucleus. This mutation specifically impaired the previously described association of the E1B protein with intranuclear structures that correspond to sites of adenoviral DNA replication and transcription (D. Ornelles and T. Shenk, J. Virol. 65:424-439, 1991) and the colocalization of the E1B and E4 Orf6 proteins. As this insertion has been shown to inhibit the interaction of the E1B with the E4 Orf6 protein in infected cell extracts (S. Rubenwolf, H. Schütt, M. Nevels, H. Wolf, and T. Dobner, J. Virol. 71:1115-1123, 1997), these phenotypes provide direct support for the hypothesis that selective viral mRNA export is determined by the functional organization of the infected cell nucleus.
Human subgroup C adenoviruses, such as adenovirus type 2 (Ad2) and Ad5, encode several proteins that exhibit transforming activity, because they disrupt host cell circuits regulating cell proliferation (33, 63). These gene products include the E1B 55-kDa protein, which can cooperate with E1A proteins in stable transformation of rodent cells and is required for efficient viral replication in permissive human cells (2, 50, 76). The former function has been ascribed to modulation of the activity and concentration of the cellular p53 protein, a sequence-specific transcriptional activator that induces cell cycle arrest or apoptosis in response to genotoxic and other forms of stress (36, 42). The E1B 55-kDa protein inhibits p53-dependent transcription by binding to the N-terminal activation domain of the cellular protein (34, 61, 80). The E1B protein can function as a general repressor of RNA polymerase II transcription (44, 80). It is therefore believed to inhibit p53-dependent transcription as a result of binding to promoter-associated p53 protein (44). Such transcriptional repression by the E1B protein correlates closely with its transforming activity (80). Indeed, phosphorylation of C-terminal serine (490 and 491) and threonine (495) residues is necessary for transcriptional repression, transformation, and inhibition of p53-dependent apoptosis induced by the viral 243R E1A protein (68), indicating that repression of p53-dependent transcription is important for survival of cells synthesizing the toxic E1A proteins. In transformed cells, the E1B 55-kDa protein also frequently relocalizes p53 from the nucleus to dense perinuclear bodies in the cytoplasm (7, 72, 82) and can do so when synthesized in the absence of other viral proteins in human cells (75). It has been demonstrated more recently that the E1B 55-kDa protein can induce accelerated degradation of p53, but only in conjunction with the E4 Orf6 protein (53, 56, 66), which also binds to p53 (19, 53) and exhibits transforming activity (19, 46). In infected human cells, such increased protein turnover overcomes the increase in p53 concentration induced by the 243R E1A protein (53, 66).
The E1B 55-kDa protein regulates viral gene expression during the infectious cycle by posttranscriptional mechanisms. In particular, it is required (2, 50, 76) for the selective export of viral mRNAs from the nucleus to the cytoplasm characteristic of the late phase of infection (23). Export of other types of cellular RNA is not inhibited in adenovirus-infected cells (13, 64), suggesting that the viral protein specifically perturbs a cellular mRNA export pathway. Consistent with this view, it has recently been reported that leptomycin B, a specific inhibitor of exportin 1-mediated export of cellular small RNAs and proteins (74), does not inhibit viral late gene expression (54). The E1B 55-kDa protein associates with the E4 Orf6 protein in infected cell extracts (60). This property and the similar phenotypes exhibited by mutant viruses defective for production of either or both of the E1B protein and the E4 protein (10, 15) originally suggested that the viral early protein complex is primarily responsible for regulation of mRNA export. The E4 Orf6 protein-dependent localization of the E1B protein to viral nuclear inclusions believed to be the sites of viral DNA transcription during the late phase of infection (49) and the ability to shuttle between the nucleus and the cytoplasm ascribed to both proteins (18, 21, 37) provide further support for this view.
Although the export of most cellular mRNAs is severely inhibited soon after the late phase of infection begins (23, 38), some host mRNAs escape this block. This set includes cellular Hsp70 mRNA made following induction of transcription of the gene by heat shock during the late phase of infection (47, 78) and several mRNAs specified by cellular genes that are transcriptionally activated during the late phase in response to viral infection (78). Efficient export of these cellular mRNAs in infected cells also requires the E1B 55-kDa protein (78). The correlation of E1B protein-dependent export with activation of transcription during the late phase of infection (78) led us to propose that activated transcription units occupy discrete nuclear microenvironments functionally specialized to promote mRNA export, that is, “gated” sites (8). This model is consistent with (i) the hypothesis that the E4 Orf6 protein-dependent localization of the E1B 55-kDa protein to peripheral zones of viral nuclear inclusion bodies, in which viral mRNA is produced, sequesters one or more limiting cellular proteins mediating mRNA export at these sites (49); (ii) the correlation of the accumulation of mature viral late mRNAs in these same sites with efficient mRNA export (1, 12); and (iii) the E1B 55-kDa protein-dependent movement of viral late mRNAs to an operationally defined “pre-export” compartment in the nucleus (41). While it is generally accepted that regulation of mRNA export in adenovirus-infected cells is coupled with mechanisms that optimize nuclear structure for efficient production and export of processed mRNAs (20), the molecular mechanism(s) of action of the E1B 55-kDa protein remains unknown.
In addition to binding to the E4 Orf6 and p53 proteins, the E1B 55-kDa protein has been reported to interact with the E4 Orf3 protein (40) and a cellular protein termed E1B-AP5 that contains an RNA-binding domain (24). It can also bind nonspecifically to RNA in vitro (32). Several studies designed to identify regions of the E1B protein necessary for its interactions with other proteins or RNA have relied on a series of mutant viruses carrying amino acid insertions at various positions within a hybrid Ad2/Ad5 E1B coding sequence (24, 34, 59, 81). The information collected has not yet allowed unambiguous conclusions to be made about the contributions of the biochemical activities of the E1B protein to regulation of mRNA export, for this step in viral late mRNA production has not been examined in cells infected with the mutant viruses. The synthesis of viral late proteins in Ad5 and mutant virus-infected cells has been compared (81), but this measure of viral late gene expression cannot be interpreted in terms of the efficiency of export of viral late mRNAs: as the late phase progresses, viruses carrying mutations that prevent production of detectable E1B 55-kDa protein exhibit defects in major late (ML) transcription and greater reductions in synthesis of viral late proteins than those in concentrations of the corresponding cytoplasmic mRNAs (39, 50, 76). These pleiotropic phenotypes seem likely to be at least in part secondary consequences of the inefficient production of viral late proteins that stimulate ML transcription (e.g., the IVa2 protein [70]) or the translation (e.g., the L4 100-kDa protein [30]) of late mRNAs. However, recent experiments suggest that the E1B 55-kDa protein may also regulate translation of viral late mRNAs more directly (28). We have therefore examined the effects of these E1B 55-kDa protein coding sequence mutations on viral mRNA metabolism in infected human cells.
MATERIALS AND METHODS
Cells and viruses.
HeLa and 293 cells were grown as monolayer cultures in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (GIBCO-BRL). The E1B mutant viruses A143, H180, H215, H224, A262, R309, H326, H354, S380, R443, H474, and F484 were generously provided by A. Berk. These mutants carry linker insertions in a hybrid Ad2/Ad5 E1B gene at the position given in their names, which indicate the sites of insertion by the residue number of the protein. With the exception of the addition of 18 amino acids at H215, 4 amino acids were inserted at each position (81). The Ad5 mutant hr6, which carries a frameshift mutation in the E1B 55-kDa protein coding sequence, was previously described (29, 76). Ad5 (wild-type 300) and E1B mutants were propagated in monolayers of 293 cells. Viruses were titrated by plaque assay on 293 cells as described previously (77), and a multiplicity of infection of 30 PFU per cell was used in all experiments.
Accumulation and intracellular distribution of E1B protein.
HeLa cells at approximately 90% confluence were infected with Ad5 or the E1B mutants described above. For immunoblotting, cells were harvested at the indicated times after infection, washed with phosphate-buffered saline (PBS), and extracted with 25 mM Tris-HCl, pH 8.0, containing 50 mM NaCl, 0.5% (wt/vol) sodium deoxycholate, 0.5% (vol/vol) Nonidet P-40 (NP-40), and 1 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. Cell debris was removed by centrifugation at 10,000 × g at 4°C for 5 min. The extracts were then analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to 0.2-μm-pore-size nitrocellulose membranes (Schleicher and Schuell) using a semidry blotting system (Owl Scientific Inc.). The nitrocellulose was blocked with 5% nonfat dry milk in PBS containing 0.05% (vol/vol) Tween 20, and proteins were detected with the anti-E1B 55-kDa protein monoclonal antibody (MAb) 2A6, which recognizes an epitope located within the N-terminal 180 amino acids of the protein (60), and peroxidase-coupled anti-mouse anti-immunoglobulin G (anti-IgG) (Boehringer Mannheim). Blots were developed using a chemiluminescent substrate as described by the manufacturer (Amersham), and the signals obtained were quantified by densitometry using NIH Image software.
For immunoprecipitation experiments, HeLa cells at approximately 90% confluence were infected with Ad5 and the E1B mutants for 12 h. Infected cells were then incubated in methionine-free Dulbecco's modified Eagle's medium for 1 h and with 100 μCi of [35S]methionine (1.175 Ci/mmol; NEN-Dupont) per 107 cells in this same medium for 4 h. Cells were harvested and washed twice with ice-cold PBS, and cytoplasmic fractions were prepared by incubation in 10 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 1.5 mM MgCl2, 0.6% (vol/vol) NP-40, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, leupeptin (2 μg/ml), and antipain (1 μg/ml) at 4°C for 5 min. Nuclei recovered by centrifugation after the second such extraction were incubated at 4°C in 50 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 0.4% (vol/vol) NP-40, 5 mM EDTA, and 1 mM dithiothreitol and sonicated twice for 10 s. Cytoplasmic and nuclear fractions were immunoprecipitated with the 2A6 monoclonal antibody at a 1:50 dilution in the same buffer used for nuclear protein extraction, but without dithiothreitol. Protein-antibody complexes were bound to protein A-Sepharose beads (Pharmacia) and washed four times before they were resuspended for SDS-polyacrylamide gel electrophoresis. Protein signals were quantified using the PhosphorImager Storm system and ImageQuant software (Molecular Dynamics).
Immunofluorescence.
HeLa cells grown on glass coverslips to approximately 90% confluence were mock infected or infected with Ad5 or E1B mutant viruses. At 14 to 20 h after infection, the cells were fixed in PBS containing 1.5 mM MgCl2 and 4% formaldehyde and permeabilized with PBS-containing 0.5% (vol/vol) Triton X-100 (Sigma Chemical Co.) as described by Ornelles and Shenk (49). They were then incubated sequentially with either the anti-E2 72-kDa DNA binding protein (DBP) monoclonal antibody B6-8 (55) diluted 1 to 50 in PBS or the anti-E4 Orf6 monoclonal antibody M45 (48) at a 1-to-10 dilution and then goat anti-mouse IgG coupled to cyanine 5 (Jackson ImmunoResearch Laboratories, Inc.) diluted 1 to 250. The E1B 55-kDa protein was then labeled with purified 2A6 IgG coupled to Alexa-Fluor 488 (Molecular Probes), prepared and purified according to the manufacturer's protocol. Nuclei were labeled with 0.001% DAP1 during the penultimate wash in PBS containing 1.5 mM MgCl2. The coverslips were mounted on glass slides in Aqua Polymount (Polysciences Inc.), and samples were examined by confocal microscopy. Images were collected with a Zeiss LSM510 confocal microscope using a C-Apochromat 1.2 NA water immersion objective and organized using Adobe Photoshop 5.0.
Rates of ML transcription.
Transcription rates were measured in isolated nuclei as described elsewhere (78). Briefly, nuclei prepared from infected HeLa cells harvested during the late phase of infection were incubated for 15 min at 30°C in the presence of 100 μCi of [32P]UTP (3,000 Ci/mmol; NEN-Dupont). Nuclear RNA was then purified and fragmented by treatment with 0.2 M NaOH for 10 min on ice. The RNA was hybridized to single-stranded DNA fragments immobilized on nylon membranes, using 3 pmol of each DNA probe. These DNA fragments, which contained sequences of the adenoviral VA RNA I gene transcribed by RNA polymerase III, the viral E1A gene, the L2 region of the ML transcription unit, or the human β-actin gene, were excised from plasmids, purified by electrophoresis in agarose gels, and denatured immediately prior to loading onto membranes. Signals were detected using the PhosphorImager Storm system and quantified using ImageQuant software (Molecular Dynamics).
Steady-state concentrations of viral late mRNA.
Cytoplasmic and nuclear RNA were prepared as described elsewhere (35). Briefly, cytoplasmic RNA was prepared from HeLa cells infected with Ad5 or E1B mutant viruses harvested during the late phase of infection by extracting the cells twice with 50 mM Tris-HCl, pH 7.5, containing 100 mM NaCl, 2 mM EDTA, and 0.6% (vol/vol) NP-40 for 5 min on ice. The pooled supernatants were treated with 40 U of RNase-free DNase I (Promega) and proteinase K (100 μg/ml; Roche), extracted with phenol-chloroform and precipitated with ethanol. Nuclei were incubated in 10 mM Tris-HCl, pH 7.4, containing 500 mM NaCl, 5 mM MgCl2, and 2 mM CaCl2 and treated sequentially with 40 U of RNase-free DNase I (Promega) for 5 min at 30°C and proteinase K (50 μg/ml) in 100 mM Tris-HCl, pH 7.4, containing 10 mM EDTA and 1% (wt/vol) SDS for 30 min at 42°C. After phenol-chloroform extraction and ethanol precipitation, samples were treated again with DNase I and proteinase K at 37°C, extracted with phenol-chloroform, and precipitated with ethanol. The RNA samples were resuspended in 10 mM Tris, pH 7.4, containing 5 mM NaCl and 2 mM EDTA and stored at −80°C until use. Poly(A)-containing mRNA was isolated from cytoplasmic RNA preparations using oligo(dT25)-coupled paramagnetic microbeads as described by the manufacturer (Dynal, S.A.). Northern blot analysis was performed with glyoxalated RNAs (45), using as DNA probes plasmids harboring viral E1A or L2 sequences or the human ribosomal protein 3 gene (American Type Culture Collection) labeled by random priming (22). Analysis of specific viral and cellular mRNAs in cytoplasmic and nuclear RNA samples by primer extension was performed as described previously (14, 35), using oligonucleotides complementary to positions 189 to 209 of the L2 penton mRNA. The cDNAs were detected and quantified using the PhosphorImager Storm system (Molecular Dynamics).
RESULTS
Accumulation and localization of altered E1B 55-kDa proteins.
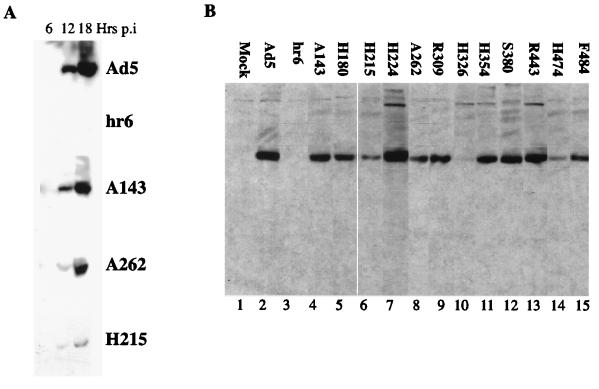
Prior to analysis of the metabolism of viral late mRNAs in cells infected with mutant viruses carrying insertions within the E1B 55-kDa protein coding sequence (81), we compared the accumulation and intracellular distribution of the wild-type and altered E1B proteins. The Ad5 mutant hr6, which carries a deletion of bp 326 in the E1B 55-kDa protein coding sequence and does not direct synthesis of detectable E1B protein (76), was also included. Soluble extracts of HeLa cells infected with Ad5 or the mutant viruses at 30 PFU/cell were prepared 6, 12, and 18 h after infection and analyzed by immunoblotting using the 2A6 anti-E1B 55-kDa protein monoclonal antibody. The wild-type E1B protein was first clearly detected 12 h after infection and increased substantially in steady-state concentration by 18 h (Fig. 1A), in agreement with previous observations (9, 58, 59, 65). All E1B 55-kDa proteins encoded by the mutant viruses exhibited a similar temporal pattern of accumulation, increasing in the same apparent proportion as the wild type as the late phase progressed. This property is illustrated in Fig. 1A for a subset of mutants representing the range of concentrations to which the altered proteins accumulated. An experiment in which the complete set of E1B proteins made in mutant virus-infected cells was examined is shown in Fig. 1B. With the exception of hr6 (Fig. 1B, lane 3), all the mutants encoded E1B 55-kDa proteins that accumulated to detectable levels by 18 h after infection. However, the altered E1B proteins varied significantly in concentration. Several exhibited no (H224, S380, and R443) (Fig. 1B, compare lanes 7, 12, and 13 with lane 2) or only minor (some 20%) (A143 and H354) (Fig. 1B, compare lanes 4 and 11 with lane 2) decreases in concentration compared to the wild type. In contrast, the steady-state concentrations of the E1B 55-kDa proteins encoded by mutants H180, H215, A262, R309, H326, H474, and F484 were more substantially decreased, by factors of 2.5 to 5 (Fig. 1B, lanes 5, 6, 8, 9, 10, 14, and 15; Table 1), indicating that these altered proteins are less stable than the wild type.
FIG. 1.
Accumulation of altered E1B 55-kDa proteins in mutant virus-infected HeLa cells. HeLa cells were mock infected or infected with Ad5 or the mutants listed for the periods indicated (A) or for 18 h (B). Total cell lysates were prepared and analyzed by immunoblotting with the anti-E1B 55-kDa protein monoclonal antibody 2A6 as described in Materials and Methods.
TABLE 1.
Properties of E1B mutant virusesa
| Virus | Concn of steady-state E1B 55-kDa protein | Cytoplasmic L2 penton mRNA/ nuclear L2 penton mRNA |
|---|---|---|
| Ad5 | 1.0 | 1.0 |
| A143 | 0.76 ± 0.37 | 0.31 ± 0.08 |
| H180 | 0.38 ± 0.13 | 0.32 ± 0.20 |
| H215 | 0.36 ± 0.03 | 0.39 ± 0.09 |
| H224 | 1.28 ± 0.19 | 0.11 ± 0.11 |
| A262 | 0.31 ± 0.16 | 0.095 ± 0.005 |
| R309 | 0.38 ± 0.06 | 0.10b |
| H354 | 0.78 ± 0.37 | 0.22 ± 0.03 |
| F484 | 0.42 ± 0.15 | 0.35 ± 0.15 |
These parameters are expressed relative to values measured in Ad5-infected cells, set at 1.0.
This value was determined in one experiment only.
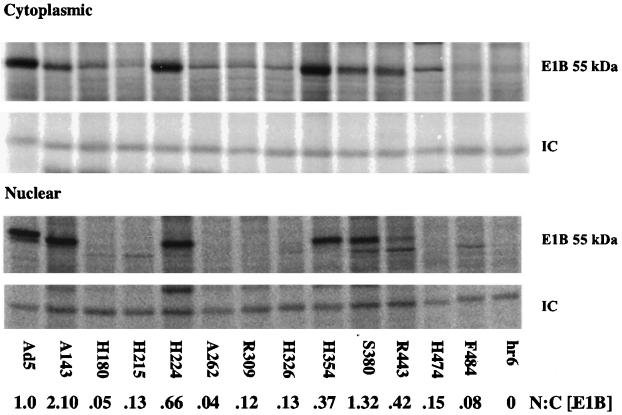
As the E1B 55-kDa protein functions in the nucleus and can shuttle from the nucleus to the cytoplasm and back (see the introduction), we also determined whether any of the amino acid insertions perturbed the partition of the protein between the nucleus and cytoplasm. The accumulation of the E1B 55-kDa proteins in infected cell nuclei was initially examined by immunoblotting of nuclear fractions with antibody 2A6. Altered E1B proteins detected at low concentrations in total infected-cell lysates (Fig. 1) exhibited similar reductions in nuclear concentration. In addition, the E1B proteins encoded by several other mutants were detected in the nuclear fraction at much lower concentrations than anticipated from the results of experiments like that shown in Fig. 1 (data not shown). To facilitate quantitative analysis, infected cells were labeled with [35S]methionine from 13 to 17 h after infection, and the E1B proteins were immunoprecipitated from nuclear and cytoplasmic fractions with monoclonal antibody 2A6. Representative results are shown in Fig. 2. The protein was detected in the cytoplasmic fraction of cells infected with all the insertion mutants (Fig. 2), in most cases at relative concentrations similar to those observed by immunoblotting of total soluble protein (Fig. 1B). However, the E1B proteins immunoprecipitated from A262-, R309-, or H326-infected cell cytoplasmic extracts were detected at concentrations three- to fourfold lower (relative to the wild-type protein) than when the proteins were examined by immunoblotting. Much greater differences were apparent in nuclear concentrations: several of the altered E1B 55-kDa proteins, notably those encoded by mutants H180, H215, A262, R309, H326, and F484, were present at much lower relative concentrations in the nucleus (Fig. 2). Quantification of the signals and calculation of the ratios of nuclear to cytoplasmic E1B 55-kDa proteins relative to that of the wild-type protein (Fig. 2) indicated that the mutations listed above induced substantial defects in the nuclear accumulation of the E1B protein during the late phase of infection. Conversely, the altered A143 E1B 55-kDa protein accumulated preferentially in the nuclear fraction (Fig. 2).
FIG. 2.
Localization of altered E1B 55-kDa proteins. HeLa cells infected with Ad5 or the mutant viruses indicated were labeled with [35S]methionine from 13 to 17 h after infection and separated into nuclear and cytoplasmic fractions as described in Materials and Methods. The E1B 55-kDa protein was immunoprecipitated from each sample with monoclonal antibody 2A6.
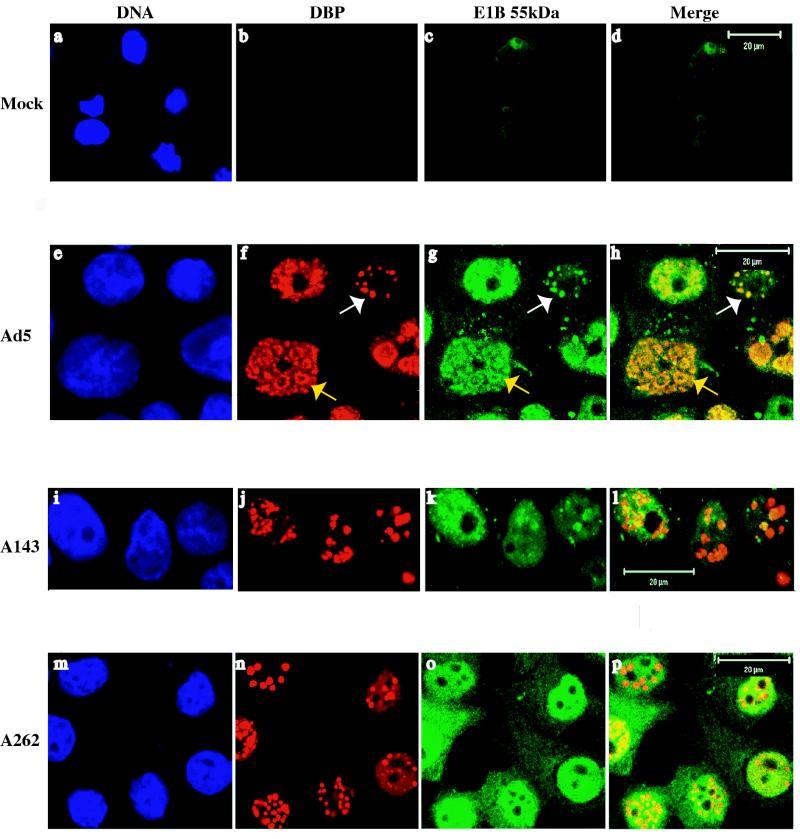
To confirm the changes in intracellular distribution of altered E1B proteins observed upon cell fractionation, protein localization in cells infected with Ad5 or mutants A143 or A262 was examined by immunofluorescence using monoclonal antibody 2A6 (Fig. 3). The results of these experiments indicated that A143 and A262 insertion altered the intranuclear localization of the E1B 55-kDa protein and its partition between nucleus and cytoplasm, respectively (data not shown). Localization of the E1B protein was therefore examined in more detail in Ad5- and mutant virus-infected cells by double labeling of the E2 72-kDa DBP and the E1B protein, as described in Materials and Methods. The DBP accumulates in viral nuclear inclusion bodies, which are the sites of viral DNA synthesis and therefore are termed replication centers or foci (51, 52, 73). Transcription of viral late genes takes place in the peripheral zones of these replication centers (11), and the E1B 55-kDa protein is also associated with them (49). At 14 to 18 h after infection with Ad5, the DBP was detected only within the nucleus, where it was present within a limited number of globular structures (Fig. 3f). These structures, which were sometimes observed on a background of diffuse nuclear staining, varied from compact spheres of various size to larger, ring-like forms with the DBP concentrated at their peripheries (white and yellow arrows, respectively, in Fig. 3f). They correspond to the replication centers described previously, which have been shown to enlarge as viral DNA accumulates within infected cell nuclei (73). Localization of the E1B 55-kDa protein in the same Ad5-infected cell populations using directly labeled MAb 2A6 revealed some fine reticular staining in the cytoplasm, as expected (49). Within the nucleus, this protein was detected in structures strikingly similar in appearance to those containing the E2 DBP (compare Fig. 3f and g). In contrast to the DBP, however, the E1B protein was observed at low concentrations throughout the nucleus in most infected cells. As illustrated in Fig. 3h, the E1B 55-kDa protein was extensively colocalized with DBP-containing replication centers, regardless of the size or morphology of the latter.
FIG. 3.
Intracellular localization of E2 72-kDa DBP and E1B 55-kDa proteins in HeLa cells infected with Ad5 or with the A143 or A262 mutants or mock-infected HeLa cells was examined 14 h after infection by immunofluorescence, using monoclonal antibodies B6-8 and 2A-6, respectively, as described in Materials and Methods.
Neither of the E1B insertion mutants examined in these experiments, A143 and A262, altered the formation of DBP-containing replication centers (Fig. 3j and n). However, in cells infected with A262, the cytoplasmic concentration of the E1B 55-kDa protein was significantly increased, and no discrete, E1B protein-containing structures could be detected in the nucleus (compare Fig. 3g and o). This steady-state distribution, in conjunction with the results of the biochemical analysis described previously, indicates that the altered A262 E1B protein not only enters the nucleus inefficiently but also is distributed nonspecifically within that organelle. In agreement with the results described previously (Fig. 2), the E1B 55-kDa protein made in A143-infected cells was concentrated in the nucleus (Fig. 3k). However, in contrast to the wild type, this altered E1B protein was observed in few discrete spheres or rings. Rather, much of the protein was diffusely distributed throughout nuclei (but excluded from nucleoli), with some forming flecks or globules. The size and shape of the latter did not match closely these features of the E1B 55-kDa protein-containing structures characteristic of Ad5-infected cells (Fig. 3g and k), and the A143 E1B protein was not closely localized with DBP-containing replication centers (Fig. 3l). The alteration in intranuclear organization of the E1B protein induced by the A143 mutation was reproducibly observed: scoring of several fields of Ad5- or A143-infected cells examined in each of three independent experiments established that the E1B protein was intimately colocalized with the DBP in 81% of Ad5-infected cells containing discrete replication centers, but some colocalization was observed in only 20% of A143-infected cells.
Effects of A143 insertion in the E1B 55-kDa protein or localization of the E4 Orf6 protein.
The A143 insertion has been reported to inhibit the binding of the E1B 55-kDa protein to the E4 Orf6 protein (59). This property accounts for the mislocalization of the E1B protein we observed in A143-infected cells: the association of the E1B protein with replication centers is eliminated in cells infected with a mutant virus that lacks E4 Orf6 protein coding sequences (49). However, whether the interaction between these two viral proteins also determines the localization of the E4 Orf6 protein in infected cell nuclei had not been determined. To address this question, we examined the distribution of this E4 protein in cells infected with Ad5 or the A143 mutant and its colocalization with the E1B 55-kDa protein.
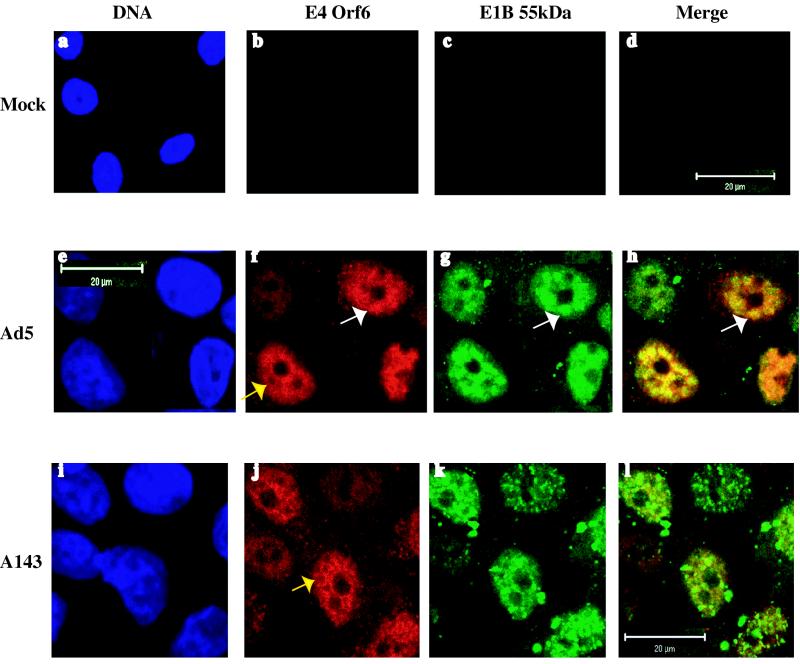
In Ad5-infected cells, the E4 Orf6 protein was detected in the cytoplasm, but concentrated into the nucleus, where it formed a reticular network within which globular or ring-like structures were evident (Fig. 4f). These structures were less discrete and well-defined than those containing the E1B 55-kDa protein (Fig. 3g and 4g). Furthermore, ring-like structures containing the E4 protein appeared connected to one another via an extensive lattice (Fig. 4f and j), whereas those in which the E1B 55-kDa protein was observed did not (Fig. 3g). The basketwork or lattice-like organization of the E4 Orf6 protein bears a striking resemblance to nuclear structures containing viral late RNAs and cellular snRNPs described by Bridge and colleagues (1, 51). However, whether this viral protein indeed localizes or associates with viral RNA or splicing components has not been investigated. Although their individual intranuclear distributions could be distinguished, the E1B and E4 proteins were observed to be extensively colocalized in nuclear structures (Fig. 4h). Detailed comparison of merged images like that shown in Fig. 4h with the individual patterns exhibited by the two viral proteins suggested that the E1B protein may be associated with specific regions of a more extensive E4 protein scaffold. The altered intranuclear distribution of the E1B protein in cells infected with mutant A143 was as described previously, but no significant change in the localization of the E4 protein was observed (Fig. 4j). Thus, extensive colocalization of the E1B and E4 proteins was not evident in A143-infected cells (Fig. 4l).
FIG. 4.
Intracellular localization of the E4 Orf6 and E1B 55-kDa proteins in HeLa cells infected with the viruses indicated or mock-infected HeLa cells was examined 14 h after infection by immunofluorescence using monoclonal antibodies M45 and 2A6, respectively.
Synthesis of viral late mRNAs.
Although defects in the synthesis of viral late proteins induced by the E1B 55-kDa protein coding sequence insertions were reported some time ago (81), the effects of these mutations on production of the corresponding mRNAs have never been examined. We therefore first examined the accumulation of poly(A)-containing, viral L2 mRNAs in the cytoplasm, using Northern blotting to measure viral mRNA concentrations as described in Materials and Methods.
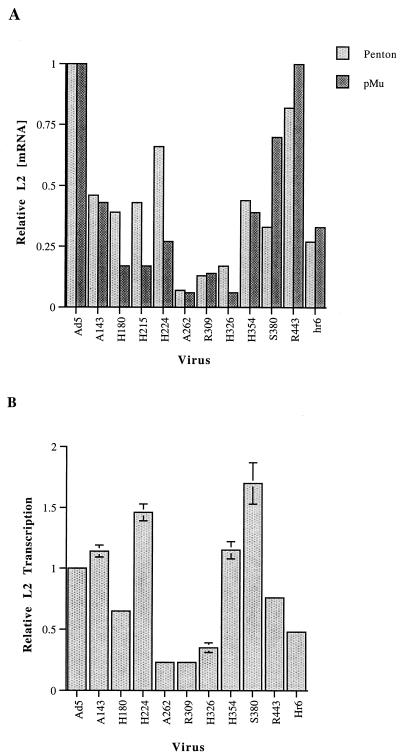
The results of such an experiment are shown for the L2 penton and pre-Mu mRNAs in Fig. 5A. Concentrations of viral L2 mRNAs lower than wild-type concentrations were observed in cells infected with the majority of the mutant viruses tested. These effects were specific to late mRNAs, as corresponding reductions in E1A mRNAs were not observed when blots were stripped and reprobed (data not shown). The individual E1B mutations altered the concentrations of the four L2 mRNAs in similar ways. For example, the H262, R309, and H326 mutations induced the greatest reductions, to steady-state concentrations 10 to 15% of that observed in Ad5-infected cells, in the accumulation of both penton and pre-Mu mRNAs (Fig. 5A and data not shown), whereas more-modest defects were observed in cells infected with mutants H180, H215, H354, and R443. These decreases in L2 mRNA concentration in mutant virus-infected cells correlated with reduced E1B 55-kDa protein stability and/or impaired accumulation of the protein in the nucleus (Fig. 1B, 2, and 5A). However, L2 mRNA concentrations were also significantly reduced in cells infected with the A143 mutant (Fig. 5A).
FIG. 5.
Cytoplasmic accumulation and transcription of viral L2 RNA. (A) Cytoplasmic L2 mRNAs were examined 18 h after infection by Northern blotting. Cells were infected with Ad5 or the E1B 55-kDa mutants listed, and the four L2 mRNAs and the human ribosomal protein 3 mRNAs were detected and quantified as described in Materials and Methods. Shown are penton and pre-Mu mRNA signals, corrected using the RpS3 mRNA internal control and expressed relative to the wild-type value, which was set at 1.0. Similar changes were observed in concentrations of the L2 V and pVII mRNAs. (B) Rates of L2 transcription were determined 18 h after infection of HeLa cells with Ad5 or the mutant viruses listed by run-on transcription in isolated nuclei and hybridization of the labeled RNA to membrane-immobilized L2 DNA. Signals were quantified and corrected as described in Materials and Methods and expressed relative to the wild-type value, which was set at 1.0. The values shown are the means of two independent experiments. Error bars show standard deviations.
It has been reported previously (39) that the cytoplasmic concentration of the shortest mRNA in each ML family is less severely depressed in the absence of the E1B 55-kDa protein than that of the longest mRNA, pre-Mu and penton mRNA, respectively, in the case of L2. This difference became increasingly pronounced as the late phase proceeded (39), a property that probably accounts for the fact that accumulation of pre-Mu mRNA was not consistently less sensitive to the E1B 55-kDa protein coding sequence mutations than that of penton mRNA under our experimental conditions (Fig. 5A).
The progressive inhibition of ML transcription characteristic of cells infected with the E1B mutant viruses dl338 or hr6 (39, 50) suggested that transcriptional defects might contribute to the reductions in cytoplasmic concentrations of viral late mRNAs. To investigate this possibility, the rates of transcription of the ML L2 region were examined in Ad5- and mutant virus-infected cells by run-on transcription in isolated nuclei (see Materials and Methods). The signals obtained were quantified by using as an internal control the viral RNA polymerase III transcript VA RNA I, whose synthesis is not affected by the absence of E1B 55-kDa protein (76). Transcription of L2 DNA was inhibited to a degree similar to or greater than that observed in hr6-infected cells for only a subset of the mutations, namely, H180, A262, R309, and H326 (Fig. 5B). Thus, in these cases, the substantial reductions in cytoplasmic L2 mRNA correlated with lower rates of transcription (Fig. 5). At an earlier point in the late phase, 13 h after infection, no such reduced rates of L2 transcription were observed in cells infected with these mutants (data not shown), consistent with previous observations (39, 50). These changes in ML transcription underscore the importance of direct examination of mRNA export efficiency in cells infected with viruses carrying potentially pleiotropic mutations in the E1B 55-kDa protein coding sequence.
Cytoplasmic and nuclear distribution of viral late mRNAs.
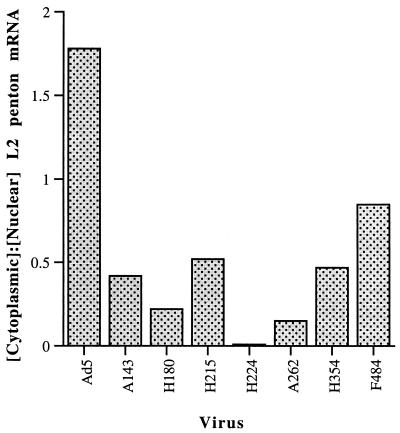
With the exception of the ML L1 mRNA specifying the 52/ 55-kDa protein, viral mRNAs processed from transcripts of the ML and other late transcription units cannot be detected in the cytoplasm until a few hours after viral DNA synthesis begins (62). Consequently, Ad5-infected cells do not contain pools of these mRNAs produced before selective export of viral mRNA is induced. Furthermore, during the initial part of the late phase in cells infected at 37 to 38.5°C, the E1B 55-kDa protein does not significantly influence the stability of viral late mRNAs in the cytoplasm (50, 76). These properties allow viral late mRNA export efficiency to be measured as the ratio of the steady-state concentration of the processed mRNA in the cytoplasm to that in the nucleus. This approach was therefore adopted to assess more directly the effects of a representative subset of the E1B 55-kDa protein coding sequence insertions on viral mRNA export, using primer extension to measure mRNA concentrations relatively early in the late phase of infection (14 h after infection). To distinguish mature mRNA from precursors present in the nucleus and to avoid hybridization of the primer to more than a single mRNA, the primers used were complementary to sequences spanning splice junctions between the tripartite leader sequence and the longest mRNAs within the ML families. We initially estimated export efficiency of the L2 penton mRNA in cells infected with Ad5 or the E1B mutants, with the stable cellular β-actin mRNA used as an internal control for quantification of both nuclear and cytoplasmic mRNAs. One set of measurements of cytoplasmic to nuclear ratios of L2 penton mRNA is shown in Fig. 6, and export efficiencies estimated in several independent infections are summarized in Table 1. The E1B insertion mutations that severely impaired nuclear accumulation of the protein, such as A262 and R309, resulted in the greatest reductions in L2 mRNA export efficiency, 5- to 10-fold (Fig. 6; Table 1). With the exception of mutant A143 (and perhaps H354 [see Discussion]), the defects in E1B 55-kDa protein stability or nuclear accumulation in cells infected with the other mutants (Fig. 1B and 2) also appeared sufficient to account for the lower efficiencies of L2 penton mRNA export observed (Fig. 6; Table 1). However, the A143 mutation led to decreased export efficiency (Fig. 6; Table 1), without corresponding reductions in either the concentration of the altered protein (Fig. 1) or its partition into the nucleus (Fig. 2).
FIG. 6.
Alterations in the ratio of cytoplasmic to nuclear L2 penton mRNA induced by E1B 55-kDa protein coding sequence mutations. Steady-state concentrations of mature L2 penton mRNA in the cytoplasm and nucleus were determined 14 h after infection of HeLa cells with Ad5 or the mutant viruses indicated by primer extension. Signals were corrected using the stable, cellular β-actin mRNA as an internal control and used to calculate the ratios of cytoplasmic to nuclear penton mRNA shown.
In parallel, we also measured the export efficiency of the longest mRNA of the ML L4 family, that encoding the 100-kDa protein, when less-substantial defects than those summarized in Table 1 for L2 penton mRNA were observed. For example, the H224 and H354 mutations induced reductions of less than 50% in the export of the L4 100-kDa mRNA. This result is consistent with a previous report that the cytoplasmic accumulation of this viral L4 mRNA is much less severely affected by the absence of the E1B 55-kDa protein than is accumulation of mRNAs belonging to the other four ML families (50).
DISCUSSION
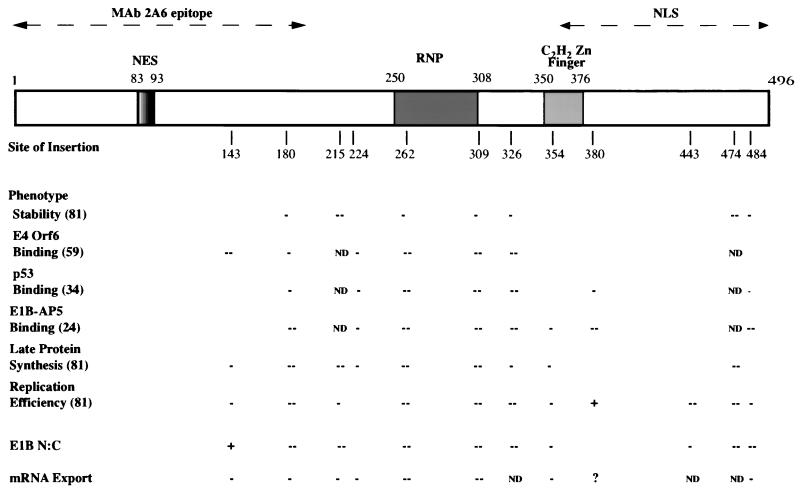
Various phenotypes, including virus yield and synthesis of viral late proteins in infected cells (81), of the recombinant Ad5/Ad2 viruses carrying small amino acid insertions in the E1B 55-kDa protein coding sequence examined in this work have been previously reported. The abilities of the altered proteins to bind to the viral E4 Orf6 and cellular p53 and E1B-AP5 proteins in infected cells have also been characterized (24, 34, 59). The results of these previous studies, as well as the new data reported here, are summarized in Fig. 7, which also shows potentially important sequences or sequence motifs of the 496-amino-acid E1B 55-kDa protein. Although all the altered E1B proteins encoded by these mutants have been reported to attain close to wild-type concentrations in infected A549 cells (59), we observed that several of the insertions significantly reduced E1B 55-kDa protein accumulation in infected HeLa cells (Fig. 1; Table 1). Our observations are in excellent agreement with those reported by Yew et al. in the initial characterization of these mutants (81). For example, in both sets of experiments, the H215 and H474 mutations reduced the steady-state concentrations of the E1B 55-kDa protein to very low levels, while clear but less severe defects were associated with the H180, A262, R309, H326, and F484 mutations (81) (Fig. 1B and 7; Table 1). Thus, insertions at these positions consistently reduce the stability of the E1B protein (at least in HeLa cells). Unexpectedly, we observed that several mutations that reduced E1B 55-kDa protein concentration more modestly were associated with substantial defects in nuclear accumulation: the total protein concentrations observed in cells infected with the H180, H215, A262, R309, A326, and F484 mutants were 2.5- to 3-fold lower than that attained in Ad5-infected cells (Fig. 1B; Table 1), but nuclear concentrations were reduced by factors of 5 to 10 (Fig. 2). Such selective decreases in the nuclear concentration of these E1B 55-kDa proteins could be the result of specific inhibition of nuclear import, abnormally rapid export from the nucleus, or accelerated degradation of the proteins within that organelle. However, several lines of evidence lead us to favor the view that the defects in nuclear accumulation reported here are a secondary consequence of misfolding of the altered E1B proteins.
FIG. 7.
Phenotypes exhibited by E1B 55-kDa protein sequence insertion mutants. The 496-residue protein is represented to scale at the top by the open box, within which are shown the positions of a leucine-rich sequence (NES) necessary and sufficient for export of E1B-GFP and glutathione S-transferase-E1B peptide-GFP fusion proteins, respectively (37); a sequence that matches the ribonucleoprotein (RNP) motif of many RNA-binding proteins in both the conserved RNP1 and RNP2 sequences and in which specific substitutions impair nonspecific RNA-binding activity of the E1B protein in vitro (32); and a potential sequence matching the consensus for C2H2 zinc fingers identified by inspection and a protein motif search in SeqWeb, version 2. The dashed, double-headed arrows drawn above indicate the region containing the epitope recognized by the monoclonal antibody 2A6 (34) that has been used in the majority of studies of the protein and a sequence required for nuclear localization (NLS) of an E1B-GFP fusion protein transiently synthesized in human cells (37). The short, vertical lines drawn immediately below the protein list the sites of insertions introduced into an Ad2/5 E1B 55-kDa protein coding sequence in the viral genome. With the exception of the 18-amino-acid addition at residue 215, these insertions comprise four amino acids (81). The effects of these alterations on the concentration of the protein and other properties listed at the left are summarized below (−− and −, severe and moderate defects, respectively; +, twofold increase in defects; ND, effect not determined; ?, effect uncertain). The sources of these data, which were collected under a variety of experimental conditions using HeLa or other lines of human tumor cells, are listed in parentheses at the left.
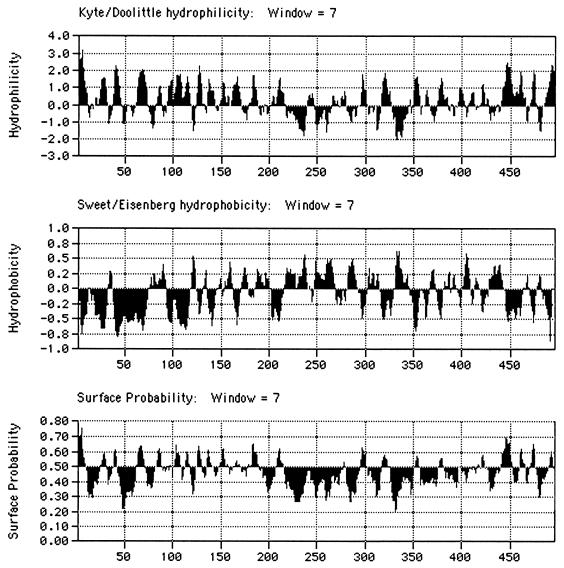
Each of the insertions that selectively impaired nuclear accumulation also reduced the steady-state concentration of the E1B protein (Fig. 1B and 2; Table 1), as would be expected were the proteins misfolded. In addition, the induction of similar nuclear accumulation phenotypes by insertions at widely spaced positions, from amino acid 180 to residue 484 in the protein, is difficult to reconcile with specific alterations in nuclear import or export. The latter process has been reported to be governed by a nuclear export signal occupying positions 83 to 93 (21, 37), a sequence not disrupted by any of the insertions that impaired nuclear accumulation. Although a nuclear localization signal has not been precisely located, the C-terminal segment comprising amino acids 364 to 496 directed the predominantly nuclear location of green fluorescent protein (GFP) when the E1B-GFP fusion protein was transiently synthesized in HeLa cells (37). This portion of the E1B protein contains only two sequences with the characteristics (short and basic) of many nuclear localization signals (17), amino acids 391 to 400 and 441 to 451, and none of the insertions that inhibited nuclear accumulation affect either of these sequences. Furthermore, when examined, each of the mutations that selectively decreased the nuclear concentration of the E1B protein has been reported to induce substantial defects in all other properties tested (Fig. 7). In particular, each of the H180, A262, R309, and H326 insertions strongly inhibited binding of the E1B protein to the viral E4 Orf6 (59) and the cellular E1B-AP5 (24) proteins in infected cell extracts and of the altered E1B proteins made in infected cells to the cellular p53 protein synthesized in vitro (34). A priori, it seems very unlikely that each of these three E1B-binding proteins contacts an identical surface and that each requires the same four regions defined by the sites of the insertions listed above. Thus, misfolding of these altered E1B proteins provides a more reasonable explanation of the pleiotropic defects observed. Indeed, it has been reported recently that the E1B 55-kDa protein sequences required for binding to the E4 Orf6 and p53 proteins can be distinguished by the effects of single-amino-acid substitutions on these interactions (62). Finally, although the structure of the E1B 55-kDa protein has not been determined, analysis of the primary sequence suggests that the central segment from approximately amino acid 215 to residue 345 is likely to make a major contribution to the interior core of the native protein: this segment contains the majority of sequences that are predicted by standard algorithms to be highly hydrophobic and largely buried (Fig. 8). All but one of the insertions that inhibited nuclear accumulation of the E1B protein lie within (H215, A262, R309, and H326) or adjacent to (H180) this largely hydrophobic region, consistent with the hypothesis that these altered proteins are misfolded (and perhaps aggregated) to such an extent that they cannot be recognized readily by the nuclear import machinery of the host cell.
FIG. 8.
Predicted properties of the E1B 55-kDa protein. The properties listed were predicted from the amino acid sequence of the 496-residue Ad5 E1B 55-kDa protein with the algorithms indicated, using the program MacVector 7.0.
It is also possible that failure of the altered F484 E1B protein to enter the nucleus efficiently is the result of improper modification. The E1B 55-kDa protein is phosphorylated in infected cells (43, 60, 79) at three residues close to the C terminus, Ser490, Ser491, and Thr495 (67-69). Substitution of alanine for these three residues eliminated the ability of the E1B protein both to block E1A-induced, p53-dependent apoptosis and to repress transcription when artificially bound to a promoter (68). Whether phosphorylation at any one of these residues governs the intracellular location of the protein is not known. However, it is well established that this modification can directly and indirectly regulate nuclear import, for example, of transcriptional activators such as NF-KB and STATS (4, 16), and the four-amino-acid insertion at position 484 might interfere with recognition of one or more of the C-terminal phosphorylation sites in the E1B protein.
The majority of the E1B mutant viruses we examined exhibited either quite modest defects in all properties assayed (H224 and R443) or substantial alterations in viral late mRNA metabolism that correlated with reduced stability and nuclear accumulation of the protein (H180, H215, A262, R309, H326, and F484). Thus, the defects in export of viral mRNA from the nucleus (Fig. 6; Table 1) induced by the latter mutations can be identified as secondary consequences of decreased E1B protein concentrations or failure of the altered proteins to enter the nucleus efficiently. A substantial reduction in L2 mRNA export efficiency was also associated with the H354 mutation (Fig. 6; Table 1), which has been reported to impair binding of the E1B 55-kDa protein to the cellular E1B-AP5 protein (24). The latter possesses sequence features and RNA-binding properties typical of RNP family proteins (24) and can bind to a human protein implicated in mRNA export, the Tap protein (3). The E1B-AP5 protein is thus an attractive candidate for a target of E1B protein-mediated regulation of mRNA export. However, the experiments reported here cannot definitively ascribe the defect in viral late mRNA export observed in cells synthesizing the altered H354 E1B protein to impaired binding to the E1B-AP5 protein: this mutation also led to reduced accumulation of the E1B 55-kDa protein in the nucleus (Fig. 2).
In contrast, the insertion at residue 143 resulted in impaired export of viral L2 penton mRNA from the nucleus (Fig. 6), but neither decreased significantly the protein's steady-state concentration (Fig. 1B) nor reduced its nuclear concentration (Fig. 2). Rather, the E1B 55-kDa protein synthesized in mutant A143-infected cells was found to be about twice as concentrated in the nuclear fraction as the wild type (Fig. 2), suggesting that it does not shuttle from nucleus to cytoplasm efficiently. As mentioned previously, the leucine-rich sequence at amino acids 83 to 93 is sufficient to mediate export of a heterologous protein via the exportin-1/Crm-1 pathway in uninfected and Ad5-infected human cells (21, 37). The four-amino-acid insertion at residue 143 lies some 50 amino acids C-terminal to this nuclear export signal and might therefore perturb secondary and/or tertiary structural features of the protein to render the sequence less accessible. This interpretation predicts that this insertion would inhibit export of an E1B-GFP fusion protein in assays for shuttling activity. Alternatively, the displacement of the E1B protein from specific intranuclear sites induced by the A143 mutation (Fig. 3) might hamper its export from the nucleus.
Several lines of evidence have implicated the E4 Orf6 protein and its interaction with the E1B 55-kDa protein in regulation of mRNA export in adenovirus-infected cells (see the introduction). However, the absence of this E4 protein results in pleiotropic phenotypes, including defects in viral DNA replication, in cytoplasmic accumulation of viral late mRNAs, and in splicing and stability of these mRNAs (20, 38). Consequently, formation of the E1B-E4 protein complex has not been unambiguously correlated with changes in mRNA export during the late phase of infection. The phenotypes exhibited by mutant A143 directly implicate the E1B-E4 protein complex in selective export of viral late mRNAs: previous studies have established that this insertion specifically inhibits interaction of the E1B 55-kDa protein with the E4 Orf6 protein in infected cell extracts (59) (Fig. 7), while the data reported here (Fig. 6; Table 1) demonstrate that this mutation impairs nuclear export of viral late mRNAs. Our conclusion that formation of the complex containing the viral E1B and E4 proteins is necessary for mRNA export regulation during adenovirus infection appears to contradict that recently reported by Shen and colleagues (62). These authors observed that replacement of Thr255 or His260 in the E1B 55-kDa protein by alanine strongly inhibited interaction with the E4 Orf6 protein in infected A549 cell extracts but did not impair viral late protein synthesis or inhibition of cellular protein synthesis (62). However, in A549 cells mutant viruses that cannot direct synthesis of the E1B 55-kDa or the E4 Orf6 protein replicate to within factors of 2 to 4 of wild-type Ad5 levels (15, 26, 28, 57, 65, 71), indicating that neither of these viral proteins is required for efficient viral late gene expression in this cell line. Indeed, Shen et al. (62) observed efficient synthesis of viral late proteins and almost complete inhibition of synthesis of cellular proteins in A549 cells infected with ONYX-015/dl1520, an Ad5 mutant that cannot express the E1B 55-kDa protein coding sequence (5).
Efficient replication of E1B null mutant viruses has been observed in a variety of other human cell lines, as well as in some primary human cells (6, 26, 28, 57, 71). The molecular basis for such host cell type dependence has not been elucidated. However, these mutant adenoviruses also exhibit both temperature- and cell cycle-dependent growth in HeLa cells, replicating more efficiently at elevated (37 to 39.5°C) than at low (32 to 33°C) temperatures and more efficiently (at 37°C) in cells infected when in S phase than in cells infected during G1 (25, 26, 31, 41). Under both conditions permissive for replication of the E1B mutants, export of viral late mRNAs to the cytoplasm is more efficient than that under the corresponding nonpermissive condition (27, 41, 76). Furthermore, it has been reported that infection at high temperature alleviates the cell cycle restriction to reproduction of E1B null mutant viruses (26). These observations suggest that both cell cycle progression and temperature modulate an mRNA export system(s) of the host cell and thus determine, at least in part, the need for the E1B 55-kDa protein to allow efficient viral late gene expression. The properties of the A143 mutant are also consistent with this hypothesis: this mutation induces defects in late mRNA export (Fig. 6; Table 1) and has been reported to restrict efficient virus growth in HeLa cells to those infected in S phase (27). It is therefore possible that differences in cellular export systems also contribute to the cell type dependence of replication of mutants that cannot direct synthesis of the E1B 55-kDa protein.
The insertion introduced by the A143 mutation also specifically eliminated association of the E1B protein with intranuclear, E2 DBP-containing replication centers (Fig. 3), in which viral DNA is replicated and transcribed during the late phase of infection (11). As the same failure of the E1B protein to localize to these structure is characteristic of infected cells in which the E4 Orf6 protein cannot be made (49), the altered intranuclear distribution of the A143 E1B protein can be attributed to its inability to interact with this E4 protein (59). In A143-infected cells, mislocalization of the E1B 55-kDa protein within the nucleus resulted in reduced efficiency of export of L2 mRNA (Fig. 6). In conjunction with the unaltered intranuclear localization of the E4 Orf6 protein observed in mutant A143-infected cells (Fig. 4), these properties provide strong support for the hypothesis that recruitment of the E1B protein by the E4 Orf6 protein to specialized nuclear sites is necessary for selective viral mRNA export. Furthermore, they indicate that the organization of the E4 Orf6 protein into specific structures within infected-cell nuclei neither requires binding to the E1B 55-kDa protein nor is sufficient for regulation of mRNA export. Additional comparisons of properties of the E1B 55-kDa proteins encoded by Ad5 and mutant A143 should provide insight into the molecular consequences of localization of the viral protein to these sites.
Acknowledgments
We thank Arnie Berk for generously providing the E1B insertion mutants; Arnie Levine, Tom Shenk, and Patrick Hearing for monoclonal antibodies and B6-8 and 2A-6, RSA-3, and M45, respectively; and Joe Goodhouse for invaluable assistance with confocal microscopy.
This work was supported by a grant from the National Institutes of Health, and R.A.G. was supported by a grant from CONACYT-SEP, Mexico.
REFERENCES
- 1.Aspegren, A., C. Rabino, and E. Bridge. 1998. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 245:203-213. [DOI] [PubMed] [Google Scholar]
- 2.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachi, A., I. C. Braun, J. P. Rodrigues, N. Pante, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Gorlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeuerle, P. A., and D. Baltimore. 1996. NF-κB: ten years after. Cell 87:13-20. [DOI] [PubMed] [Google Scholar]
- 5.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373-376. [DOI] [PubMed] [Google Scholar]
- 7.Blair-Zajdel, M. E., and G. E. Blair. 1988. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene 2:579-584. [PubMed] [Google Scholar]
- 8.Blobel, G. 1987. How proteins move across the endoplasmic reticulum membrane. Hepatology 7:26S-29S. [DOI] [PubMed] [Google Scholar]
- 9.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 11.Bridge, E., and U. Pettersson. 1996. Nuclear organization of adenovirus RNA biogenesis. Exp. Cell Res. 229:233-239. [DOI] [PubMed] [Google Scholar]
- 12.Bridge, E., K. U. Riedel, B. M. Johansson, and U. Pettersson. 1996. Spliced exons of adenovirus late RNAs colocalize with snRNP in a specific nuclear domain. J. Cell Biol. 135:303-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castiglia, C. L., and S. J. Flint. 1983. Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol. Cell. Biol. 3:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., R. Vinnakota, and S. J. Flint. 1994. Intragenic activating and repressing elements control transcription from the adenovirus IVa2 initiator. Mol. Cell. Biol. 14:676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences--a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 18.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 20.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-54. [DOI] [PubMed] [Google Scholar]
- 21.Dosch, T., F. Horn, G. Schneider, F. Krätzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55k oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg, A. P., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 23.Flint, S. J. 1986. Regulation of adenovirus mRNA formation. Adv. Virus Res. 31:169-228. [DOI] [PubMed] [Google Scholar]
- 24.Gabler, S., H. Schutt, P. Groitl, H. Wolf, T. Shenk, and T. Dobner. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada, J. N., and A. J. Berk. 1999. p53-independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J. Virol. 73:5333-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison, T. J., F. Graham, and J. F. Williams. 1977. Host range mutants of adenovirus type 5 defective for growth on HeLa cells. Virology 77:319-329. [DOI] [PubMed] [Google Scholar]
- 30.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, Y. S., R. Galos, and J. F. Williams. 1982. Isolation of type 5 adenovirus mutants with a cold-sensitive phenotype: genetic evidence of an adenovirus transformation maintenance function. Virology 122:109-110. [DOI] [PubMed] [Google Scholar]
- 32.Horridge, J. J., and K. N. Leppard. 1998. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J. Virol. 72:9374-9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, N. C. 1990. Transformation by the human adenoviruses. Semin. Cancer Biol. 1:425-435. [PubMed] [Google Scholar]
- 34.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179:806-814. [DOI] [PubMed] [Google Scholar]
- 35.Kasai, Y., H. Chen, and S. J. Flint. 1992. Anatomy of an unusual RNA polymerase II promoter containing a downstream TATA element. Mol. Cell. Biol. 12:2884-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 37.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 38.Leppard, K. N. 1998. Regulated RNA processing and RNA transport during adenovirus infection. Semin. Virol. 8:301-307. [Google Scholar]
- 39.Leppard, K. N. 1993. Selective effects on adenovirus late gene expression of deleting the E1B 55K protein. J. Gen. Virol. 74:575-582. [DOI] [PubMed] [Google Scholar]
- 40.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 41.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 43.Malette, P., S. P. Yee, and P. E. Branton. 1983. Studies on the phosphorylation of the 58000 dalton early region 1B protein of human adenovirus type 5. J. Gen. Virol. 64:1069-1078. [DOI] [PubMed] [Google Scholar]
- 44.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMaster, G. K., and G. G. Carmichael. 1977. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA 74:4835-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, R., M. Dixon, R. Smith, G. Peters, and C. Dickson. 1986. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J. Virol. 61:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ornelles, D., and T. Shenk. 1991. Location of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stablization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puvion-Dutilleul, F., E. Puvion, C. Icard-Liepkalns, and A. Macieira-Coelho. 1984. Chromatin structure, DNA synthesis and transcription through the lifespan of human embryonic lung fibroblasts. Exp. Cell Res. 151:283-298. [DOI] [PubMed] [Google Scholar]
- 53.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabino, C., A. Aspegren, K. Corbin-Lickfett, and E. Bridge. 2000. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 74:6684-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognise native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 56.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rothmann, T., A. Hengstermann, N. J. Whitaker, M. Scheffner, and H. zur Hausen. 1998. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J. Virol. 72:9470-9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowe, D. T., P. E. Branton, and F. L. Graham. 1984. The kinetics of synthesis of early viral proteins in KB cells infected with wild-type and transformation-defective host-range mutants of human adenovirus type 5. J. Gen. Virol. 65:585-597. [DOI] [PubMed] [Google Scholar]
- 59.Rubenwolf, S., H. Schütt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the Ad5 E1B-58K tumor antigen in adenovirus-infected and transformed cells. Virology 120:387-394. [DOI] [PubMed] [Google Scholar]
- 62.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shenk, T. 1996. Adenoviridae and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 64.Smiley, J. K., M. A. Young, C. L. Bansbach, and S. J. Flint. 1995. The metabolism of small cellular RNA species during productive subgroup C adenovirus infection. Virology 206:100-107. [DOI] [PubMed] [Google Scholar]
- 65.Smiley, J. K., M. A. Young, and S. J. Flint. 1990. Intranuclear location of the adenovirus type 5 E1B 55-kilodalton protein. J. Virol. 64:4558-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 67.Takayesu, D., J. G. Teodoro, S. G. Whalen, and P. E. Branton. 1994. Characterization of the 55K adenovirus type 5 E1B product and related proteins. J. Gen. Virol. 75:789-798. [DOI] [PubMed] [Google Scholar]
- 68.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teodoro, J. G., T. Halliday, S. G. Whalen, D. Takayesu, F. L. Graham, and P. E. Branton. 1994. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J. Virol. 68:776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcription activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turnell, A. S., R. J. Grand, and P. H. Gallimore. 1999. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J. Virol. 73:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van den Heuvel, S. J., T. van Laar, W. M. Kast, C. J. Melief, A. Zantema, and A. J. van der Eb. 1990. Association between the cellular p53 and the adenovirus 5 E1B-55kd proteins reduces the oncogenicity of Ad-transformed cells. EMBO J. 9:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voelkerding, K., and D. F. Klessig. 1986. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J. Virol. 60:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weis, K. 1998. Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci. 23:185-189. [DOI] [PubMed] [Google Scholar]
- 75.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams, J., B. D. Karger, Y. S. Ho, C. L. Castiglia, T. Mann, and S. J. Flint. 1986. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells 4:275-284. [Google Scholar]
- 77.Williams, J. F. 1973. Oncogenic transformation of hamster embryo cells in vitro by adenovirus type 5. Nature 243:162-163. [DOI] [PubMed] [Google Scholar]
- 78.Yang, U.-C., W. Huang, and S. J. Flint. 1996. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 70:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yee, S. P., D. T. Rowe, M. L. Tremblay, M. McDermott, and P. E. Branton. 1983. Identification of human adenovirus early region 1 products by using antisera against synthetic peptides corresponding to the predicted carboxy termini. J. Virol. 46:1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 81.Yew, P. R., C. C. Kao, and A. J. Berk. 1990. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker-insertional mutagenesis. Virology 179:795-805. [DOI] [PubMed] [Google Scholar]
- 82.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]