Abstract
Progenitor cells undergo T cell receptor (TCR) gene rearrangements during their intrathymic differentiation to become T cells. Rearrangements of the variable (V), diversity (D), and joining (J) segments of the TCR genes result in deletion of the intervening chromosomal DNA and the formation of circular episomes as a byproduct. Detection of these extrachromosomal excision circles in T cells located in the peripheral lymphoid tissues has been viewed as evidence for the existence of extrathymic T cell generation. Because all of the T cells in chickens apparently are generated in the thymus, we have employed this avian model to determine the fate of the V(D)J deletion circles. In normal animals we identified TCR Vγ-Jγ and Vβ-Dβ deletion circles in the blood, spleen, and intestines, as well as in the thymus. Thymectomy resulted in the gradual loss of these DNA deletion circles in all of the peripheral lymphoid tissues. A quantitative PCR analysis of Vγ1-Jγ1 and Vβ1-Dβ deletion circles in splenic γδ and Vβ1+ αβ T cells indicated that their numbers progressively decline after thymectomy with a half-life of approximately 2 weeks. Although TCR deletion circles therefore cannot be regarded as reliable indicators of in situ V(D)J rearrangement, measuring their levels in peripheral T cell samples can provide a valuable index of newly generated T cells entering the T cell pool.
During their intrathymic T cell differentiation in birds and mammals, thymocyte progenitors undergo a complex series of developmental events that can be monitored by the sequential gene rearrangement and acquisition of cell surface differentiation markers (1, 2). The genes that encode the variable regions of the T cell receptor (TCR)-α, -β, -γ, and -δ chains are assembled through recombination of the variable (V), diversity (D), and joining (J) gene segments (3). V(D)J recombination is initiated by the recognition of recombination signal sequences (RSS) that immediately flank the coding segments and consist of conserved heptamer and nonamer sequences separated by a nonconserved spacer of 12 or 22/23 bp (4, 5). The lymphocyte-specific recombinase-activating gene products, RAG-1 and RAG-2, work together to recognize the RSS and introduce site-specific cleavages at the signal-coding borders of a given pair of coding segments (6, 7). The cleavage occurs on both strands of DNA, generating two types of ends: blunt signal ends and covalently sealed coding ends. The ubiquitously expressed DNA repair machinery of the cell is then recruited to complete the reaction (8–10). The joining of coding ends often is accompanied by nucleotide addition or trimming, leading to imprecise coding joints and further TCR diversity (3, 11). The joining of the two RSS ends is precise and results in the formation of a circular episome (5, 12). The excised DNA circles derived by TCR rearrangement initially were thought to be degraded rapidly in the cells in which they are generated, and their detection in T cells therefore was considered an indication of in situ TCR gene rearrangement (13). A more recent analysis of the rearrangement status of the TCR-α/δ locus, which indicated the presence of excised chromosomal DNA in murine thymocytes and mature T cells, suggested that TCR DNA deletion circles could be relatively stable (14).
Evidence favoring the idea that T cells can be generated in extrathymic sites has been obtained in mice (15–17) and humans (18, 19), whereas the thymus appears to be the sole source of T cells in birds (20–23). In the chicken, we observed an enrichment of the TCR deletion circles in recent T cell emigrants marked by their transient expression of the chT1 thymocyte antigen (23). The levels of chT1+ T cells in the periphery declined in parallel with age-related thymic involution, and these cells disappeared completely after early thymectomy. The numbers of chT1+ T cells in the circulation were found to correlate directly with the functional thymic mass. The chT1 antigen thus provides a convenient marker for recent thymic emigrants in birds.
Because a cell surface marker for recent thymic emigrants in mammals has not been identified, the present experiments exploited the chicken model system to determine whether or not the TCR deletion circles themselves could constitute a reliable marker of newly formed T cells. In these studies we employed a quantitative PCR-based assay to identify and calibrate the TCR rearrangement deletion circles in peripheral T cells. Thymectomy resulted in a steady decline in the DNA deletion circles over the succeeding weeks. The results of this analysis indicate that the entry of newly formed T cells into the T cell pool can be monitored by measuring the deletion circle levels in peripheral T cell samples.
MATERIALS AND METHODS
Animals and Cell Lines.
SC strain chicken eggs (Hyline International, Dallas City, IA) were incubated at 41°C in a humidified incubator with intermittent rotation. Hatched chicks were maintained in our vivarium under conventional conditions. The UG-9 αβ T cell line (L. Schierman, University of Georgia), the P5 γδ T cell line (W. McCormack, University of Florida), and the BM2 macrophage cell line (R. Dietert, Cornell University) were maintained in a humidified atmosphere at 41°C and 7% CO2 in RPMI 1640 medium adding 10% fetal calf serum/1 mM glutamine/100 units penicillin/100 μg streptomycin per ml.
Thymectomy.
Surgical thymectomy or sham thymectomy was performed on 1- and 4-week-old chickens anesthetized by i.m. or i.v. injection of Nembutal Sodium Solution (Abbott). The thymic lobes were removed through a dorsal incision in the neck as described previously (24).
Cell Isolation and Cell Sorting.
Peripheral blood leukocytes were isolated by low-speed centrifugation (60 × g, 20 min) of heparinized blood samples. Single-cell suspensions were prepared by passage of thymus and spleen cells through fine-mesh screen followed by centrifugation over a Ficoll/Hypaque density gradient (25). Intestinal epithelial lymphocytes were derived from small intestine segments by incubation in 0.1 mM EDTA/0.1 mM DTT with constant stirring at 37°C for 30 min, followed by passage of the cells through a glass-wool column and density gradient centrifugation. γδ and Vβ1+ αβ T cells were purified by fluorescence-activated cell sorting (FACStar; Becton Dickinson) after staining splenocytes with phycoerythrin-conjugated TCR1 mAb (anti-γδTCR) and fluorescein isothiocyanate-conjugated TCR2 mAb (anti-Vβ1TCR) (Southern Biotechnology Associates). Purity of the sorted cells in each experiment was >98%.
RNA Isolation and cDNA Synthesis.
Total RNA was isolated from various tissues or cells by using the TRI-reagent according to the method provided by the supplier (Molecular Research Center, Cincinnati). The DNase-treated RNA samples were subjected to first-strand cDNA synthesis using oligo(dT)(15) primer and avian myeloblastosis virus reverse transcriptase (GIBCO/BRL).
Genomic DNA Preparation.
The DNA preparation method (22) involved washing the cells in lysis buffer (10 mM Tris, pH 7.4/10 mM NaCl/5 mM MgCl2), lysis with 0.5% Nonidet P-40, pelleting of the nuclei by centrifugation at 250 × g, and treatment with 250 μg/ml proteinase K at 50°C for 3–4 hr. DNA was precipitated in 2.5 M ammonium acetate and an equal volume of isopropanol. After centrifugation, the pellet was washed twice with 70% of ethanol and resuspended in water.
PCR and Southern Blot Analysis.
PCR conditions to amplify RAG and actin transcripts included 3-min incubation at 94°C followed by 35 reaction cycles (1 min at 94°C, 1 min at 56°C, 45 sec at 72°C) and a final 7-min extension at 72°C. Amplification of the deletion circles was performed under the same conditions except that the annealing temperature was 65°C. PCR products separated on 1% agarose gels were denatured, neutralized, and transferred onto nitrocellulose membranes (Biotechnology Systems, Boston). The membranes were hybridized overnight with [γ-32P]ATP-labeled internal probes at 42°C and washed with 3× SSC at 62°C for 30–40 min and exposed to x-ray film. The PCR primers and hybridization probes are listed in Table 1.
Table 1.
Sequences of the PCR primers and internal oligonucleotides probes
| PCR product | Primer sequence | |
|---|---|---|
| Vγ1-Jγ1 circles | Up | 5′-Bio-TCCTCAGTGTAAGCCTGTGG-3′ |
| Down | 5′-CCAAACAGCACTTACACTCT-3′ | |
| Internal | 5′-CAGAGCAGGTGATCTTCAGAGGGTCCT-3′ | |
| Vγ2-Jγ1 circles | Up | 5′-Bio-CTTCTGACTGAGCAGCTCAT-3′ |
| Down | 5′-CCAAACAGCACTTACACTCT-3′ | |
| Internal | 5′-CAGAGCAGGTGATCTTCAGAGGGTCCT-3′ | |
| Vγ3-Jγ1 circles | Up | 5′-Bio-TCACAGTAACAGCAAAGTCACC-3′ |
| Down | 5′-CCAAACAGCACTTACACTCT-3′ | |
| Internal | 5′-CAGAGCAGGTGATCTTCAGAGGGTCCT-3′ | |
| Vβ1-Dβ circles | Up | 5′-Bio-GCAGAGAACAAGCAGAAGAC-3′ |
| Down | 5′-ACAGAGCAGTGGCAGAGGAA-3′ | |
| Internal | 5′-GTGACTGGATGACAAAATGAATGGAGTGGA-3′ | |
| Actin | Up | 5′-TACCACAATGTACCCTGGC-3′ |
| Down | 5′-CTCGTCTTGTTTTATGCGC-3′ | |
| Internal | 5′-TGGATCAGCAACAGGAGTA-3′ | |
| Rag-1 | Up | 5′-GTGGCAGTTGACTCTTGACA-3′ |
| Down | 5′-GTTCCACTGAATCATTGCTT-3′ | |
| Internal | 5′-GCTGTGCCAGTACAGCTATA-3′ | |
| Rag-2 | Up | 5′-GGTACTGGATCATCTGCTGT-3′ |
| Down | 5′-TTCACTGGTGTGGTTAACTT-3′ | |
| Internal | 5′-GATCTGTCAGAGAGCATGCT-3′ | |
Construction of Competitor for Quantitative PCR.
Sense and antisense oligonucleotides corresponding to the relevant primer sequences for the Vγ1-Jγ1, Vβ1-Dβ1 rearrangement deletion circles (Table 1) were synthesized as indicated in Fig. 1B. Ten micrograms of each complementary oligonucleotide pair was annealed in 100 μl annealing buffer (10 mM Tris, pH 7.4/1 mM EDTA). The 5′ and 3′ primer templates were cloned into the pQPCR1 vector (ref. 26; Fig. 1B), which contains a 230-bp “stuffer” to separate the 5′ and 3′ primer templates. Sequence analysis of miniprep DNA confirmed that the correct templates were inserted. The copy numbers of the competitor were calculated as:
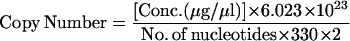 |
Figure 1.
(A) Schematic representation of V(D)J recombination and the formation of DNA deletion circles. Joining of two coding segments in the same transcriptional orientation on the chromosome results in the deletion of intervening DNA and its reconfiguration as a circular episome by joining of the two RSS, indicated by open and solid triangles. The deletion circles can be detected by PCR by using primers that anneal to sequences adjacent to the RSS flanking the coding segments, which are represented by rectangles. PCR primers used to detect TCR deletion circles in these studies are indicated by short arrows, indicating the direction of strand synthesis. (B) Construction of the competitor for quantitative PCR. Vector pQPCR1 (26), which contained a “stuffer” (230-bp nontranscribed murine genomic DNA fragment), was used for cloning. Sense and antisense oligonucleotide corresponding to the sequence of 5′ and 3′ primers for Vβ1-Dβ1 and Vγ1-Jγ1 deletion circles were synthesized, annealed, and cloned into the 5′ and 3′ ends of the stuffer, respectively.
Competitive PCR.
Lymphocyte DNA samples were digested with BamHI and EcoRI (neither enzyme cuts the fragments to be amplified), and the competitor was linearized with KpnI. DNA from 104 cells was coamplified with various copy numbers of the competitor (range of 10–104 copies), using primers (3′ primers were biotinylated) specific for Vγ1-Jγ1 or Vβ1-Dβ deletion circles. The PCR conditions were as described above.
ELISA.
The PCR products were quantified by ELISA as described (26). In brief, the PCR products were diluted 1:30 in PBS/Tween solution and added to four separate avidin-coated microtiter wells, with 30 μl added per well. The plates were incubated at 42°C for 1–3 hr. The DNA bound to the wells was denatured with denaturing solution (50 mM NaOH/2 mM EDTA). Two of the wells were hybridized with digoxigenin-labeled detection oligonucleotides specific for deletion circles, and the other two were hybridized with digoxigenin-labeled detection oligonucleotides specific for the stuffer. Alkaline phosphatase-conjugated antidigoxigenin Fab fragment Ab (1:5,000 dilution in 1% PBS/BSA) then was added and incubated at 37°C for 1–2 hr. After the antibody reaction, ρ-nitrophenyl phosphate solution (1 mg/ml in diethanolamine buffer) was added. The plates then were incubated at 37°C, and the OD405 determined at various time points. Between each step, the plates were washed two to four times with PBS/Tween solution.
RESULTS
Detection of TCR V(D)J Rearrangement Deletion Circles in the Peripheral Lymphoid Tissues.
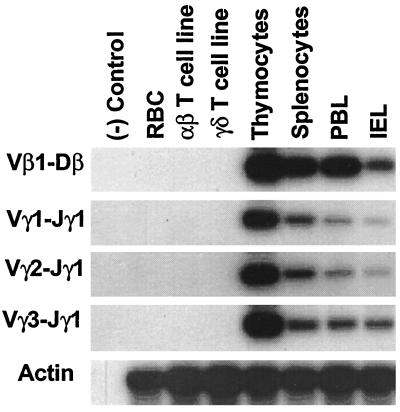
The genes encoding the chicken αβ and γδ TCR and their mode of diversification resemble their mammalian counterparts, although the chicken TCR gene loci are relatively simple (27). The chicken TCR-γ locus includes three Vγ subfamilies, three Jγ segments, and a single Cγ segment (28), and the TCR-β locus includes only two Vβ subfamilies, one Dβ, four Jβ, and one Cβ segment (29, 30). In this experiment we surveyed central and peripheral lymphoid tissues for the deletion circles resulting from TCRVγ1, Vγ2, Vγ3 rearrangements to Jγ1 and from Vβ1 recombination with Dβ. For this analysis, specific PCR primers were used to identify the Vγ1-Jγ1, Vγ2-Jγ1, Vγ3-Jγ1, and Vβ1-Dβ deletion circles (Table 1 and Fig. 1). All of the different types of V(D)J deletion circles were found in the blood, spleen, and intestine as well as in the thymus, whereas they could not be detected in red blood cells or in αβ and γδ T cell lines (Fig. 2).
Figure 2.
Analysis of the tissue distribution of the deletion circles created by TCR V(D)J rearrangement. Lymphocytes were isolated from thymus, spleen, peripheral blood lymphocyte (PBL), and intestinal epithelial lymphocyte (IEL) compartments of a 4-week-old chicken and examined for extrachromosomal deletion circles by the PCR strategy illustrated in Fig. 1. PCR products separated in 1% agarose gels and transferred onto nitrocellulose membranes were hybridized with specific 32P-labeled internal oligonucleotide probes (Table 1) before membrane exposure to x-ray films for 12 hr.
Analysis of TCR Deletion Circles in Peripheral T Cells from Thymectomized Animals.
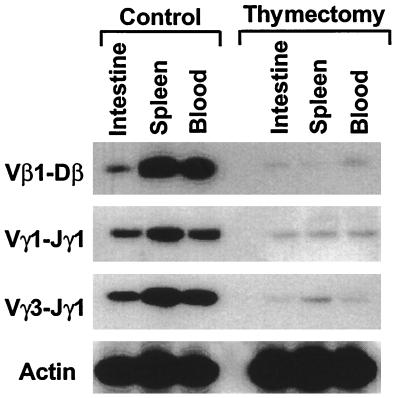
The above findings and the enrichment of TCR deletion circles observed in recent thymic emigrants (23) suggested that excision circles generated in the thymus are relatively stable and can be retained by thymus-derived T cells in the periphery. Because V(D)J recombination deletion circles lack replication origins (14), those retained in mature T cells should be depleted with time through cell division, cell death, and, possibly, by intracellular degradation, unless they were renewed subsequently by reactivation of the recombinase-mediated V(D)J rearrangement process. In an initial experiment designed to assess the fate of TCR V(D)J deletion circles, we analyzed the T cells in the different lymphoid compartments of thymectomized chickens. When 4-week-old chickens were thymectomized and examined 10 weeks later, the Vγ1-Jγ1, Vγ1-Jγ3, and Vβ1-Dβ deletion circles in circulating, splenic, and intestinal T cells were greatly reduced (Fig. 3). Employment of this semiquantitative assay for evaluation of the deletion circles in the peripheral lymphoid compartments at various intervals after thymectomy indicated their progressive loss over time (data not shown).
Figure 3.
Comparison of the levels of TCR gene deletion circles in normal and thymectomized chickens. Surgical thymectomy was performed at 4 weeks of age, and the thymectomized birds and age-matched controls were analyzed 10 weeks later for V(D)J deletion circles by PCR analysis of lymphoid cells in peripheral lymphoid tissues.
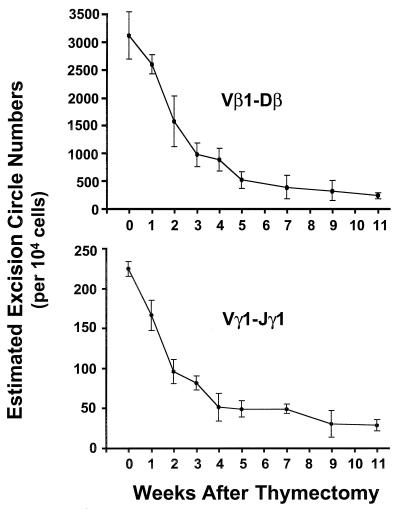
To determine more precisely the half-life of the DNA deletion circles in peripheral T cells, a quantitative PCR assay (see Materials and Methods and Fig. 1B) was used to estimate the numbers of Vγ1-Jγ1 and Vβ1-Dβ deletion circles in TCR-γδ+ and the TCRVβ1+ T cells as a function of time after thymectomy. The chicks in this experiment were thymectomized at 1 week of age, and their splenic γδ and Vβ1+ αβ cells were purified by fluorescence-activated cell sorting at serial time intervals after thymectomy. When DNA samples from the TCR-γδ+ and TCRVβ1+ cells were analyzed by competitive PCR, the number of Vγ1-Jγ1 deletion circles before thymectomy was found to be ≈220/104 γδ T cells, whereas the number of Vβ1-Dβ1 deletion circles was ≈2,800/104 Vβ1+ αβ T cells. By 2 weeks after thymectomy, the numbers of Vγ1-Jγ1 and Vβ1-Dβ1 deletion circles had decreased by ≈50% (Fig. 4). The mean level of Vβ1-Dβ1 deletion circles at 11 weeks postthymectomy was 7.3% of the prethymectomy level, and the level of Vγ1-Jγ1 deletion circles was 12.5% of the initial value.
Figure 4.
Serial measurement of TCR gene deletion circles in αβ and γδ splenic T cells after thymectomy. Chicks were thymectomized at 1 week of age, and spleen samples were collected at regular intervals for purification of TCRVβ1+ and TCR-γδ+ cells by fluorescence-activated cell sorting. DNA from the purified subpopulations was used to determine the numbers of deletion circles created by Vβ1-Dβ and Vγ1-Jγ1 rearrangements in each T cell subpopulation by using the quantitative PCR-based strategy illustrated in Fig. 1B. Three animals were examined at each time point.
Analysis of RAG-1 And RAG-2 Gene Transcripts in Peripheral Lymphoid Tissues.
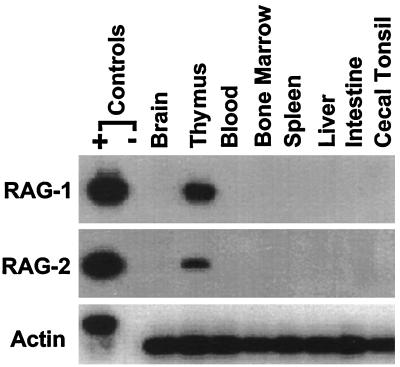
The residual DNA deletion circles found in T cells in the circulation and other peripheral lymphoid compartments 11 weeks after thymectomy (Figs. 3 and 4) could reflect the retention of excision circles in a subpopulation of nondividing, long-lived T cells (31, 32). Alternatively, this finding could be explained by the occurrence of TCR rearrangements outside the thymus. To evaluate the possibility that TCR deletion circles might be generated by V(D)J recombination occurring in peripheral lymphoid tissues, we used a reverse transcriptase–PCR assay to search for recombinase gene expression in the different lymphoid compartments. Although RAG-1 and RAG-2 transcripts were detected easily in the thymus, these could not be identified in the peripheral lymphoid tissues of normal (Fig. 5) or thymectomized animals (not shown). The sensitivity of this assay was evaluated by adding variable numbers of CD4+CD8+ thymocytes to a fixed number of RAG-negative macrophages and using the reverse transcriptase–PCR to identify RAG-1 and RAG-2 transcripts in the mixture. In this experiment the RAG transcripts could be detected in as few as 10 thymocytes among 106 RAG-negative macrophages. Finally, we sought RAG transcripts in 106 cells from the intestinal epithelial lymphocyte compartment, because this may represent an extrathymic site for T cell generation in mice (15–17). No RAG transcripts were detected in the intestinal epithelial lymphocyte sample, thereby further mitigating against the possibility of T cell generation in peripheral lymphoid tissues of the chicken.
Figure 5.
Analysis of the tissue distribution of V(D)J RAG expression. Total RNA prepared from different tissues of a 4-week-old chicken was subjected to DNase treatment before synthesizing cDNA by using an oligo(dT)(15) primer and avian myeloblastosis virus reverse transcriptase. PCR products obtained with RAG-1 and RAG-2 specific primers (Table 1) were separated in 1% agarose gels, transferred onto nitrocellulose membranes, and hybridized with 32P-labeled internal oligonucleotides. Genomic DNA was used as a positive PCR control. Membranes being examined for control actin products were exposed to x-ray films for 12 hr, whereas a 24-hr exposure time was used for the evaluation of RAG-1 and RAG-2 expression.
DISCUSSION
TCR V(D)J gene recombination in lymphoid precursors is essential for their differentiation to become T cells (2, 33, 34). During the V(D)J gene rearrangement process that occurs in the thymus, the intervening DNA between the two coding segments in the same transcriptional orientation is deleted from the chromosome and ligation of the recombination signal sequences leads to the formation of DNA circles (5, 12). Although it had been presumed that the DNA excision circles would be found only in T lineage cells that recently have undergone V(D)J recombination, the present findings indicate the presence of Vγ-Jγ and Vβ-Dβ deletion circles not only in the thymus but in thymus-derived T cells in all of the peripheral lymphoid compartments as well. After surgical removal of the thymus, however, we observed a progressive decline in the levels of V(D)J deletion circles in all of the peripheral lymphoid compartments. These findings thus indicate that the TCR deletion circles carried by T cell emigrants from the thymus into the peripheral T cell pool are relatively stable.
To determine the ultimate fate of the deletion circles in thymus-derived T cells more precisely, we determined their copy numbers in peripheral T cells as a function of time after thymectomy. The numbers of Vγ1-Jγ1 deletion circles in γδ T cells and Vβ1-Dβ deletion circles in Vβ1+ αβ T cells steadily declined after thymectomy with a half-life of approximately 2 weeks. By 11 weeks postthymectomy, the levels of residual deletion circles were ≈10% of their prethymectomy levels. Although the high mortality rate of thymectomized animals prevented longer follow-up, the absence of any detectable recombinase gene expression in the peripheral lymphoid tissues suggests that the residual TCR excision circles represented the products of intrathymic V(D)J recombination events. Cell death and dilution of the deletion circles by T cell proliferation could account for the diminishing levels in the peripheral lymphocyte pool of the thymectomized birds. In this regard, murine αβ T cells have an estimated life span of approximately 8 weeks (31) and γδ T cells have an average life span of around 4 weeks (32). Furthermore, an enhanced expansion rate has been observed for the αβ T cells in athymic mice (35).
Our estimate of ≈2,800 Vβ1-Dβ deletion circles in 104 splenic Vβ1+ T cell suggests that, depending on whether one or both alleles undergo Vβ-Dβ rearrangement, 14–28% of the Vβ1+ cells contain one or two Vβ1 deletion circles. Approximately 220 Vγ1-Jγ1 deletion circles were found in 104 γδ T cells from the spleen. This somewhat lower estimate reflects the fact that we examined the entire γδ T cell population, and the chicken TCR-γ locus contains three Vγ subfamilies and three Jγ gene segments. Members of each Vγ subfamily may recombine with each of the three Jγ gene segments without apparent bias (28), so that the Vγ1-Jγ1 rearrangements should account for approximately one-ninth of the Vγ-Jγ rearrangements. This leads to the estimate that around 10–20% of the γδ T cells contain Vγ-Jγ deletion circles.
The numbers of αβ and γδ T cells in the periphery estimated to contain TCR V(D)J excision circles correspond roughly to the frequency of recent thymic emigrants marked by their expression of the chT1 antigen in the peripheral tissues (23). On leaving the avian thymus, T cells express the chT1 thymocyte antigen for only a limited period of time. In young birds, 10–20% of the T cells in the periphery can be identified as recent T cell emigrants by their expression of this cell surface marker (23). The numbers of chT1+ T cells in the circulation thus provides a convenient index of thymic function in chickens. A comparable marker in humans would be invaluable for evaluating the capacity of the thymus to restore the peripheral T cell pool after accidental irradiation, immunosuppressive therapy, viral infections, and other disease states resulting in temporary or persistent T cell depletion. Regardless of the cause, gaps created in the T cell repertoire would require repair from a source of newly formed T cells. Although a suitable chT1 surrogate has not been identified in mammals, our data in this avian model suggest that the enumeration of TCR deletion circles in circulating T cells also may provide an accurate indicator of newly formed T cells in mammals.
Acknowledgments
We thank Drs. Peter D. Burrows and Casey Weaver for their helpful comments and manuscript review, Dr. Larry Gartland for help with cell sorting, and Mrs. Ann Brookshire for help in preparing the manuscript. This work was supported by grants from the National Institutes of Health (AI39561) and the Human Frontier Science Program (RG-366/96). M.D.C. is a Howard Hughes Medical Institute Investigator.
ABBREVIATIONS
- TCR
T cell receptor
- V
variable
- D
diversity
- J
joining
- RSS
recombination signal sequences
- RAG
recombinase-activating gene
References
- 1.Cooper M D, Chen C H, Bucy R P, Thompson C B. Adv Immunol. 1991;50:87–117. doi: 10.1016/s0065-2776(08)60823-8. [DOI] [PubMed] [Google Scholar]
- 2.Kisielow P, von Boehmer H. Adv Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 3.Davis M M, Bjorkman P J. Nature (London) 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 4.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 5.Lewis S M. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 6.Oettinger M A, Schatz D G, Gorka C, Baltimore D. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 7.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 8.Taccioli G E, Alt F W. Curr Opin Immunol. 1995;7:436–440. doi: 10.1016/0952-7915(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 9.Gellert M. Adv Immunol. 1996;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 10.Steen S B, Zhu C, Roth D B. Curr Top Microbiol Immunol. 1996;217:61–77. doi: 10.1007/978-3-642-50140-1_5. [DOI] [PubMed] [Google Scholar]
- 11.Lafaille J J, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 12.Okazaki K, Davis D D, Sakano H. Cell. 1987;49:477–485. doi: 10.1016/0092-8674(87)90450-8. [DOI] [PubMed] [Google Scholar]
- 13.Makino Y, Yamagata N, Sasho T, Adachi Y, Kanno R, Koseki H, Kanno M, Taniguchi M. J Exp Med. 1993;177:1399–1408. doi: 10.1084/jem.177.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak F, Schatz D G. Mol Cell Biol. 1996;16:609–618. doi: 10.1128/mcb.16.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poussier P, Julius M. Annu Rev Immunol. 1994;12:521–531. doi: 10.1146/annurev.iy.12.040194.002513. [DOI] [PubMed] [Google Scholar]
- 16.Lefrancois L, Puddington L. Immunol Today. 1995;16:16–21. doi: 10.1016/0167-5699(95)80065-4. [DOI] [PubMed] [Google Scholar]
- 17.Lin T, Matsuzaki G, Kenai H, Nomoto K. Immunol Cell Biol. 1995;73:469–473. doi: 10.1038/icb.1995.73. [DOI] [PubMed] [Google Scholar]
- 18.McVay L D, Carding S R. J Immunol. 1996;157:2873–2882. [PubMed] [Google Scholar]
- 19.Howie D, Spencer J, DeLord D, Pitzalis C, Wathen N C, Dogan A, Akbar A, MacDonald T T. J Immunol. 1998;161:5862–5872. [PubMed] [Google Scholar]
- 20.Coltey M, Bucy R P, Chen C H, Cihak J, Lösch U, Char D, Le Douarin N M, Cooper M D. J Exp Med. 1989;170:543–557. doi: 10.1084/jem.170.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunon D, Cooper M D, Imhof B A. J Exp Med. 1993;177:257–263. doi: 10.1084/jem.177.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickel J M, McCormack W T, Chen C H, Cooper M D, Thompson C B. Int Immunol. 1993;5:919–927. doi: 10.1093/intimm/5.8.919. [DOI] [PubMed] [Google Scholar]
- 23.Kong F-k, Chen C H, Cooper M D. Immunity. 1998;8:97–104. doi: 10.1016/s1074-7613(00)80462-8. [DOI] [PubMed] [Google Scholar]
- 24.Cooper M D, Peterson R D A, South M A, Good R A. J Exp Med. 1966;123:75–102. doi: 10.1084/jem.123.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara Y, Chen C H, Cooper M D. Eur J Immunol. 1993;23:2230–2236. doi: 10.1002/eji.1830230927. [DOI] [PubMed] [Google Scholar]
- 26.Hockett R T, Janowski K M, Bucy P R. J Immunol Methods. 1995;187:273–285. doi: 10.1016/0022-1759(95)00195-5. [DOI] [PubMed] [Google Scholar]
- 27.Chen C L H, Six A, Kubota T, Tsuji S, Kong F-k, Gobel T, Cooper M D. Curr Top Microbiol Immunol. 1995;212:37–53. doi: 10.1007/978-3-642-80057-3_5. [DOI] [PubMed] [Google Scholar]
- 28.Six A, Rast J P, McCormack W T, Dunon D, Courtois D, Li Y, Chen C H, Cooper M D. Proc Natl Acad Sci USA. 1996;93:15329–15334. doi: 10.1073/pnas.93.26.15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tjoelker L, Carlson L, Lee K, Lahti J, McCormack W, Leiden J, Chen C H, Cooper M D, Thompson C B. Proc Natl Acad Sci USA. 1990;87:7856–7860. doi: 10.1073/pnas.87.20.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahti J M, Chen C H, Tjoelker L W, Pickel J M, Schat K A, Calnek B W, Thompson C B, Cooper M D. Proc Natl Acad Sci USA. 1991;88:10956–10960. doi: 10.1073/pnas.88.23.10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Boehmer H, Hafen K. J Exp Med. 1993;177:891–896. doi: 10.1084/jem.177.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tough D, Sprent J. J Exp Med. 1998;187:357–365. doi: 10.1084/jem.187.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;69:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 34.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 35.Mackall C L, Granger L, Sheard M A, Cepeda R, Gress R E. Blood. 1993;82:2585–2594. [PubMed] [Google Scholar]