Abstract
Enteroviral persistence has been implicated in the pathogenesis of several chronic human diseases, including dilated cardiomyopathy, insulin-dependent diabetes mellitus, and chronic inflammatory myopathy. However, these viruses are considered highly cytolytic, and it is unclear what mechanisms might permit their long-term survival. Here, we describe the generation of a recombinant coxsackievirus B3 (CVB3) expressing the enhanced green fluorescent protein (eGFP), which we used to mark and track infected cells in vitro. Following exposure of quiescent tissue culture cells to either wild-type CVB3 or eGFP-CVB3, virus production was very limited but increased dramatically after cells were permitted to divide. Studies with cell cycle inhibitors revealed that cells arrested at the G1 or G1/S phase could express high levels of viral polyprotein and produced abundant infectious virus. In contrast, both protein expression and virus yield were markedly reduced in quiescent cells (i.e., cells in G0) and in cells blocked at the G2/M phase. Following infection with eGFP-CVB3, quiescent cells retained viral RNA for several days in the absence of infectious virus production. Furthermore, RNA extracted from nonproductive quiescent cells was infectious when transfected into dividing cells, indicating that CVB3 appears to be capable of establishing a latent infection in G0 cells, at least in tissue culture. Finally, wounding of infected quiescent cells resulted in viral protein expression limited to cells in and adjacent to the lesion. We suggest that (i) cell cycle status determines the distribution of CVB3 during acute infection and (ii) the persistence of CVB3 in vivo may rely on infection of quiescent (G0) cells incapable of supporting viral replication; a subsequent change in the cell cycle status may lead to virus reactivation, triggering chronic viral and/or immune-mediated pathology in the host.
Coxsackieviruses are members of the picornavirus family and Enterovirus genus, which is subdivided into coxsackieviruses A and B, polioviruses, echoviruses, and other unclassified enteroviruses. Acute coxsackievirus infection can cause diseases ranging from mild (rash and myalgia) to severe (pancreatitis, meningitis, and myocarditis). Unsuspected acute viral myocarditis may lead to the collapse and death of young and vigorous individuals, especially during exertion, from catastrophic dysfunction of the electrical pathways in the heart (5, 62). Although the majority of symptomatic patients recover well from acute myocarditis, inflammatory events may continue or recur and can have serious long-term sequelae; some 10 to 20% of patients with symptomatic enteroviral myocarditis (∼20,000 to 40,000/year in the United States) will develop chronic disease, progressing over time (usually years) to dilated cardiomyopathy (DCM) (38, 54), where one or both ventricles dilate and decompensate, with resulting cardiac failure. The prevalence of DCM in the general population is much lower (∼0.005%), and a large study showed a strong correlation (P < 0.001) between prior coxsackievirus infection and DCM (51).
The enterovirus most commonly associated with myocarditis is coxsackievirus B3 (CVB3), but the mechanisms underlying viral pathogenicity—especially the ongoing myocarditis sometimes seen long after the clearance of infectious virus—remain obscure. Coxsackieviruses are usually considered highly cytolytic, both in tissue culture and in vivo. However, several enteroviruses can establish long-term persistent infections in tissue culture, perhaps by the emergence of viral variants (8, 50, 58), and some researchers hypothesize that persistent enteroviral infections may underlie several chronic human diseases. Although this idea remains quite controversial for humans (30, 35), slot blot hybridization studies have shown positive signal for coxsackievirus RNA in myocardial biopsy specimens from approximately 45% of patients with myocarditis or DCM compared with none of the controls (29), and some 43% of patients with healed myocarditis or DCM remained positive for CVB signal (2). In addition, unequivocal data from animal models indicate that coxsackieviral RNA may persist for months or years in vivo (1, 22, 59), and ultrastructural changes occur in cells which contain CVB RNA (22).
These data, although suggesting that CVB may persist in vivo, do not prove it, because infectious coxsackievirus was not isolated from these human or murine tissues, and defective CVB RNA molecules can—in the absence of infectious virus production—cause cytopathology when introduced into cells by transfection (63) or via transgenic procedures (64). However, the CVB RNA which persists in vivo does not carry mutations and appears to persist, in the absence of replication, as a stable double-stranded viral RNA complex (58). Therefore, persistent CVB RNA might represent a latent form of the virus which, although difficult to reactivate experimentally, could act as the nidus of productive infection under appropriate conditions in vivo.
It is important to understand how a lytic virus such as coxsackievirus can suspend replication and, perhaps, establish latency in susceptible cells. As a first step, in this study we have investigated the interaction between the virus and the host cell in tissue culture. We have focused on (i) the role of the cell cycle in regulating the host cell's ability to support productive CVB3 infection and (ii) the capacity of infected cells to retain potentially infectious RNA when they do not produce detectable infectious virus. These studies of CVB3 tropism and persistence have been facilitated by our construction of a recombinant CVB3 expressing enhanced green fluorescent protein (eGFP) (eGFP-CVB3).
MATERIALS AND METHODS
Cells and virus.
HeLa RW cells were obtained from R. Wessely (then at University of California-San Diego); CVB infection of these cells results in large plaques. Cells are maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml (complete DMEM). These cells were used for the preparation of viral stocks and plaque assays. wtCVB3 was produced by transfection of HeLa RW cells with plasmid pH3 (24) (kindly provided by Kirk Knowlton, University of California, San Diego), which encodes a myocarditic strain of CVB3.
Virus titration.
Six-well dishes were plated with 3.75 × 105 HeLa RW cells/well 48 h prior to infection. Serial dilutions (10-fold dilutions with DMEM) were incubated on HeLa RW monolayers for 1 h and rocked every 15 min. After absorption, cells were overlaid with 4 ml of 1× DMEM in 0.6% agar (1:1 mixture of 2× DMEM at 37°C and 1.2% agar at 50°C). At 40 to 48 h following infection, cells were fixed by addition of 2 ml of methanol-acetic acid (3:1, vol/vol) to each well for 10 min, after which the fixative and plugs were removed. Cells were stained with 1 ml of a crystal violet solution (0.5% crystal violet in 20% ethanol) for 1 h and rinsed in tap water, and plaques were counted.
Recombinant eGFP-CVB3 cDNA construction and virus isolation.
Our laboratory has engineered a unique SfiI restriction enzyme site into the backbone of the infectious CVB3 viral clone pH3. The resulting plasmid, pMKS1 (52), facilitates the insertion of a foreign sequence into the CVB3 genome and the subsequent isolation of recombinant virus following transfection of the plasmid into HeLa RW or COS cells. A fragment encoding eGFP was amplified by PCR from plasmid pEGFP (Clontech, Palo Alto, Calif.) and cloned into pMKS1. COS cells were transfected, and virus was harvested after 48 h and freeze-thawed three times on dry ice to release virions. HeLa RW cells were incubated with freeze-thawed supernatants for 1 h, and the recombinant eGFP-CVB3 virus was harvested from these cells after 48 h. The recombinant virus was plaque purified, and viral stocks were prepared in HeLa RW cells.
Induction of cellular quiescence by serum depletion.
A total of 5 × 104 HeLa RW cells were added to each well of a 24-well plate and grown in DMEM plus 10% fetal bovine serum (FBS) (complete medium) overnight. The medium was replaced with DMEM (no FBS) for 24 h to induce quiescence, and then the cells were infected with 104 PFU of eGFP-CVB3 (approximate multiplicity of infection [MOI] of 0.1) for 1 h at 37°C and washed with saline solution. Infected cells were processed as described in the text and figures.
Cell cycle arrest using inhibitors of the cell cycle.
A total of 5 × 104 HeLa RW cells were added to each well of a 24-well plate and grown in DMEM plus 10% FBS overnight in a 37°C incubator with 5% CO2. At this time, wells were ∼75% confluent. The indicated cell cycle inhibitor (300 μM mimosine, 25 nM rapamycin, 1 mM hydroxyurea, 5 μg of aphidicolin per ml, 1 μg of nocodazole per ml, or 200 nM taxol) was added, and cells were incubated for 16 h (concentrations of all inhibitors were taken from reference 6). All inhibitors (and control wells) contained dimethyl sulfoxide (DMSO) at a final concentration of 0.1%. Cells were infected with wtCVB3 or eGFP-CVB3 (MOI = 0.1) for 1 h and washed with saline solution. Infected cells were then processed as described above.
Evaluation of cell status by propidium iodide staining.
Cells were analyzed by propidium iodide staining and flow cytometry to confirm that the cell cycle inhibitors or serum starvation was having the intended effects. HeLa RW cells were treated with the various cell cycle inhibitors in complete medium for 16 h as described above. Alternatively, HeLa RW cells were grown for 16 h in the presence of DMEM with 0, 0.1, 1.0, or 10% FBS. The cells were trypsinized, washed twice with saline solution, and fixed with 1 ml of 2% neutral-buffered formaldehyde for 30 min at room temperature. Fixed cells were incubated in a propidium iodide-phosphate-buffered saline (PBS) solution (containing propidium iodide at 40 μg/ml and RNase A at 100 μg/ml) for 30 min at 37°C. After being washed with PBS, cells were analyzed by flow cytometry to measure DNA content under the FL2 channel. Cells inhibited at G1 and G1/S were expected to have diploid DNA (2N) content, and cells inhibited at G2/M were expected to have tetraploid DNA (4N) content. A normal cycling population (i.e., cells grown in complete medium) was expected to have ∼5 to 10% of cells in G2 (4N).
Evaluation of viral polyprotein expression by flow cytometry.
Cells were infected as described in the text, and at the indicated times, the cells were trypsinized, centrifuged, washed, and fixed with 2% neutral-buffered formaldehyde prior to analysis of eGFP expression by flow cytometry.
Identification of CVB3 RNA by RT-PCR.
All work was carried out in a designated PCR-clean area. RNA was extracted from infected cells using Trizol reagent (Gibco-BRL, Rockville, Md.) and isolated as specified by the manufacturer. RNA was isolated from uninfected HeLa RW cells as a negative control. RNA samples were resuspended in diethyl pyrocarbonate (DEPC)-treated water, quantified, and stored at −80°C.
Primers were designed to amplify the 5′ untranslated leader of CVB3 (CVB3 forward primer, 5′-GCTAGTTGGTAATCCTCCGGCCCCTGAATG-3′; CVB3 reverse primer, 5′-AATAAAATGAAACACGGACACCCAAAGTAG-3′). Reverse transcription (RT) of eGFP-CVB3 RNA was performed using SuperScript II reverse transcriptase (Gibco-BRL), as specified by the manufacturer. Briefly, approximately 1 μg of total RNA was added to 10 μl of DEPC-treated water plus 30 pmol of CVB3 forward or reverse primer (to detect antigenomic or genomic RNA, respectively). The mixture was heated to 70°C for 10 min and chilled on ice. Next, 4 μl of 5× first-strand buffer, 0.01 M dithiothreitol, and 5 nmol of each deoxynucleotide were added to bring the reaction to 20 μl total volume, and the contents were incubated at 42°C for 2 min. Then 200 U of Superscript II RT enzyme was added, and the mixture was incubated at 42°C for an additional 50 min.
The β-actin primers (Gibco-BRL) were used in parallel for RT-PCR for each sample to ensure the integrity of the isolated RNA. PCR was used to amplify the RT products using a DNA Engine (MJ Research, Watertown, Mass.). Prior to PCR amplification, the reaction mix was incubated at 70°C for 15 min to inactivate the Superscript II RT enzyme. A 50-μl PCR mix contained 2.5 U of Taq polymerase (Gibco-BRL), primers (30 pmol of each), 10 nmol of each deoxynucleotide, 1.0 mM MgCl2, and one-tenth of the RT product (2 μl). The amplification was carried out for 40 cycles under the following conditions: an initial denaturation of 95°C for 3 min plus 40 cycles of 95°C for 45 s, 65°C for 45 s, and 72°C for 1 min. This was followed by a final extension at 72°C for 10 min. The PCR amplification products (140 bp for CVB3 and eGFP-CVB3 products) were detected by electrophoresis on a 1.0% agarose gel.
Detection of infectious RNA by transfection of cellular RNA from infected cells.
HeLa RW cells were grown to 75% confluence in six-well plates. Transfection of cellular RNA was done using Lipofectamine Plus (Gibco-BRL). Plus reagent (6 μl) was mixed with 5 μl (approximately 250 ng) of cellular RNA and 100 μl of OptiMEM medium (Gibco-BRL) in a polystyrene snapcap tube. In a separate tube, 4 μl of Lipofectamine reagent was added to 100 μl of OptiMEM. After 15 min, both samples were mixed and incubated a further 15 min. The entire volume was added to prewashed (with OptiMEM) HeLa RW cells and brought to 1 ml with OptiMEM. The cultures were incubated at 37°C for 5 h. A further 3 ml of complete medium was added to each well, and incubation was continued for 12 more hours at 37°C. Every day until day 4, a 1-ml aliquot of supernatant was harvested (for titration) and replaced with 1 ml of fresh complete medium. Cells were also observed for cytopathic effect until day 4.
Cell wounding on chamber slides.
A total of 5 × 105 HeLa RW cells in complete medium were added to the wells of a four-chamber slide (Becton Dickinson Labware, Franklin Lakes, N.J.) and incubated for 48 h, by which time the cells had formed overconfluent monolayers. Thereafter, two experimental approaches were used.
(i) Wounding cells after infection.
A total of 105 PFU of eGFP-CVB3 (MOI, approximately 0.1) were added to three chambers, and 1 h later the inoculum was removed and the infected cells were washed with saline solution. In one chamber, the cell monolayer was left intact; in the second chamber, the monolayer was wounded immediately postinfection by scratching with a sterile pipette tip; and in the third chamber, the monolayer was scratched 12 h after infection. In all cases, the infected cells were incubated for a total of 24 h postinfection in complete DMEM and then fixed, and eGFP expression was detected by fluorescence microscopy.
(ii) Infecting cells after wounding.
The monolayer in one chamber was scratched, and 12 h later, 106 PFU of eGFP-CVB3 were added to the chamber. Immediately postinfection, the infected cells were overlaid with 1.5 ml of complete DMEM with 0.6% agarose and observed under a fluorescence microscope at the indicated times postinfection.
RESULTS
Isolation and characterization of a recombinant CVB3 expressing eGFP.
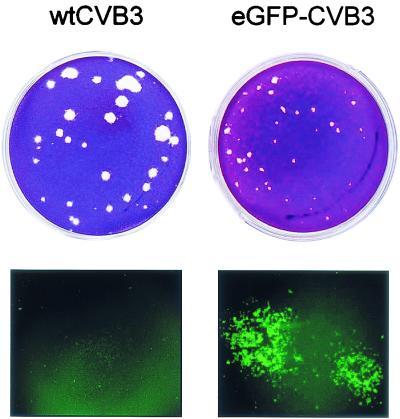
Recently, stable recombinant coxsackieviruses have been successfully prepared by several labs, including our own (7, 14, 15, 52). As described in Materials and Methods, the eGFP gene was cloned into an infectious cDNA, and virus was generated. Compared to wtCVB3, eGFP-CVB3 displayed slightly delayed growth kinetics (as described for other recombinant CVB3 in reference 52; data not shown). eGFP-CVB3 plaques were smaller, and fluorescence microscopy revealed extensive eGFP expression in cells within and surrounding the plaques (Fig. 1). The eGFP insert was stably maintained in eGFP-CVB3 to at least five passages in vitro and can be detected in the hearts of B-cell-knockout mice for at least 30 days postinfection (data not shown).
FIG. 1.
Comparison of plaques formed by wtCVB3 and eGFP-CVB3. HeLa RW cells were infected with the indicated viruses, and 24 h later the cells were evaluated by fluorescence microscopy (lower row). At 48 h postinfection the monolayers were fixed, and plaques were visualized by crystal violet staining (upper row).
While growing the recombinant virus in HeLa RW cells, we noted that even at high multiplicities of infection (MOI = 5), only a portion of the cell population expressed high levels of eGFP early after infection (12 h). When more time was allowed to pass (∼20 h) under routine tissue culture conditions, all cells either expressed eGFP or displayed cytopathic effects. These initial observations were serendipitous and could not be considered quantitative but led us to hypothesize that the cell cycle status might be affecting viral polyprotein synthesis; perhaps, in an asynchronous tissue culture population, only those cells at the appropriate stage could express eGFP soon after infection, but at later times, all of the cells would have completed at least one full cell cycle, and thus all cells could make eGFP. All subsequent experiments reported here were designed to test this hypothesis.
Viral polyprotein expression is enhanced in actively dividing cells.
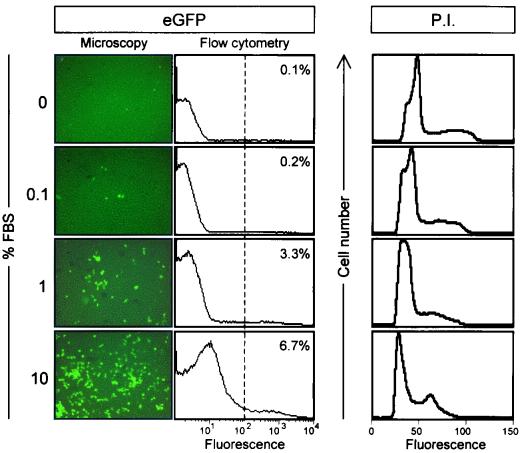
Cells can be made quiescent by serum removal, so our first approach was to maintain cells in various concentrations of FBS and then to infect them with eGFP-CVB3 (MOI = 0.1). After 16 h, eGFP expression was determined by fluorescence microscopy. As shown in Fig. 2, cells driven into quiescence by low concentrations of FBS expressed very low levels of eGFP, indicating poor expression of the viral polyprotein. Expression increased at 1% FBS, and in complete medium (10% FBS), expression was much more readily detected. To quantitate the proportion of cells expressing eGFP, cells were fixed, and analyzed by flow cytometry; the resulting data confirmed the visual impression. To ensure that serum starvation resulted in cellular quiescence, cells were stained with propidium iodide and analyzed by flow cytometry. As shown in the right-hand panels of Fig. 2, a clear peak of dividing cells was present only in the population maintained in 10% FBS.
FIG. 2.
Serum-depleted cells are quiescent and fail to express CVB3 polyprotein. HeLa RW cells were grown for 24 h in medium with the indicated concentrations of FBS and then infected with eGFP-CVB3 (MOI = 0.1). The infected cells were incubated for 16 h in the indicated medium, fixed, and observed under a fluorescence microscope (left panels). All images are shown at ×31 magnification. For each sample, duplicate cells were harvested and evaluated by flow cytometry (middle panels). The intensity of fluorescence (arbitrary units, log10 scale) is shown, and the dotted line represents a level of fluorescence 10-fold greater than the mean fluorescence of cells grown in 10% FBS. In each population, the percentage of cells showing fluorescence above this level is indicated. To identify the cell cycle status of the cells grown in the different serum concentrations, cells were stained with propidium iodide (P.I.) and analyzed by flow cytometry; the results are shown at the right-hand side of the figure.
eGFP expression varies markedly with cell cycle status.
A trivial explanation for reduced viral expression in FBS-depleted cells would be that starvation, rather than the cell cycle, had a direct inhibitory effect on the virus replication complex. However, the observed expression of eGFP in only a subset of dividing cells led us to hypothesize that CVB3 expression might depend on the cell cycle status of infected cells; cells forced into quiescence (G0) by serum starvation might be unable to express the viral polyprotein, but eGFP would be expressed in cycling cells which were at a particular stage of the cell cycle. In support of this idea, early studies on the replication of other picornaviruses indicated that the kinetics of viral RNA synthesis in synchronized cells differed depending on the phase of the cell cycle (10, 25, 28, 56).
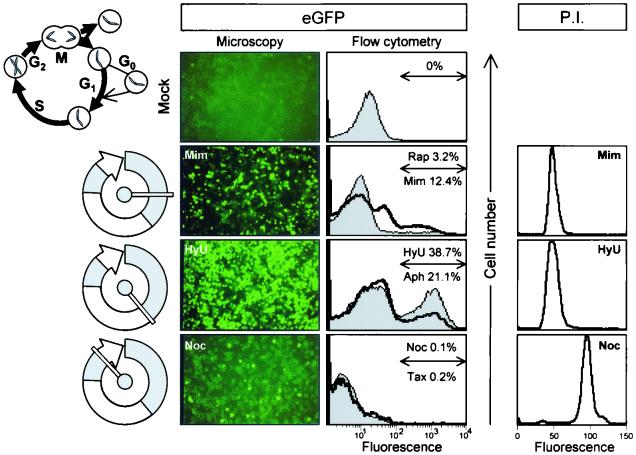
To address the question of cell cycle dependence, cells were arrested at various stages of the cell cycle using inhibitors which block replicating cells at either the G1 phase (mimosine and rapamycin), the G1/S phase (hydroxyurea and aphidicolin), or the G2/M phase (nocodazole and taxol). After incubation for 16 h in the presence of inhibitor, cells were infected with eGFP-CVB3 (MOI = 0.1), and medium (plus inhibitors) was restored. At 16 to 20 h later, eGFP expression (as an indicator of viral polyprotein expression) was evaluated visually and quantitated by flow cytometry (Fig. 3). eGFP expression was detected in ∼3 to 12% of cells arrested in the G1 phase and in ∼21 to 39% of cells arrested at the G1/S junction, but was almost undetectable in cells arrested at the G2/M phase. It is noteworthy that the proportion of cells expressing eGFP in a nonsynchronized actively dividing population (∼7%, Fig. 2) can be increased by enriching for cells at the G1/S stage (Fig. 3), consistent with the hypothesis that only a subset of cells in a dividing population can support viral gene expression. Note that, since an MOI of ∼0.1 was used, the presence of eGFP in >10% of cells indicates that secondary infection must have occurred during the 20-h incubation in the presence of inhibitors. Production of infectious virus by inhibitor-treated cells is described below.
FIG. 3.
CVB3 polyprotein expression varies markedly depending on cell cycle status. HeLa RW cells were treated for 16 h with the indicated cell cycle inhibitor [rapamycin (Rap), mimosine (Mim), hydroxyurea (HyU), aphidicolin (Aph), nocodazole (Noc), or taxol (Tax)] and then infected with eGFP-CVB3 (MOI = 0.1). Medium (plus inhibitors) was restored, and the infected cells were incubated for a further 20 h, fixed, and observed under a fluorescence microscope. A representative field is shown for one inhibitor at each stage of cell cycle arrest (left panels). All images are shown at ×31 magnification. In addition, for each sample, cells were harvested at 16 h and evaluated by flow cytometry. Histograms are shown for all inhibitors used (middle panels). At each stage of the cell cycle, the drug named above the arrow correlates with the thin line/filled curve, and the drug named below the arrow corresponds to the thick line; and for each inhibitor, the percentage of fluorescent cells is indicated. A diagram of the cell cycle (∼24 h for human cells) is included, and the stages at which the cells are arrested are shown to the left of each row. To ensure that the cell cycle inhibitors were having the expected effects, inhibitor-treated cells were stained with propidium iodide (P.I.) and analyzed by flow cytometry; the results for one inhibitor of each class are shown at the right-hand side of the figure. Similar results were obtained for the other drug of each pair.
To ensure that the cell cycle inhibitors were having the expected effects, inhibitor-treated cells were stained with propidium iodide and analyzed by flow cytometry. As shown in Fig. 3, cells blocked at G1 or G1/S (i.e., prior to DNA synthesis) showed tight peaks and had similar levels of fluorescence, while cells blocked at G2/M (i.e., after DNA synthesis) also showed a tight peak and fluoresced twice as strongly. Thus, the inhibitors had the intended effect.
Production of infectious virus varies with cell cycle status and can be rescued by allowing nonproductive cells to enter the cell cycle.
In the above experiment, cell cycle dependence was inferred by using eGFP as a surrogate marker of viral polyprotein expression. Next, we expanded this analysis in three ways. First, to ensure that the effects were not limited to recombinant viruses, we used both eGFP-CVB3 and wtCVB3; second, we evaluated infectious virus production by arrested cells; and, third, we determined whether “nonexpressing” cells (arrested at the G0 or G2/M stage) could produce virus if they were permitted to cycle after exposure to CVB3.
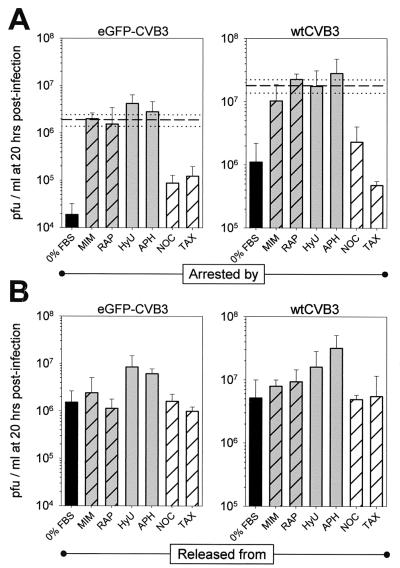
Cells were incubated with inhibitors for 16 h and then infected. Alternatively, cells were placed in serum-free medium and infected immediately. Twenty hours postinfection, supernatants were harvested and virus was counted (Fig. 4A). For both eGFP-CVB3 and wtCVB3, the highest level of virus production occurred in cells arrested at the G1/S phase; virus production was only slightly lower in cells arrested in G1 and was ∼10- to ∼100-fold lower in cells arrested at the G2/M or G0 phase. For comparative purposes, both viruses were also grown for 20 h in a cycling cell population (i.e., with 10% FBS). As shown in Fig. 4A, for both eGFP-CVB3 and wtCVB3, virus yields from cycling cells were similar to those obtained from cells blocked at the G1 phase, marginally lower than those for cells held at G1/S, and substantially higher than the yields from quiescent cells or from cells blocked at G2/M. Thus, the effect of the cell cycle on the production of infectious virus (Fig. 4A) correlates very well with its effect on eGFP expression (Fig. 3), and it affects not only recombinant CVB but also wild-type virus.
FIG. 4.
Production of infectious CVB3 is cell cycle dependent. HeLa RW cells were grown to 70% confluence in complete medium and then for 16 h in the presence of the indicated cell cycle inhibitor. G0 cells were obtained by growing cells in medium with 0% FBS. Cells were infected with eGFP-CVB3 or wtCVB3 (MOI = 0.1) for 1 h, washed with saline solution, and given either appropriate medium to maintain their arrested cell cycle status (A) or complete medium without inhibitors to allow cycling (B). Then, 20 h later, viral titers were determined from supernatants. All experiments were carried out in triplicate for each drug and time point, and error bars represent the standard deviation. Bars are coded according to the stage of cell cycle arrest: black, G0; grey hatched, G1; grey, G1/S; hatched, G2/M. For comparative purposes, both viruses were also grown in cycling cells (i.e., with 10% FBS) and harvested after 20 h. The average titers (dashed lines) and standard deviations (dotted lines) are included in the charts in panel A. See the legend to Fig. 3 for abbreviations.
What happens if, immediately after infection, the arrested cells are “released” by the provision of complete medium? As shown in Fig. 4B, in cells which had been arrested at the G0 or G2/M stage, infected, and then grown for 16 h in complete medium (without inhibitors), virus production increased substantially (5- to 80-fold). Thus, virus production is severely limited in cells infected and held at the G0 or G2/M stage (Fig. 4A), but these cells can produce substantial quantities of virus if allowed to divide (Fig. 4B). The release data show that the low production of virus from cells arrested at, for example, the G2/M stage cannot be due to lack of virus binding because, after the virus inoculum has been removed, the cells produced virus if permitted to cycle, indicating that virus must have bound to cells arrested at the G2/M stage. By similar reasoning, the limited virus production by cells held continually in G0 or G2/M (Fig. 4A) cannot be due to the inhibitors' having caused irrevocable damage to the cells. Indeed, related studies (data not shown) indicated that the opposite was true; after infection, the viability of cells maintained at the G0 or G2/M phase was higher than the viability of cells permitted to enter the cell cycle. For example, after infection of G0 cells, the viability of cells maintained for 20 h postinfection at the G0 stage was ∼95%, but the viability of cells permitted to cycle for 20 h after infection was ∼25%. We suggest that the continual arrest of G0 or G2/M cells protects against the virus-induced cytolysis which occurs if the cells are permitted to divide.
Virus can be reactivated for several days after infection of quiescent cells.
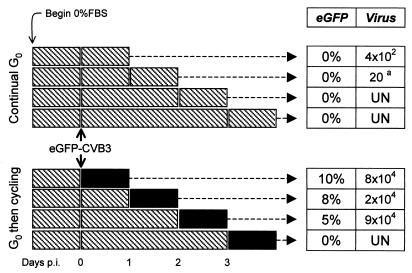
The above data show that quiescent tissue culture cells do not support extensive viral polyprotein synthesis or virus production unless encouraged to enter the cell cycle. We next wished to determine for how long after infection such cells could retain viral materials. HeLa RW cells were rendered quiescent by growth for 24 h in FBS-free medium and then exposed to eGFP-CVB3 (MOI = 0.1) for 1 h. Cells were washed and further incubated in serum-free medium for 0 to 3 days postinfection. At each of these four time points, the serum-free medium was replaced either by complete medium (to permit the cells to cycle) or by fresh serum-free medium (negative controls), and 24 h later, the cells were evaluated for viral protein expression (eGFP) and virus production (plaque assay). The experimental protocol and resulting data are shown in Fig. 5.
FIG. 5.
CVB latency in and reactivation from quiescent tissue culture cells. Cells were grown for 24 h in serum-free medium, infected with eGFP-CVB3 (MOI = 0.1), and washed. The infected cells were then grown in serum-free medium for 0, 1, 2, or 3 days (the four rows in each data set), after which times the serum-free medium was replaced by either fresh serum-free medium (upper data set) or complete medium (lower data set). Then, 24 h later, the cells were evaluated for eGFP expression and for virus production. For clarity, the experimental procedures are shown in diagrammatic form. Each rectangle represents the addition of fresh medium. Hatched rectangles indicate incubation in serum-free medium, and solid rectangles indicate complete medium. For each time point in both data sets, the percentage of cells expressing eGFP and the virus titers (PFU per milliliter) are tabulated. All samples were evaluated in triplicate; values shown are the means. For sample a, virus was detected in only one of the triplicates. UN, undetectable.
When infected cells were held continually in G0 (Fig. 5, upper data set), eGFP expression could not be detected at any of the four selected time points. This is consistent with the low expression observed at 16 h postinfection in Fig. 2 and indicates that virus polyprotein expression is very limited in quiescent cells even after prolonged culture. In addition, when cells were not allowed to cycle, infectious virus production was extremely limited. Under these conditions, only a few hundred PFU were produced at the earliest time point assayed, and after 2 days in serum-free medium, virus could be found in only one of the triplicate samples; thereafter, infectious virus could not be detected. In contrast, if complete medium was restored to the infected cells for 24 h (Fig. 5, lower data set), either immediately postinfection or after 1 or 2 additional days in serum-free medium, eGFP expression was initiated in ∼5 to 10% of the cells, a number consistent with the idea that gene expression of the input virus (MOI = 0.1) had been activated, and infectious virus was produced. However, if the infected cells were maintained in serum-free medium for 3 days, the subsequent addition of complete medium did not lead to eGFP expression or virus production.
Taken together, these data indicate that CVB3 can be maintained for at least 2 days in quiescent tissue culture cells in the absence of extensive viral polyprotein expression or infectious virus production and that the virus can be reactivated by the restoration of complete medium. However, at least in this in vitro system, when cells are forced to remain quiescent (i.e., in G0) for 3 days after infection, neither viral polyprotein expression nor the production of infectious virus can be detected even after the cells are permitted to enter the cell cycle.
CVB3 RNA is present at 3 days postinfection in quiescent cells which neither express viral proteins nor produce infectious virus.
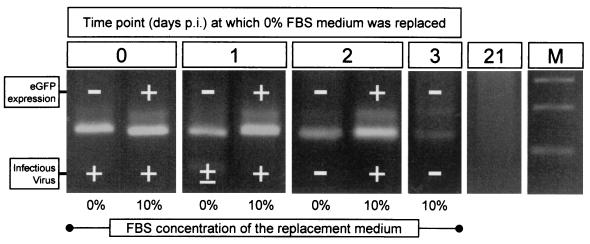
The absence of detectable eGFP expression and virus production could most obviously be attributed to the loss of all viral materials from the cells. However, an alternative explanation is that viral materials are retained, but in a nonexpressed form. To distinguish between these possibilities, the triplicate cell samples at each time point shown in Fig. 5 were pooled, and RNA was prepared and analyzed by RT-PCR with CVB-specific primers (Fig. 6); RT-PCR was also done with β-actin primers to ensure that all samples contained RNA. To facilitate comparison between the presence and absence of CVB3 RNA (Fig. 6) and gene expression and virus production (Fig. 5), each RT-PCR lane is marked (+ or −) to indicate the eGFP and infectious virus status of the source cells.
FIG. 6.
CVB3 RNA is present 3 days after infection of quiescent cells despite lack of eGFP expression or virus production. RT-PCR was performed on RNA isolated from cells infected with eGFP-CVB3 and maintained in DMEM without FBS for the indicated amount of time before replacement of medium (addition of complete DMEM or serum-free DMEM for an additional 24 h). Triplicate samples of cells from each time point of the experiment diagrammed in Fig. 5 were pooled, RNA was prepared, and an aliquot was analyzed by RT-PCR. Products are shown from RT-PCRs using primers to detect genomic CVB RNA. For each RNA sample, the eGFP expression and virus production in the source cells (data from Fig. 5) are indicated by + or −. The day 21 RNA was derived from cells which were infected, grown for 3 days in serum-free medium, then given complete medium, and passaged twice until day 21 postinfection. Lane M, DNA size markers.
As shown in Fig. 6, genomic CVB3 RNA could be detected in all samples analyzed up to 3 days postinfection; most importantly, viral RNA was present in cell populations in which eGFP expression and/or infectious virus production could not be detected. Parallel cultures were held in serum-free medium for 3 days postinfection and then passaged in complete medium until 21 days postinfection. As shown in Fig. 6, viral RNA could not be detected at this time point; these cells also showed no visible cytopathic effect and no detectable eGFP expression (data not shown). However, the cells could be reinfected by eGFP-CVB (data not shown), excluding the possibility that we had selected for cells that were fundamentally resistant to this virus. In all cases where genomic RNA was detected, the antigenomic material was also present at similar levels (data not shown). However, because our RT-PCR studies were not designed to precisely quantitate the RNAs, we cannot draw any conclusions about the relative abundances of genomic and antigenomic materials. No β-actin RT-PCR signal was detected in the RNA from cells harvested at day 3 and replaced with 0% FBS (which were negative for both eGFP and virus; see Fig. 5), so this sample was omitted from subsequent analyses.
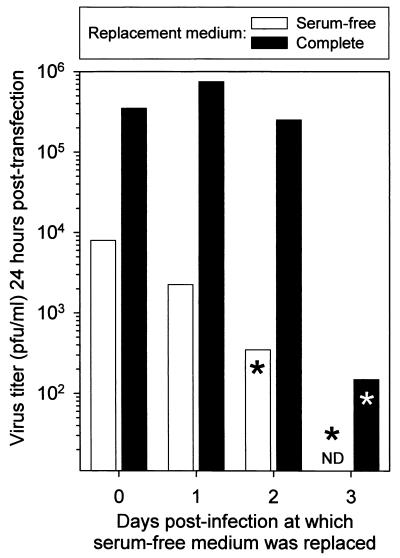
CVB3 RNA extracted from nonproductively infected cells is infectious when transfected into cycling cells.
From the above, we conclude that viral RNA may be present even in the absence of detectable protein expression or virus production. Is this RNA in some way defective, or is it viable but not expressed in quiescent cells? We evaluated the infectivity of the RNA samples described in the preceding paragraph by transfecting the purified RNA samples into a subconfluent (actively dividing) population of HeLa RW cells. The transfected cells were harvested 24 h later, and virus titers were determined (Fig. 7). As would be expected, all cell populations known to produce infectious virus also scored positive for infectious RNA in this transfection assay. Importantly, RNA extracted from cells which were not themselves producing infectious virus (indicated by an asterisk in Fig. 7) was infectious when transfected into cycling cells. These data strongly suggest that quiescent cells can harbor CVB3 RNA which is not expressed in those cells but is viable when introduced into cells capable of supporting its expression; thus, in quiescent cells, CVB3 appears to be able to persist in a form similar or identical to viral latency.
FIG. 7.
RNA extracted from nonproductive quiescent cells is infectious when transfected into dividing cells. An aliquot of each of the pooled RNA samples described for Fig. 6 was transfected into cycling tissue culture cells. Then, 24 h later, supernatants were collected, and virus titers were determined. Asterisks indicate RNA samples purified from cell populations which did not produce detectable quantities of infectious virus (as determined by plaque assay, Fig. 5). ND, not determined (see text).
Viral protein expression in contact-inhibited cells is upregulated by wounding.
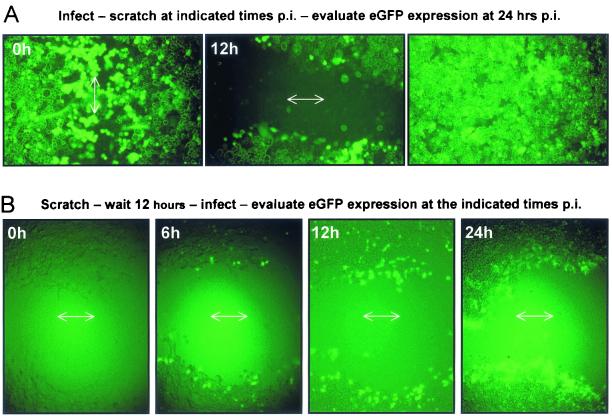
Most cells in adult animal tissues are nondividing (G0), but, of course, they are held at that stage not by serum starvation but instead by contact-dependent inhibition of cell growth. Therefore, to more closely approximate the in vivo situation, we evaluated CVB3 growth and reactivation in a contact-inhibited cell monolayer. Cells were grown to confluence in complete medium on a chamber slide. The confluent monolayers were infected with eGFP-CVB3, and then one area was wounded with a pipette tip, either immediately postinfection or 12 h later. eGFP expression was evaluated at 24 h postinfection.
As shown in Fig. 8A, in contact-inhibited cells which had not been wounded, very little eGFP expression was seen 24 h later (right-hand photograph). In contrast, in cells which had been scraped immediately after infection, eGFP-expressing cells were abundant at 24 h postinfection, at the wound margin, and in cells which had migrated into the wound (left-hand photograph). These findings are consistent with the idea that cellular activation is required for CVB3 gene expression. Furthermore, viral activation does not require that the wound be made immediately postinfection; cells incubated for 12 h after infection and then wounded expressed eGFP at the wound edges.
FIG. 8.
Viral protein expression in contact-inhibited cells is upregulated by wounding. HeLa RW cells were plated at high density on a chamber slide and grown for 2 days. (A) The confluent monolayer was infected with eGFP-CVB3 for 1 h, washed with saline solution, and either wounded immediately after infection by drawing a pipette tip across the cells (0 h, left photograph) or wounded 12 h after infection (center photograph). eGFP expression in the two scratched areas and in a region located distant from a scratched area (right photograph) was recorded at 24 h postinfection. (B) The confluent monolayer was scratched and 12 h later was infected with eGFP-CVB3. After infection, an agarose-complete medium overlay was added to the chamber, and at the indicated time points thereafter, eGFP expression was assessed. Arrows indicate the direction of wounding.
In the above cases, the cells all were infected before being scratched. As an alternative approach, the protocol was reversed; a confluent cell monolayer was scratched, and the cells were incubated in complete medium for 12 h before being exposed to eGFP-CVB3. Immediately after infection, the cells were covered with an agarose overlay, and the kinetics of eGFP expression were evaluated by fluorescence microscopy. In this case (Fig. 8B), eGFP expression was readily detected as early as 6 h postinfection, and, as before, infection was restricted to cells at the wound margin. These results demonstrate that contact-inhibited cells infected with eGFP-CVB support viral replication rather poorly and that cellular insult can permit viral gene expression.
DISCUSSION
The molecular interactions between viruses and their host cells play an important part in determining the outcome of acute and persistent virus infections, but they remain very poorly understood. For example, a cell cycle effect on picornaviral replication was suggested by studies carried out some two to three decades ago (10, 25, 28, 56), but it has not, until now, been clearly delineated, and—judging from its omission from recent reviews on virus-cell cycle interactions (40, 57)—appears not to be widely appreciated. Furthermore, coxsackieviruses are considered highly lytic, but persistent RNA can readily be detected in vivo long after infectious virus has been eradicated.
How can RNA persistence be explained, and what are the implications for the pathogenesis of acute and chronic myocarditis? Our initial approach to these issues has been to analyze CVB-host cell interactions in tissue culture. For CVB and other picornaviruses, persistent tissue culture infections can be readily established by the in vitro passage of actively dividing cell lines which are not susceptible to viral cytopathic effect; under these circumstances, viral persistence results from the selection of nonlytic viral variants (27, 42, 50). However, serial passage of infected cells does not parallel the environment in vivo, where most cells are thought to be in a nondividing (G0) state. We considered that a tissue culture model that relied on the infection of highly susceptible cells at various stages of the cell cycle might be more revealing for both acute and persistent CVB infections.
Here we describe such a model and a recombinant virus, eGFP-CVB3, which we used to evaluate viral polyprotein expression. We show (Fig. 2) that quiescent cells do not support viral gene expression or virus production and that these aspects of virus replication are exquisitely responsive to the cell cycle; CVB protein synthesis is dramatically higher in cells arrested at the G1 or G1/S stage (Fig. 3), as is virus production (Fig. 4A). Cells infected at the G0 or G2/M stage express less protein and make less infectious virus, but when permitted to cycle, both protein synthesis and virus production are increased (Fig. 4B).
How might the relationship between CVB3 and the cell cycle be explained? Translation of most eukaryotic mRNAs is facilitated by a 5′ cap, a structure absent from picornavirus mRNA, which instead contains (in the 5′ untranslated leader sequence) an internal ribosome entry site (IRES). Ribosomes bind directly to the IRES, permitting cap-independent translational initiation and polyprotein synthesis. Cap-dependent translation of cellular proteins is most robust during the G1 phase but is impaired at mitosis; interestingly, at least some of the cellular proteins synthesized during mitosis are encoded by mRNAs which contain an IRES (44, 47, 48), and in one case the encoded protein plays a pivotal role in regulating the cell cycle (12). Therefore, the IRES appears to be a cellular element which has evolved in concert with the eukaryotic cell cycle, and one might predict that the viral equivalent would be able to respond to changes in cell cycle status.
Consistent with this idea, recent studies indicate that cellular proliferation-associated proteins may regulate the IRES of some picornaviruses (44) and hepatitis C virus (16). However, cellular IRESs are used in the G2/M phase—at which point, as we have shown here, eGFP expression is very low. Why has the coxsackievirus IRES evolved to operate in the G1 phase, a time at which cap-dependent translation is dominant? During picornaviral infection, host protein synthesis is rapidly shut off due to virus-mediated modification of cellular proteins required for cap-dependent translation (11, 13, 21, 48). Thus, we suggest that, at the G1 stage, coxsackievirus attacks host translation during its most cap-dependent phase and that the viral IRES has evolved to allow CVB to circumvent this translational blockade.
An IRES-mediated interaction with the cell cycle could have profound in vivo effects. For example, we have shown that CVB-infected B cells appear to undergo a burst of viral replication as the cells enter the splenic germinal center, an area of active B-cell proliferation (32). Furthermore, others have shown that CVB3 replication in vitro and in vivo in T cells is dependent on the kinase p56lck (26), an enzyme known to be required for T-cell activation (34). In addition, an effect of cell status might help explain the extremely focal distribution of virus revealed by in situ hybridization studies of acute CVB3 myocarditis, in which the infection appears to be localized to individual cells, leaving adjacent cells unaffected (see, for example, references 19 and 32); DNA synthesis can be detected in occasional myocardiocytes in normal adult mouse hearts (55), and we suggest that only these few cells can interact appropriately with the viral IRES, with consequent viral replication.
Coxsackievirus infections are common and severe in human neonates (20, 33, 60), perhaps because of the high level of cellular proliferation occurring during growth and development. Consistent with the IRES being a determining factor in regulating CVB replication in the heart, CVB cardiovirulence has been mapped to the 5′ untranslated region (9, 61). Interactions between the host cell and the viral IRES may also explain some of the results of an elegant transgenic mouse study in which a defective CVB genome was expressed under the control of a myocardial promoter (64). Both positive- and negative-sense CVB RNAs could be found in the heart, indicating that viral protein synthesis must have occurred to direct the synthesis of the negative strand from the positive-sense primary transcript. However, in situ hybridization analyses revealed a focal distribution of CVB-specific signal.
Why was CVB RNA detectable in only a limited number of myocardiocytes when, presumably, the primary transcript was present throughout the heart? The primary transcript in these mice contained the viral IRES, and it is tempting to speculate that viral protein expression was regulated by the IRES and took place only in cells which were at a particular phase of the cell cycle; as a result, the defective genomic RNA was amplified to detectable levels only in those few cells. Finally, the focal nature of acute CVB myocarditis may be, in part, attributable to the infection of other cell types in the heart tissue. Ultrastructural and morphological analyses have demonstrated beyond doubt that myocardiocytes can be infected by CVB, but the heart also contains fibroblasts, mesenchymal stem cells and endothelial cells, which may undergo cell division more often than myocardiocytes and could act as targets for acute but focal virus infection.
Our observations also have implications for chronic or recurrent disease. Many studies in mice and humans have demonstrated CVB RNA in host tissues months or years after infection, during which time viral gene expression is extremely restricted (23). A recent publication compared CVB persistence in serially passaged tissue culture cells with RNA persistence in vivo, and the authors concluded that the two proceeded by very different mechanisms; the tissue culture virus contained many mutations, while the RNA which persisted in vivo had no detectable sequence changes (although infectious virus was not isolated [58]). In several analyses of persistent CVB RNA, the ratio of genomic to antigenomic materials has been used to infer the in vivo replication status of CVB. However, the results are open to interpretation: some studies imply that the presence of genomic RNA alone indicates viral persistence and that detection of antigenomic material is indicative of replication (43), while other workers view an excess of genomic material as indicative of replication and the presence of equal amounts of genomic and antigenomic material as evidence of viral persistence (22). Furthermore, although the detection and quantitation of genomic and antigenomic RNA is of great interest, it does not necessarily reflect the existence of persistent (or latent) virus; such a conclusion should be reached only when infectious virus has been isolated.
Nevertheless, the presence of persistent (or latent) CVB RNA in vivo has several pathological implications. First, CVB gene products, even in the absence of virus production, are directly cytopathic in tissue culture (63), and their expression in the hearts of transgenic mice leads to interstitial fibrosis (64). Second, viral gene products could induce immunopathological responses, leading to focal inflammation and fibrosis. Third—although so far not demonstrated in humans or in animal models of CVB infection—persistent CVB RNA might lead to the sporadic production of infectious virus, with consequent exacerbation of disease. All three mechanisms would support the concept of viral and immune-mediated pathogenesis rather than the induction of autoimmunity, which has been proposed to explain ongoing myocarditis in the absence of detectable infectious virus. However, little is known about the mechanisms which allow viral materials to remain in host cells after the apparent clearance of infectious virus.
We show here that quiescent cells infected with CVB3 can harbor viral materials for several days without producing detectable levels of viral protein or infectious virus (Fig. 5). Importantly, when these cells resume division, viral protein synthesis is detected, and infectious virus is released into the supernatant. Furthermore, nondividing tissue culture cells can retain viral RNA in the absence of virus production (Fig. 6), and RNA extracted from those cells is infectious (Fig. 7). These findings suggest to us that, in nondividing cells, CVB RNA may exist in a latent form—perhaps being held in this state by an interaction with a cellular protein(s)—and that Trizol extraction and transfection into dividing cells restores the infectivity of this viral RNA.
We designed in vitro wounding experiments as a first step toward testing the hypotheses that tissue damage could (i) lead to viral gene expression in cells which were already infected (Fig. 8A) and (ii) increase the number of cells which were susceptible to infection if they were subsequently exposed to the virus (Fig. 8B). In both cases, wounding led to activation of eGFP expression, visible only in cells bordering the wound. Cellular activation is known to reactivate latent herpesviruses and human immunodeficiency virus (53, 65), and we speculate that infectious CVB may be reactivated in vivo if cells containing persistent viral RNA can be appropriately stimulated, perhaps in response to local trauma. This would release the latent viral RNA from its cellular restraints, and the burgeoning infection may be amplified by the presence, in the immediate proximity, of dividing cells which would be susceptible to infection.
Recent studies have shown that human myocardiocytes divide following myocardial infarction (4), and—by analogy with the eGFP expression seen after wounding of infected cells (Fig. 8A)—localized myocardiocyte division may be sufficient to upregulate viral polyprotein expression and/or production of infectious virus in cells which carry persistent viral RNA. Moreover, following myocardial damage in mice, bone marrow stem cells can enter the myocardium to repair the damaged tissue (41); such actively dividing cells—like the cells which have migrated into the wound prior to virus exposure (Fig. 8B)—may provide an additional platform for virus replication. Together, these data suggest an explanation for chronic coxsackievirus myocarditis. Our recombinant eGFP-CVB3 will help us to determine whether other cells in the heart, in addition to myocardiocytes, are susceptible to infection and whether such cells can harbor viral RNA long enough to allow reactivation after the appropriate cellular signal is received.
Although we show here a profound effect of the cell cycle, it appears unlikely that cell division is an absolute requirement for CVB to replicate in a host cell in vivo. In addition to myocarditis, CVB3 causes severe pancreatitis, with profound destruction of acinar cells (31, 49). These cells are not known to divide frequently; however, they are metabolically highly active, producing large amounts of enzymes. Thus, CVB3 replication may be determined by interactions with a regulatory protein(s) which varies not only with the cell cycle, but also with the metabolic activity of certain cells. Alternatively, it has been demonstrated that some picornaviruses may carry with them proteins having homology to cell proliferation genes (18), and coxsackievirus infections may result in the cleavage of certain cellular proteins (3, 17), suggesting that some picornaviruses may be able to directly influence the cell cycle status, a strategy employed by other viruses (36, 37, 39, 45, 46).
Regardless of the underlying mechanisms, our observations show that coxsackieviruses respond to the cell cycle; that they may establish latent infections; and that reactivation may depend on appropriate stimulation of cells carrying the viral genome. We are currently attempting to test these hypotheses in vivo to determine the effects of cell status on viral latency and reactivation in host tissues.
Acknowledgments
We are grateful to Annette Lord for excellent secretarial support, to Michael Buchmeier for advice on fluorescence microscopy, and to Fei Liu, Dan Hassett, and Jens Leifert for helpful discussions.
This work was supported by NIH grant AI-42314 (to J.L.W.) and by NIH postdoctoral fellowship HL-10490 (to R.F.).
Footnotes
This is manuscript number 14295-NP from the Scripps Research Institute.
REFERENCES
- 1.Adachi, K., A. Muraishi, Y. Seki, K. Yamaki, and M. Yoshizuka. 1996. Coxsackievirus B3 genomes detected by polymerase chain reaction: evidence of latent persistency in the myocardium in experimental murine myocarditis. Histol. Histopathol. 11:587-596. [PubMed] [Google Scholar]
- 2.Archard, L. C., N. E. Bowles, L. Cunningham, C. A. Freeke, E. G. Olsen, M. L. Rose, B. Meany, H. J. Why, and P. J. Richardson. 1991. Molecular probes for detection of persisting enterovirus infection of human heart and their prognostic value. Eur. Heart J. 12(Suppl. D):56-59. [DOI] [PubMed] [Google Scholar]
- 3.Badorff, C., G.-H. Lee, B. J. Lamphear, M. E. Martone, K. P. Campbell, R. E. Rhoads, and K. U. Knowlton. 1999. Enteroviral protease 2A cleaves dystrophin: Evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat. Med. 5:320-326. [DOI] [PubMed] [Google Scholar]
- 4.Beltrami, A. P., K. Urbanek, J. Kajstura, S. M. Yan, N. Finato, R. Bussani, B. Nadal-Ginard, F. Silvestri, A. Leri, C. A. Beltrami, and P. Anversa. 2001. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 344:1750-1757. [DOI] [PubMed] [Google Scholar]
- 5.Bendig, J. W. A., P. S. O'Brien, P. Muir, H. J. Porter, and E. O. Caul. 2001. Enterovirus sequences resembling coxsackievirus A2 detected in stool and spleen from a girl with fatal myocarditis. J. Med. Virol. 64:482-486. [DOI] [PubMed] [Google Scholar]
- 6.Bird, J. J., D. R. Brown, A. C. Mullen, N. H. Moskowitz, M. A. Mahowald, J. R. Sider, T. F. Gajewski, C. R. Wang, and S. L. Reiner. 1998. Helper T cell differentiation is controlled by the cell cycle. Immunity 9:229-237. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, N. M., K. S. Kim, S. Tracy, J. Jackson, K. Hofling, J. S. Leser, J. Malone, and P. Kolbeck. 2000. Coxsackievirus expression of the murine secretory protein interleukin-4 induces increased synthesis of immunoglobulin G1 in mice. J. Virol. 74:7952-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colbere-Garapin, F., G. Duncan, N. Pavio, I. Pelletier, and I. Petit. 1998. An approach to understanding the mechanisms of poliovirus persistence in infected cells of neural or non-neural origin. Clin. Diagn. Virol. 9:107-113. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, J. J., N. M. Chapman, S. Tracy, and J. R. Romero. 2000. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: localization to the 5′ nontranslated region. J. Virol. 74:4787-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eremenko, T., A. Benedetto, and P. Volpe. 1972. Virus infection as a function of the host cell life cycle: replication of poliovirus RNA. J. Gen Virol. 16:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 12.Giraud, S., A. Greco, M. Brink, J. J. Diaz, and P. Delafontaine. 2001. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 276:5668-5675. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J., D. Etchison, J. W. Hershey, and E. Ehrenfeld. 1982. Association of cap-binding protein with eucaryotic initiation factor 3 in initiation factor preparations from uninfected and poliovirus-infected HeLa cells. J. Virol. 42:200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henke, A., R. Zell, G. Ehrlich, and A. Stelzner. 2001. Expression of immunoregulatory cytokines by recombinant coxsackievirus B3 variants confers protection against virus-caused myocarditis. J. Virol. 75:8187-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofling, K., S. Tracy, N. Chapman, K. S. Kim, and L. J. Smith. 2000. Expression of an antigenic adenovirus epitope in a group B coxsackievirus. J. Virol. 74:4570-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 17.Huber, M., K. A. Watson, H. C. Selinka, C. M. Carthy, K. Klingel, B. M. McManus, and R. Kandolf. 1999. Cleavage of RasGAP and phosphorylation of mitogen-activated protein kinase in the course of coxsackievirus B3 replication. J. Virol. 73:3587-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes, P. J., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen Virol 81:201-207. [DOI] [PubMed] [Google Scholar]
- 19.Kandolf, R., M. Sauter, C. Aepinus, J. J. Schnorr, H. C. Selinka, and K. Klingel. 1999. Mechanisms and consequences of enterovirus persistence in cardiac myocytes and cells of the immune system. Virus Res. 62:149-158. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan, M. H., S. W. Klein, J. McPhee, and R. G. Harper. 1983. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev. Infect. Dis. 5:1019-1032. [DOI] [PubMed] [Google Scholar]
- 21.Kleijn, M., C. L. Vrins, H. O. Voorma, and A. A. Thomas. 1996. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology 217:486-494. [DOI] [PubMed] [Google Scholar]
- 22.Klingel, K., C. Hohenadl, A. Canu, M. Albrecht, M. Seemann, G. Mall, and R. Kandolf. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. USA 89:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowlton, K. U., and C. Badorff. 1999. The immune system in viral myocarditis: maintaining the balance. Circ. Res. 85:559-561. [DOI] [PubMed] [Google Scholar]
- 24.Knowlton, K. U., E. S. Jeon, N. Berkley, R. Wessely, and S. A. Huber. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70:7811-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lake, R. S., D. C. Winkler, and E. H. Ludwig. 1970. Delay of mengovirus-induced cytopathology in mitotic L-cells. J. Virol. 5:262-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, P., K. Aitken, Y. Y. Kong, M. A. Opavsky, T. Martino, F. Dawood, W. H. Wen, I. Kozieradzki, K. Bachmaier, D. Straus, T. W. Mak, and J. M. Penninger. 2000. The tyrosine kinase p56lck is essential in coxsackievirus B3-mediated heart disease. Nat. Med. 6:429-434. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd, R. E., and M. Bovee. 1993. Persistent infection of human erythroblastoid cells by poliovirus. Virology 194:200-209. [DOI] [PubMed] [Google Scholar]
- 28.Mallucci, L., V. Wells, and D. Beare. 1985. Cell cycle position and expression of encephalomyocarditis virus in mouse embryo fibroblasts. J. Gen. Virol. 66:1501-1506. [DOI] [PubMed] [Google Scholar]
- 29.Martino, T. A., P. Liu, M. Petric, and M. J. Sole. 1995. Enteroviral myocarditis and dilated cardiomyopathy: a review of clinical and experimental studies, p. 291-351. In H. A. Rotbart (ed.), Human enterovirus infections. ASM Press, Washington, D.C.
- 30.Melchers, W., J. Zoll, F. van Kuppeveld, C. Swanink, and J. Galama. 1994. There is no evidence for persistent enterovirus infections in chronic medical conditions in humans. Rev. Med. Virol. 4:235-243. [Google Scholar]
- 31.Mena, I., C. Fischer, J. R. Gebhard, C. M. Perry, S. Harkins, and J. L. Whitton. 2000. Coxsackievirus infection of the pancreas: evaluation of receptor expression, pathogenesis, and immunopathology. Virology 271:276-288. [DOI] [PubMed] [Google Scholar]
- 32.Mena, I., C. M. Perry, S. Harkins, F. Rodriguez, J. R. Gebhard, and J. L. Whitton. 1999. The role of B lymphocytes in coxsackievirus B3 infection. Am. J. Pathol. 155:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modlin, J. F. 1988. Perinatal echovirus and group B coxsackievirus infections. Clin. Perinatol. 15:233-246. [PubMed] [Google Scholar]
- 34.Molina, T. J., K. Kishihara, D. P. Siderovski, W. van Ewijk, A. Narendran, E. Timms, A. Wakeham, C. J. Paige, K. U. Hartmann, A. Veillette, D. Davidson, and T. W. Mak. 1992. Profound block in thymocyte development in mice lacking p56lck. Nature 357:161-164. [DOI] [PubMed] [Google Scholar]
- 35.Muir, P., and L. C. Archard. 1994. There is evidence for persistent enterovirus infections in chronic medical conditions in humans. Rev. Med. Virol. 4:245-250. [Google Scholar]
- 36.Muszynski, K. W., D. Thompson, C. Hanson, R. Lyons, A. Spadaccini, and S. K. Ruscetti. 2000. Growth factor-independent proliferation of erythroid cells infected with Friend spleen focus-forming virus is protein kinase C dependent but does not require Ras-GTP. J. Virol. 74:8444-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naniche, D., S. I. Reed, and M. B. Oldstone. 1999. Cell cycle arrest during measles virus infection: a G0-like block leads to suppression of retinoblastoma protein expression. J. Virol. 73:1894-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell, J. B. 1987. The role of myocarditis in end-stage dilated cardiomyopathy. Tex. Heart Inst. J. 14:268-275. [PMC free article] [PubMed] [Google Scholar]
- 39.Oleksiewicz, M. B., and S. Alexandersen. 1997. S-phase-dependent cell cycle disturbances caused by Aleutian mink disease parvovirus. J. Virol. 71:1386-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Op De Beeck, A., and P. Caillet-Fauquet. 1997. Viruses and the cell cycle. Prog. Cell Cycle Res. 3:1-19. [DOI] [PubMed] [Google Scholar]
- 41.Orlic, D., J. Kajstura, S. Chimenti, D. M. Bodine, A. Leri, and P. Anversa. 2001. Transplanted adult bone marrow cells repair myocardial infarcts in mice. Ann. N. Y. Acad. Sci. 938:221-229. [DOI] [PubMed] [Google Scholar]
- 42.Pardoe, I. U., K. K. Grewal, M. P. Baldeh, J. Hamid, and A. T. Burness. 1990. Persistent infection of K562 cells by encephalomyocarditis virus. J. Virol. 64:6040-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pauschinger, M., A. Doerner, U. Kuehl, P. L. Schwimmbeck, W. Poller, R. Kandolf, and H. P. Schultheiss. 1999. Enteroviral RNA replication in the myocardium of patients with left ventricular dysfunction and clinically suspected myocarditis. Circulation 99:889-895. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 45.Poggioli, G. J., C. Keefer, J. L. Connolly, T. S. Dermody, and K. L. Tyler. 2000. Reovirus-induced G2/M cell cycle arrest requires sigma1s and occurs in the absence of apoptosis. J. Virol. 74:9562-9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon, B., K. Grovit-Ferbas, S. A. Stewart, and I. S. Chen. 1998. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science 281:266-269. [DOI] [PubMed] [Google Scholar]
- 47.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 48.Pyronnet, S., and N. Sonenberg. 2001. Cell cycle-dependent translational control. Curr. Opin. Genet. Dev. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 49.Ramsingh, A. I. 1997. Coxsackieviruses and pancreatitis. Front. Biosci. 2:e53-e62. [DOI] [PubMed] [Google Scholar]
- 50.Reagan, K. J., B. Goldberg, and R. L. Crowell. 1984. Altered receptor specificity of coxsackievirus B3 after growth in rhabdomyosarcoma cells. J. Virol. 49:635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riecansky, I., Z. Schreinerova, A. Egnerova, A. Petrovicova, and O. Bzduchova. 1989. Incidence of Coxsackie virus infection in patients with dilated cardiomyopathy. Cor Vasa 31:225-230. [PubMed] [Google Scholar]
- 52.Slifka, M. K., R. R. Pagarigan, I. Mena, R. Feuer, and J. L. Whitton. 2001. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells in controlling picornaviral infection. J. Virol. 75:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 54.Sole, M. J., and P. Liu. 1993. Viral myocarditis: a paradigm for understanding the pathogenesis and treatment of dilated cardiomyopathy. J. Am. Coll. Cardiol. 22:99A-105A. [DOI] [PubMed] [Google Scholar]
- 55.Soonpaa, M. H., and L. J. Field. 1997. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. 272:H220-H226. [DOI] [PubMed] [Google Scholar]
- 56.Suarez, M., G. Contreras, and B. Fridlender. 1975. Multiplication of Coxsackie B1 virus in synchronized HeLa cells. J. Virol. 16:1337-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanton, C., and N. Jones. 2001. Strategies in subversion: de-regulation of the mammalian cell cycle by viral gene products. Int. J. Exp. Pathol. 82:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam, P. E., and R. P. Messner. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tam, P. E., A. M. Schmidt, S. R. Ytterberg, and R. P. Messner. 1994. Duration of virus persistence and its relationship to inflammation in the chronic phase of coxsackievirus B1-induced murine polymyositis. J. Lab. Clin. Med. 123:346-356. [PubMed] [Google Scholar]
- 60.Tracy, S., K. Hofling, S. Pirruccello, P. H. Lane, S. M. Reyna, and C. J. Gauntt. 2000. Group B coxsackievirus myocarditis and pancreatitis: connection between viral virulence phenotypes in mice. J. Med. Virol 62:70-81. [DOI] [PubMed] [Google Scholar]
- 61.Tracy, S., Z. Tu, N. Chapman, and G. Hufnagel. 1995. Genetics of coxsackievirus B3 cardiovirulence. Eur. Heart J. 16:15-17. [DOI] [PubMed] [Google Scholar]
- 62.Ward, C. 1978. Severe arrhythmias in Coxsackievirus B3 myopericarditis. Arch. Dis. Child 53:174-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wessely, R., A. Henke, R. Zell, R. Kandolf, and K. U. Knowlton. 1998. Low-level expression of a mutant coxsackieviral cDNA induces a myocytopathic effect in culture: an approach to the study of enteroviral persistence in cardiac myocytes. Circulation 98:450-457. [DOI] [PubMed] [Google Scholar]
- 64.Wessely, R., K. Klingel, L. F. Santana, N. Dalton, M. Hongo, L. W. Jonathan, R. Kandolf, and K. U. Knowlton. 1998. Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J. Clin. Investig. 102:1444-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]