Abstract
Human cytomegalovirus (HCMV) ie1 deletion mutant CR208 is profoundly growth deficient after low-multiplicity infection of primary fibroblasts. Previously, we showed that many fewer cells infected with CR208 at low multiplicity accumulated the delayed-early (DE) protein ppUL44 than accumulated the immediate-early 2 (IE2) p86 protein, indicating a high frequency of abortive infections. We now demonstrate that accumulation of all DE proteins tested was defective after low-multiplicity infection in the absence of IE1 p72. Accumulation of the DE proteins pUL57, pUL98, and pUL69 followed a pattern very similar to that of ppUL44 during low-multiplicity CR208 infection. Accumulation of the ppUL112-113 proteins occurred in a greater proportion of cells than other DE proteins during low-multiplicity CR208 infection, but was still deficient relative to wild-type virus. We also show for the first time that steady-state levels of many DE RNAs were reduced during low-multiplicity CR208 infection and that by in situ hybridization of the abundant cytoplasmic 2.7-kb TRL4 DE (β2.7) RNA, a viral DE RNA followed a defective pattern of accumulation similar to that of ppUL44. Furthermore, transfected DE promoter-reporter constructs were found in transient assays to be considerably less responsive to CR208 infection than to infection by wild-type Towne virus. Our results indicate a general defect in DE gene expression following low-multiplicity HCMV infection in the absence of functional IE1 p72, most probably mediated by reduced transcription of DE genes and by the reduced accumulation of DE RNAs.
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus which infects between 50 and 100% of the human population, depending on the population group tested. Primary infection is generally asymptomatic or not diagnosed, with the virus persisting lifelong in the host. However, HCMV becomes a significant medical problem in individuals with immature or compromised immune systems (11). As in other herpesviruses, viral gene expression during productive infection occurs in an ordered temporal cascade, with immediate-early (IE or α), delayed-early (DE or β), and late (L or γ) kinetics (50). Expression of immediate-early genes is a prerequisite for progression into the delayed-early phase of virus growth and the subsequent replication of viral DNA, which is in turn required for late-phase entry and virion assembly and release.
Two major IE proteins (IE1 p72 and IE2 p86) are expressed, their messages generated by alternate splicing of a single RNA transcript from the major immediate-early (MIE) locus. IE2 p86 is thought to be the major transcription-activating protein of HCMV and is also responsible for negative autoregulation of the major immediate-early promoter (MIEP) (51). Transient-cotransfection assays have shown the specific activation of viral and cellular promoters by IE2 p86 (37, 50, 75). Specific DNA binding sites for IE2 p86 have been found in the upstream regions of promoters activated by IE2 p86 (6, 9, 69, 70) and at the cap site of the MIEP (32, 41, 45). IE2 p86 interacts directly with components of the basal transcription machinery (i.e., TBP, TFIIB, and hTAFII130) (13, 25, 33, 43), activating a variety of promoters in a TATA-box-dependent manner. Specific interaction with a number of cellular transcription factors such as CREB, CBP, SP-1, and c-Jun has also been recorded (40, 44, 68, 70).
In comparison, the more abundant IE1 p72 protein activates a limited number of viral and cellular genes. IE1 p72 activates the MIEP via NF-κB sites in the MIEP repeat region (65), and specific activation of the cellular DNA polymerase α, dihydrofolate reductase (DHFR), and prointerleukin-1β genes has been shown (26, 28, 48, 60), as well as the ability to activate the human immunodeficiency virus long terminal repeat (77).
Whereas IE1 p72 physically interacts with the cellular transcription factors E2F-1 and CTF-1 (26, 48), interaction with the basal transcription machinery is limited to the binding of hTAFII130 (43). During activation of the cellular DHFR promoter, IE1 p72 may mediate its function either by a direct physical interaction with E2Fs (48), by phosphorylation of E2Fs and pocket proteins (55), or by release of active E2Fs from repression via binding and displacement of the cellular pocket protein p107 (60). Although HCMV infection of resting fibroblasts results in a cell cycle block prior to cellular DNA synthesis (10, 18, 42), these functions of IE1 p72 may be involved in the induction of an S-phase-like environment conducive to viral DNA replication (12).
Although IE1 p72 is necessary during low-multiplicity infections, the exact functional role of the protein during the viral life cycle remains unclear. A number of activities have been observed for this protein, whose functional significance remains to be determined. For example, IE1 p72 interacts with cellular chromatin both during infection and when expressed exogenously within cells (39), a feature shared with the Epstein-Barr virus latent antigen EBNA-1. Independent exogenous expression of IE1 p72 can inhibit HeLa cell apoptosis induced by tumor necrosis factor alpha (82), but fails to protect against Fas-mediated apoptosis (21). Exogenous expression of IE1 p72 disrupts nuclear ND10 domains (3, 38, 78), and IE1 p72 is required for the disruption of ND10 during infection (2). Furthermore, IE1 p72 is required in conjunction with IE2 p86 and five other auxiliary factors for the efficient replication of DNA driven by core viral replication proteins at the HCMV lytic origin (56), although IE1 p72's involvement may be limited to the stimulation of expression of other components (66).
One of the principal functions assigned to IE1 p72 is its ability to augment transcriptional activation mediated by IE2 p86. Expression of IE1 p72 enhances IE2 p86-mediated transactivation at a number of viral and cellular promoters, including a wide variety of DE viral promoters (46, 74). This includes the genes encoding the core viral DNA replication proteins (35), although other factors may also be required for maximal expression at these loci (30, 73).
Deletion of IE1 p72-specific sequences from the HCMV genome resulted in a virus (CR208) that was able to replicate in primary fibroblasts (HFs) after high-multiplicity input but has a pronounced low-multiplicity growth defect (23). Therefore, although strictly nonessential during infection in vitro, IE1 p72 function is probably required during the low-multiplicity infection events that are expected to predominate in vivo. During high-multiplicity infection of HFs by CR208, the IE2 p86 protein and the DE ppUL44 protein accumulated with wild-type kinetics. Low-multiplicity infections of HFs in the absence of IE1 p72 were characterized by abortively infected cells which accumulated the IE2 p86 protein but failed to establish viral DNA replication compartments. This defect was associated with the failure to accumulate the DE protein ppUL44, one of the core viral DNA replication proteins.
Further studies of CR208 were made by Ahn et al. (2), using semipermissive U373 cells. These studies repeated the observation that ppUL44 accumulated to approximately wild-type levels during high-multiplicity infection by CR208. However, the accumulation of lower-molecular-weight forms of ppUL44 and the late pp28 (ppUL99) protein was delayed during such infections by CR208, suggesting regulatory defects even in a high-multiplicity scenario. During low-multiplicity infections of U373s by CR208, Ahn et al. also showed an increased proportion of cells that accumulated IE2 p86 but failed to accumulate ppUL44 or form replication compartments, once again supporting results obtained using permissive HF cells.
Altogether, these results suggested the possibility of a general block to DE protein accumulation during low-multiplicity infection in the absence of IE1 p72, which might reflect a deficiency in DE gene transcription in the presence of IE2 p86 alone. To establish whether such a general block to DE gene expression occurs in the absence of IE1 p72, we investigated the accumulation of other viral DE proteins during low-multiplicity infection by CR208. To test whether a failure in DE gene transcription was responsible, we investigated the accumulation of viral DE RNAs and tested the activity of viral DE gene promoters in cells infected by CR208. Our results demonstrate a broad block to DE protein accumulation during low-multiplicity infection in the absence of IE1 p72, associated with defective DE promoter activation and defective DE RNA accumulation.
MATERIALS AND METHODS
Plasmids.
Plasmid DNAs were maintained and propagated in Escherichia coli strain DH5α and prepared by alkaline lysis followed by CsCl density gradient centrifugation as previously described (22). To generate pKS+β2.7, sequences specific for HCMV 2.7-kb DE RNA were amplified by PCR from HCMV Towne strain viral DNA using primers B2.7-FOR (5′-CGAAGAATTCAAAGTCGACGATTCC-3′) and B2.7-BACK (5′-GGGGATCCTAAGAGGTTTCAAGTGC-3′). The resulting 1,626-bp EcoRI-BamHI fragment (AD169 equivalent nucleotides [nt] 185495 to 187120) (15) was ligated into the EcoRI and BamHI sites of the pBluescriptII KS+ vector (Stratagene).
The pKS+HSVtk plasmid consisted of a 456-bp NruI-PstI fragment of pRB103 (61) ligated into the EcoRV and PstI sites of the pBluescriptII KS+ vector. A 5.7-kb ApaI-EcoRI fragment of cosmid pCM1029 (19) containing the UL69 open reading frame and flanking sequences from HCMV AD169 (nt 97292 to 103025) (15) was subcloned into pBluescriptII SK+ (Stratagene) to give plasmid pON2522. A 436-bp XhoI-NcoI fragment of pON2522 containing consensus promoter sequences immediately 5′ to the UL69 start codon (AD169 nt 100871 to 100435) (15) was subcloned between the XhoI and NcoI sites of pGL3Prom (Promega) to give luciferase reporter plasmid pRG298.
Sequences contained within the UL44 open reading frame were amplified by PCR from HCMV Towne strain viral DNA using primers UL44-F (5′-TAGAATTCTACAAGACGGCTATCCAGC-3′) and UL44-B (5′-TAGGATCCTCAGGATAAACTTGATGGC-3′). The 369-bp EcoRI fragment (AD169 equivalent nt 56088 to 56455) was then inserted into the EcoRI site of pBluescriptII KS+ to yield plasmid pKS+UL44. A 592-bp fragment of HCMV AD169 (nt 57103 to 56511) (15) containing previously characterized UL44 promoter sequences (74) was PCR amplified from cosmid pCM1049 (19) using primers UL44FORB (5′-ATCGCTCGAGCATGCTCGAGTTCGGATTGCG-3′) and UL44REV (5′-ATCGAAGCTTATCCCGGACAGCGTGCAAGTC-3′) and subcloned into XhoI- and HindIII-cut pGL3Prom to give luciferase reporter plasmid pRG295.
A 441-bp fragment (AD169 nt 142229 to 142669) (15) containing previously characterized UL98 promoter sequences (1) was PCR amplified from cosmid pCM1007 (19) using primers UL98FORWARD (5′-ATCGCTCGAGGTTGCTCTTTAAGCACGCC-3′) and UL98REV (5′-ATCGAAGCTTCGGTGGGTTTGTACCTTCTC-3′) and subcloned into XhoI- and HindIII-cut pGL3Prom to give luciferase reporter plasmid pRG296. A 5.1-kb XbaI-MunI fragment of cosmid pCM1058 (19) (AD169 nt 159095 to 164178) was subcloned into XbaI- and EcoRI-cut pGEM3ZF+ (Promega) to give plasmid pRG309. A PstI-BamHI fragment of pRG309 (AD169 nt 160856 to 161289) was then subcloned into pBluescript KS+ to give pKS+UL112.
A 387-bp fragment of HCMV AD169 (nt 160163 to 160549) (15) containing previously characterized UL112-113 promoter sequences (6, 69) was PCR amplified from cosmid pCM1058 (19) using primers UL112FOR (5′-GTCGAGATCTCACAGAGGTAACAACGTG-3′) and UL112BACK (5′-GTATAAGCTTGGCCGTGGAGCGAGTG-3′) and subcloned into BglII- and XbaI-cut pGL3Prom to give luciferase reporter plasmid pRG311. Plasmids pAI13 and pLHB2 (30), containing the UL54 and UL57 promoter, respectively, linked to a luciferase reporter gene, were the gift of Dave Anders.
Cell line and virus culture.
All cell lines were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with penicillin, streptomycin, nonessential amino acids, and 10% (vol/vol) NuSerum (Collaborative Research). All procedures were carried out at 37°C with 5% CO2. Primary human foreskin fibroblast (HFF) GMO3468A cells were obtained from the National Institute for General Medical Sciences (Camden, N.J.). Immortalized fibroblast lines ihf-2 and ihf-ie1.3 were prepared as previously described (23). ihf-2 cells are polyclonal immortalized human fibroblasts transduced by a retroviral expression vector expressing the human papillomavirus type 16 E6 and E7 genes. ihf-ie1.3 cells are a polyclonal human fibroblast population transduced initially by a retroviral expression vector expressing the HCMV IE1 p72 protein and subsequently immortalized by transduction with a retroviral expression vector expressing the human papillomavirus type 16 E6 and E7 genes.
The ie1 null mutant CR208 is an HCMV Towne strain mutant carrying a 1.4-kb deletion which completely removes exon 4 of the major immediate-early gene, excising all IE1-specific coding sequences (23). CRQ208 is a rescued derivative of CR208 with wild-type genome structure and growth characteristics (23). Cell-free Towne and CRQ208 virus stocks were prepared in HFF cells. Cell-free CR208 stocks were prepared in ihf-ie1.3 cells. Cells were infected with virus at approximately 0.01 PFU/cell, cultured until cytopathic effect was observed in 100% of the cells, and then refed with fresh growth medium. After 5 days, the cell supernatant was harvested, passed through a 0.45-μm filter, and stored in aliquots at −70°C.
Virus titers were determined by plaque dilution assay performed on ihf-ie1.3 cells, as previously described (23).
Immunofluorescence analysis of viral antigens.
Hybridoma supernatants of mouse monoclonal antibodies SMX (59) and BS510 (67), which recognize IE2 p86 and ppUL44, respectively, were gifts from Bodo Plachter. Mouse monoclonal antibody I2 ascites (1), which recognizes pUL98, was a gift from Richard Stenberg. Mouse monoclonal antibody CH167 ascites, which recognizes pUL57 (35), was a gift from Lenore Pereira. Hybridoma supernatant of the mouse monoclonal antibody α-UL69-66 raised against pUL69 was a gift from Thomas Stamminger, and mouse monoclonal antibody hybridoma supernatant M23 (32), which recognizes the ppUL112-113 proteins, was a gift from Kanji Hirai. Fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody CCH2, which recognizes ppUL44, was purchased from Dako.
Immunocytochemical staining of single antigens in infected cells on coverslips was carried out as previously described (23). The sodium salt of phosphonoformic acid (PFA) was used in cultures at a concentration of 200 μg/ml. To dual stain for IE2 and ppUL44, fixed and permeablized cells were incubated with monoclonal antibody SMX (undiluted hybridoma supernatant) for 1 h at room temperature. Cells were then washed and incubated for 1 h at room temperature with goat anti-mouse immunoglobulin (Ig)-Texas Red conjugate (Serotec), diluted 1:50 in phosphate-buffered saline (PBS) with 10% fetal calf serum (PBS-FCS). Cells were then PBS washed and transferred to 10% mouse serum in PBS-FCS to block free Ig binding sites. After 30 min at room temperature, cells were transferred to FITC-conjugated CCH2 antibody, diluted 1:12 in PBS-FCS-10% mouse serum. After a 1-h incubation, cells were washed, counterstained with Hoechst dye, and mounted.
RNA analysis by Northern blotting.
Whole-cell RNA was prepared at the indicated time points after infection from 5 × 106 human fibroblasts using RNeasy mini columns (Qiagen). Then 10 μg of each purified RNA was electrophoretically separated on agarose-glyoxal gels (Ambion Northern-Max Gly kit) and blotted to Hybond N+ filters (Amersham). Filters were hybridized to radiolabeled DNA probes overnight at 42°C in Ultrahyb buffer (Ambion). Washed filters were exposed to film, and bound radiation was quantitated by phosphorimager (Molecular Dynamics Storm 860). Probes were radiolabeled by Klenow extension from double-stranded DNA fragments and random decamer primers (Ambion Decaprime kit).
To detect the 2.7-kb DE TRL4 RNA, an SpeI-BamHI probe fragment was purified from plasmid pKS+β2.7, representing AD169 equivalent nucleotides 186007 to 187120. For UL44 RNA analysis, an EcoRI probe fragment was purified from plasmid pKS+UL44, representing AD169 equivalent nucleotides 56088 to 56455. To detect UL69 RNA, a BglII fragment was purified from pON2522, representing AD169 nucleotides 98548 to 99460. To detect UL112-113 RNAs, a Pst-BamHI fragment was purified from plasmid KS+pUL112, representing AD169 nucleotides 160856 to 161289.
FISH analysis of viral RNA.
For fluorescent in situ hybridization (FISH) analysis of viral RNA, specific fluorescein-labeled riboprobes were generated by in vitro transcription using a RiboMax II kit (Promega) in the presence of 350 nM fluorescein-12-UTP (Roche) with 650 nM UTP (Promega). To generate antisense riboprobes, the pKS+β2.7 and pKS+HSVtk plasmids were linearized with DraI and EcoRV, respectively, for use as in vitro transcription templates for T7 polymerase. To generate the TRL4/β2.7 sense probe, pKS+β2.7 was digested with ScaI for use as an in vitro transcription template for T3 polymerase. Riboprobes were purified using High-Pure PCR purification columns (Roche) and eluted in diethyl pyrocarbonate (DEPC)-treated sterile water, according to the manufacturer's instructions.
For FISH detection of viral RNA, 5 × 104 HFF cells grown on 19-mm-diameter coverslips in 12-well dishes were infected with virus diluted in serum-free medium at various multiplicities, and after a 1-h adsorption step, the inoculum was replaced with standard growth medium. After 48 h, the cells were washed in DEPC-treated PBS and subsequently fixed with 4% paraformaldehyde in PBS for 30 min. Cells were then dehydrated and rehydrated by sequential 10-min incubations in 70% ethanol, 100% ethanol, 70% ethanol, and finally DEPC-treated PBS. Prepared monolayers were incubated with approximately 100 ng of fluorescence-labeled probe in hybridization solution (60% deionized formamide, 300 mM NaCl, 30 mM sodium citrate, 25 mM NaH2PO4 [pH 7.4], 5% dextran sulfate, and 500 ng of sheared salmon sperm DNA per ml) for 16 h at 37°C. Stained cells were then washed in wash buffer (60% deionized formamide, 300 mM NaCl, 30 mM sodium citrate) three times at room temperature and once at 37°C. Cell nuclei were counterstained with Hoechst 33258 at 100 ng/ml in PBS for 15 min, subsequently rinsed three times in PBS, mounted in glycerol-PBS (Citifluor), and visualized by confocal microscopy using a Leica TCS-SP microscope system.
To dual stain for ppUL44 protein, PBS-washed FISH-stained cells were incubated with BS510 monoclonal antibody (diluted in 10% FCS in PBS) for 1 h. Stained cells were then rinsed three times in PBS and incubated with Texas Red-conjugated secondary antibody (Harlan Serolabs) diluted 1:50 in 10% FCS in PBS for 1 h. Cell nuclei were counterstained with Hoechst 33258 at 100 ng/ml during the final incubation step. Stained cells were subsequently rinsed three times in PBS, mounted in glycerol-PBS (Citifluor), and visualized by confocal microscopy.
Reporter gene analysis of delayed-early promoter activity.
Luciferase reporter constructs (1 μg) were transfected into 5 × 104 cells grown in 12-well dishes using Lipofectin reagent (Invitrogen) for approximately 16 h, according to the manufacturer's instructions. Following transfection, cells were returned to normal growth medium and incubated at 37°C. At 24 h, cells were infected with virus diluted in serum-free medium, and, after adsorption for 1 h, centrifuged at 2,500 rpm (IEC Centra 8 benchtop centrifuge) for 30 min to enhance the efficiency of infection. Subsequently, the inoculum was replaced with standard growth medium. At 24 h postinfection, cells were washed in PBS, lysed in lysis buffer (25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, 1% [vol/vol] Triton X-100, 1 mM dithiothreitol [DTT]) and harvested. Following clarification by centrifugation, cell lysate supernatants were assayed for luciferase activity (17) using an injection luminometer (EG&G Berthold Autolumat LB953). Final assay conditions were 21 mM glycylglycine (pH 7.8), 13 mM MgSO4, 3.4 mM EGTA, 0.87 mM KH2PO4, 9.7 mM K2HPO4, 0.18% Triton X-100, 2.65 mM DTT, 1.8 mM ATP, and 35 μM d-luciferin.
RESULTS
Accumulation of several delayed-early proteins is defective during low-multiplicity infection in the absence of IE1 p72.
To investigate the extent of the block to DE gene expression during low-multiplicity CR208 infection, we analyzed three DE proteins: the single-stranded DNA-binding protein pUL57 (34); pUL69, the regulatory protein homologous to herpes simplex virus type 1 ICP27 and Epstein-Barr virus BMLF1 (80); and the alkaline exonuclease pUL98 (1, 71). Together with ppUL112-113 (discussed later), these are proteins known to accumulate with DE kinetics, for which it is possible to perform unequivocally specific antibody staining during virus infection.
Each of these DE proteins accumulates in the nucleus of infected cells, and all except pUL98 show prominent staining of viral DNA replication compartments during productive infection (data not shown) (57). Although pUL69 is present in HCMV virions (80), our detection procedure did not produce any staining indicative of virion-delivered pUL69 in newly infected cells. This may reflect the low abundance of virion pUL69 rather than a failure of delivery. The nuclear signal we detected at 48 h therefore represented de novo-synthesized pUL69.
In our previous work, the increased frequency of abortive infections during low-multiplicity infection in the absence of IE1 p72 was most readily apparent when viral antigens were analyzed at the single-cell level by immunocytochemical staining (23). We therefore conducted this broader analysis using the same techniques. Monolayers of HFF cells were infected with Towne or CR208 at various multiplicities and stained with antibodies specific for IE2, ppUL44, pUL57, pUL69, and pUL98 at 48 h postinfection (p.i.).
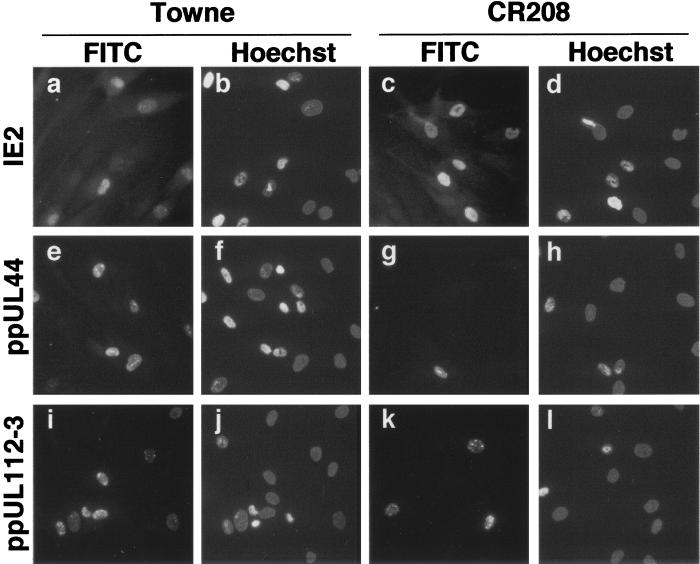
At 48 h p.i., viral DNA replication compartments are beginning to form and are manifest as discrete patches within the nucleus. Although cell staining with antibody SMX cannot distinguish between IE2 p86 and shorter late forms of IE2 (59), accumulation of IE2 proteins at 48 h p.i. and earlier is expected to be limited to IE2 p86 (58, 62). At an input multiplicity of 10 PFU/cell, high-level accumulation of the IE2 protein and the DE protein ppUL44 was observed in a high percentage of cells during both Towne and CR208 infections (data not shown). At an input multiplicity of 0.4, similar proportions of cells expressing IE2 protein were observed in both Towne and CR208 infections (Fig. 1a and c, respectively). In contrast, whereas comparable numbers of cells expressed ppUL44 following infection with Towne strain, many fewer cells expressed this protein after infection with CR208 at this multiplicity (Fig. 1e and g, respectively). These results are all in accordance with our previously published data (23).
FIG. 1.
Photomicrographs of immunofluorescence analysis of viral DE antigens expressed at 48 h following infection with Towne and CR208 viruses at a multiplicity of 0.4 PFU/cell. Panels a, c, e, g, i, k, m, o, q, and s are immunostained cells; panels b, f, j, n, r, d, h, l, p, and t are the same respective fields visualized using Hoechst nuclear counterstain; panels a, b, e, f, i, j, m, n, q, and r are HFFs infected with Towne strain HCMV; panels c, d, g, h, k, l, o, p, s, and t are HFFs infected with CR208. Cells were stained for IE2 protein with SMX antibody diluted 1:2 (a and c), ppUL44 DE protein with BS510 antibody diluted 1:25 (e and g), pUL57 DE protein using CH167 antibody diluted 1:10 (i and k), pUL69 DE protein with anti-UL69-66 antibody undiluted (m and o), or pUL98 DE protein using I2 antibody diluted 1:10 (q and s). Color-positive transparencies were digitized with a Nikon Coolscan III slide scanner and converted to grey scale using Adobe Photoshop software. Images obtained of cells stained with the same antibody were exposed and treated identically. Each panel represents a field 150 by 150 μm.
Patterns of expression similar to that of ppUL44 were seen with the other DE proteins studied. Whereas the majority of cells accumulated DE proteins during both Towne and CR208 infections at a multiplicity of 10 PFU/cell (data not shown), greatly reduced numbers of cells expressed pUL57, pUL69, and pUL98 in CR208 infections at an input multiplicity of 0.4 PFU/cell (Fig. 1k, o, and s, respectively) compared to Towne infection (Fig. 1i, m, and q, respectively). At 48 h p.i. the IE2 staining pattern differed between Towne and CR208 (Fig. 1a versus c). We have previously shown this to reflect the localization of IE2 p86 to nascent viral DNA replication compartments, which do not form during abortive infection by CR208 (23). Rare nuclear DE protein staining in cells infected with CR208 at low multiplicity of infection (Fig. 1g, k, and o) was generally reduced in intensity and more diffuse compared to low multiplicity of infection Towne infection (Fig. 1e, i, and m). This probably reflects a delay of replication compartment formation by CR208 even in those cells which accumulate detectable DE proteins.
Similar experiments were undertaken after 48 h of infection in the presence or absence of the viral DNA replication inhibitor phosphonoformic acid (PFA). The results in Fig. 1 were essentially repeated, and staining patterns in the presence and absence of drug were numerically indistinguishable. However, DNA replication compartment staining was not seen in the presence of PFA, and DE protein staining was weaker and more diffuse (data not shown). This result is similar to data already published for ppUL44 (23). As many HCMV DE proteins continue to accumulate throughout infection, this result shows that the failure of accumulation of each DE protein by CR208 did not occur as a downstream result of a failure in DNA replication. Furthermore, experiments undertaken at time points between 24 and 72 h p.i. demonstrated no significant increases in the proportions of CR208-infected cells staining positively for DE antigens over time (data not shown).
The implication of all of these data together is that there is a block rather than a delay to DE protein accumulation in the absence of IE1 p72. As a high proportion of CR208-infected cells clearly expressed the IE2 protein (Fig. 1c), the majority of these infection events were, by inference, abortive and did not result in accumulation of the DE antigens ppUL44, pUL57, pUL98, or pUL69.
To quantify the observations shown in Fig. 1, the numbers of specifically stained nuclei in multiple representative fields of infected cells were counted and expressed as a percentage of the total number of cells, as determined by counting Hoechst-stained nuclei for each of the proteins studied (Table 1). The results show that at a high multiplicity of infection of 10 PFU/cell, similar proportions of cells expressed IE2, ppUL44, pUL57, pUL69, and pUL98 during both Towne and CR208 infections. In contrast, when cells were infected at a multiplicity of infection of 0.4 PFU/cell, similar proportions of cells expressed IE2; however, the proportions of cells expressing DE proteins during CR208 infections were profoundly reduced compared to Towne infection. Furthermore, the proportions of cells expressing DE proteins in low multiplicity of infection CR208 infections were considerably lower than would be predicted from an infection at 0.4 PFU/cell.
TABLE 1.
Frequencies of nuclear staining for IE2 and DE proteins at 48 or 24 h p.i.
| Time (h p.i.) | Protein | Towne
|
CR208
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 10 PFU/cell
|
0.4 PFU/cell
|
10 PFU/cell
|
0.4 PFU/cell
|
||||||
| No. of stained nuclei/ total no. of cells | % Positive cells | No. of stained nuclei/ total no. of cells | % Positive cells | No. of stained nuclei/ total no. of cells | % Positive cells | No. of stained nuclei/ total no. of cells | % Positive cells | ||
| 48 | IE2 | 178/220 | 81 | 80/206 | 39 | 103/120 | 76 | 98/240 | 41 |
| ppUL44 | 190/210 | 90 | 115/226 | 51 | 96/114 | 84 | 7/220 | 3.2 | |
| pUL69 | 120/135 | 89 | 119/268 | 44 | 120/143 | 84 | 2/271 | 0.74 | |
| pUL98 | 170/184 | 92 | 70/219 | 32 | 129/150 | 86 | 4/203 | 2.0 | |
| pUL57 | 165/190 | 87 | 112/249 | 45 | 78/119 | 66 | 2/216 | 0.93 | |
| 24 | IE2 | 79/84 | 94 | 81/153 | 53 | 93/106 | 88 | 58/114 | 51 |
| ppUL44 | 66/79 | 84 | 54/120 | 45 | 83/102 | 82 | 6/207 | 2.9 | |
| ppUL112-113 | 92/98 | 94 | 65/139 | 47 | 70/75 | 93 | 35/215 | 16 | |
Similar experiments were conducted comparing CR208 and its rescued derivative (CRQ208) (23). The behavior of the rescue virus CRQ208 in these experiments was indistinguishable from that of wild-type Towne virus (data not shown), indicating that the broad failure in DE protein accumulation was directly due to the absence of IE1 p72 during low-multiplicity CR208 infection.
To illustrate the abortive nature of the majority of infections by CR208 at low multiplicity, monolayers of HFs were infected at low multiplicity (0.1 PFU/cell) with CR208 or Towne virus. A coverslip from each infection was fixed at each of five time points and dual stained for IE2 and ppUL44 antigens. Several fields of cells were randomly selected, and numbers of nuclei staining positively for IE2 or ppUL44 were counted. Total numbers of nuclei were quantitated by Hoechst counterstaining.
As anticipated, all nuclei staining positively for ppUL44 also stained IE2 positive, though not all IE2-positive nuclei stained for ppUL44. The results are shown in Table 2. During the Towne infection, the majority of IE2-positive cells at 48 h were also ppUL44 positive. After this time point, there was significant spread of the infection, culminating in 88% of cells staining positively for IE2 at the day 5 time point. ppUL44 staining was also observed to spread, lagging slightly behind the spread of IE2 staining.
TABLE 2.
Frequencies of nuclear staining for IE2 and ppUL44 at various times after infection by Towne or CR208 at 0.1 PFU/cell
| Time (h p.i.) | Towne
|
CR208
|
||||||
|---|---|---|---|---|---|---|---|---|
| IE2
|
ppUL44
|
IE2
|
ppUL44
|
|||||
| No. of stained nuclei/total no. of cells | % Positive cells | No. of stained nuclei/total no. of cells | % Positive cells | No. of stained nuclei/total no. of cells | % Positive cells | No. of stained nuclei/total no. of cells | % Positive cells | |
| 24 | 42/590 | 7.12 | 31/590 | 5.25 | 48/577 | 8.32 | 4/577 | 0.693 |
| 48 | 45/622 | 7.23 | 39/622 | 6.27 | 53/551 | 9.62 | 4/551 | 0.725 |
| 72 | 127/896 | 14.2 | 103/896 | 11.5 | 54/826 | 6.54 | 2/826 | 0.242 |
| 96 | 258/780 | 33.1 | 211/780 | 27.1 | 36/845 | 4.26 | 3/845 | 0.355 |
| 120 | 621/706 | 88.0 | 549/706 | 77.8 | 38/732 | 5.19 | 4/732 | 0.546 |
During CR208 infection, despite significant numbers of cells (8 to 9%) staining positively for IE2 at early time points, numbers of ppUL44-positive cells remained low (lower than 1%) throughout the time course. IE2-positive cells persisted through the time course, though numbers declined slightly. Thus, a large proportion of cells initially staining positively for IE2 failed to progress to ppUL44 expression and failed to replicate virus. These infection events were therefore apparently abortive. Upon further examination of stained monolayers, rare foci of productive infection by CR208 were observed at the 120 h time point, with clear spread of IE2 and ppUL44 antigen staining. The rarity of these foci is consistent with our previous suggestion that such productive events derive from multiple-hit infections.
Accumulation of UL112-113 family of delayed-early proteins observed in a proportion of cells infected abortively with ie1 mutant CR208.
During our analyses, it was observed that the accumulation of the DE family of phosphoproteins derived from the UL112-113 open reading frames, although still defective, occurred in higher proportions of cells than accumulation of other DE proteins during low-multiplicity infections with CR208. These data are shown in Fig. 2.
FIG. 2.
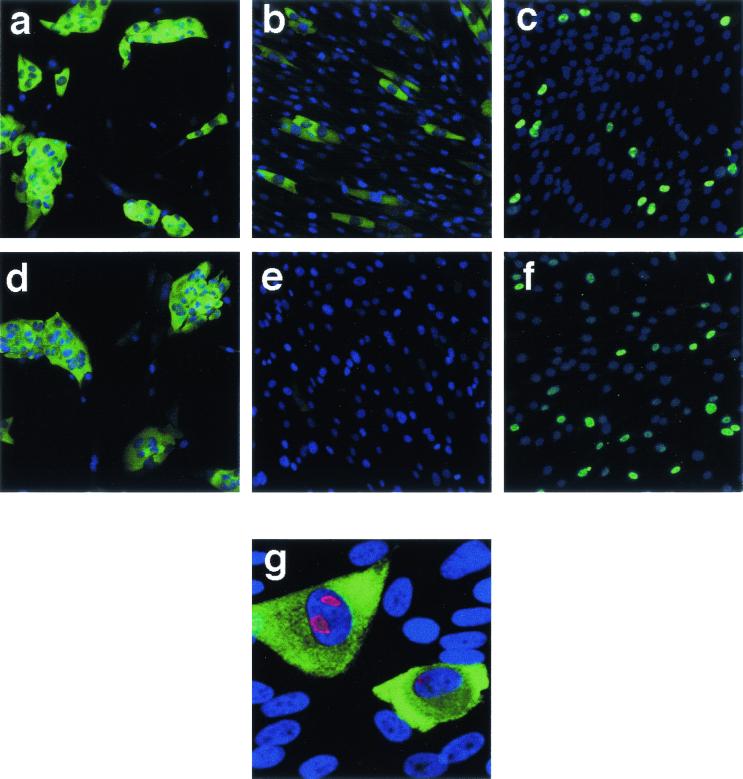
Photomicrographs of immunofluorescence analysis of viral DE antigens expressed at 24 h following infection with Towne and CR208 at a multiplicity of 0.4 PFU/cell. Panels a, c, e, g, i, and k are immunostained cells; panels b, d, f, h, j, and l are the same respective panels visualized using Hoechst nuclear counterstain; panels a, b, e, f, i, and j are HFFs infected with Towne strain HCMV; panels c, d, g, h, k, and l are HFFs infected with CR208. Cells were stained for IE2 protein using SMX antibody diluted 1:2 (a and c), ppUL44 DE protein using BS510 antibody diluted 1:25 (e and g), or ppUL112-113 DE protein using M23 antibody diluted 1:10 (i and k). Color-positive transparencies were processed as described for Fig. 1. Images obtained of cells stained with the same antibody were exposed and treated identically. Each panel represents a field 200 by 200 μm.
Monolayers of HFF cells were infected with either Towne or CR208 at various multiplicities and stained with antibodies specific for IE2, ppUL44, or ppUL112-113 at 24 h pi. At an input multiplicity of 10 PFU/cell, accumulation of IE2, ppUL44, and ppUL112-113 was observed in a high proportion of cells and at similar relative levels (data not shown). Following an input multiplicity of 0.4 PFU/cell, comparable numbers of cells expressed IE2, ppUL44, and ppUL112-113 proteins in Towne infection (Fig. 2a, e, and i, respectively). Infection with CR208 at a multiplicity of infection of 0.4 PFU/cell demonstrated a profound defect in the number of cells accumulating ppUL44 compared to the number accumulating IE2, as expected (Fig. 2c and g, respectively). There was also a deficiency in the number of cells accumulating the DE protein ppUL112-113 (Fig. 2k), although this deficiency was less pronounced than that seen with ppUL44 and the other DE proteins studied.
These data were quantified by counting the number of cells specifically stained for IE2, ppUL44, or ppUL112-113 in representative fields of cells and expressing this as a percentage of the total number of cells, as determined by counting Hoechst-stained nuclei (Table 1). The results showed that at a multiplicity of infection of 10 PFU/cell, similar proportions of cells expressed IE2, ppUL44, and ppUL112-113 in both Towne and CR208 infections. In contrast, when cells were infected at a multiplicity of infection of 0.4 PFU/cell, similar proportions of cells expressed IE2, whereas the proportions of cells expressing ppUL44 and ppUL112-113 in CR208 infections were reduced compared to Towne infection. Although there was a deficiency in the proportion of cells expressing ppUL112-113 after low-multiplicity CR208 infection compared to Towne infection (16% versus 47%, respectively), considerably more cells expressed ppUL112-113 protein than pUL44 protein after low-multiplicity CR208 infection (16% versus 2.9%, respectively). Similar results were seen at 48 h p.i., and also at 24 h p.i. when CR208 infection was compared to CRQ208 infection (data not shown).
Taken together with our observation that the proportion of cells expressing ppUL44, pUL57, pUL69, and pUL98 did not increase after 48 h p.i., these data show that a significant proportion of cells infected abortively with CR208 accumulated ppUL112-113 proteins while failing to accumulate other DE proteins. This suggests that the accumulation of ppUL112-113 proteins was less dependent on IE1 p72 at low multiplicity than was the accumulation of many other DE proteins. This may indicate that, in the context of infection, transcription from the UL112-113 promoter is less dependent on IE1 p72 function than is transcription from other DE promoters.
Defective accumulation of viral delayed-early transcripts during low-multiplicity infection in the absence of IE1 p72.
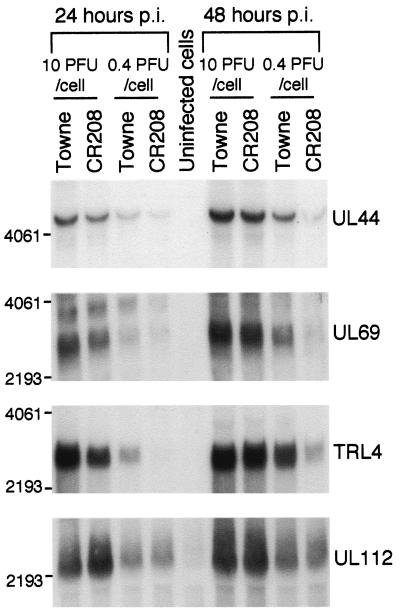
Given the enhancing effect of IE1 p72 on IE2-mediated transcriptional activation of viral DE gene promoters, one possible explanation for the apparent defect in DE protein accumulation is a deficiency in DE gene transcription and hence DE RNA accumulation during low-multiplicity infection in the absence of IE1 p72. In order to investigate this possibility, we analyzed the accumulation of several DE RNAs by Northern blot. RNA samples were prepared from cells infected with either CR208 or Towne virus at high (10 PFU/cell) or low (0.4 PFU/cell) multiplicity of infection. Whole-cell RNA samples were prepared at 24 h and 48 h p.i. time points. The results of Northern blotting using a variety of DE gene probes are shown in Fig. 3.
FIG. 3.
Northern blot analysis of 10 μg of whole-cell RNA, derived from cells infected at 10 or 0.4 PFU/cell with Towne or CR208 virus and harvested at 24 or 48 h p.i. Each panel shows an autoradiograph after separate hybridization of replica blots with different DNA probes derived from HCMV DE genes. Sizes (in nucleotides) of RNA markers are shown to the left of each panel. The specificity of the radiolabeled probe used for each blot (detailed in Materials and Methods) is shown to the right of each panel.
A unique 4.8-kb UL44 transcript was detected, in agreement with previous studies (20). This transcript accumulated to 78% of wild-type levels during high-multiplicity infection by CR208 at 24 h and 85% at 48 h. However, during low-multiplicity CR208 infection, there was a 2.8-fold deficit in UL44 RNA accumulation at 24 h and a 4.9-fold deficit at 48 h.
We detected several transcripts from the UL69 region, including the 2.7-kb and 3.5-kb DE transcripts characterized by Winkler et al. (79). Our analysis concentrated on the 2.7-kb transcript, which derives entirely from UL69 and is thought to code for the pUL69 protein (79). During high-multiplicity infection, the 2.7-kb transcript accumulated to 75% of wild-type levels at 24 h and 89% at 48 h. During low-multiplicity infection, a pronounced accumulation defect was again observed (2.3-fold deficit at 24 h and 3.7-fold deficit at 48 h). The 3.5-kb transcript evident at 24 h postinfection again followed a similar pattern of accumulation.
Probing for TRL4 produced a unique 2.7-kb band, in agreement with previous reports (24). This highly abundant DE transcript also accumulated defectively during low-multiplicity CR208 infection (9.0-fold deficit at 24 h and 3.9-fold deficit at 48 h). Even at high multiplicity, a clear defect in TRL4 RNA accumulation was apparent at 24 h postinfection by CR208, reaching only 60% of normal levels, though by 48 h this had reached 90%.
We detected a prominent 2.2-kb transcript using a UL112 probe, in agreement with previous studies (72). In marked contrast to the other DE transcripts studied, this transcript accumulated to wild-type levels or higher during high- and low-multiplicity CR208 infections, providing a partial explanation for the significant accumulation of ppUL112-113 proteins that we had observed previously during low-multiplicity CR208 infection. In summary, RNA accumulation data for the UL44, UL69, and TRL4 DE transcripts were consistent with the bulk of our DE protein accumulation data, showing a pronounced defect during low-multiplicity infection in the absence of IE1. UL112-113 transcripts and proteins were exceptions to this observation.
For these results to be fully comparable to our viral antigen analysis, we also conducted staining and analysis at the single-cell level, using FISH. We chose to study the major 2.7-kb TRL4 DE transcript of HCMV, also termed the β2.7 RNA. This transcript is highly abundant, constituting up to 20% of the total viral RNA expressed during infection (24, 49). Furthermore, it is expressed with DE kinetics, and its transcription in cotransfection assays is both driven by IE2 p86 and enhanced by IE1 p72, as are many other DE promoters (36).
The 2.7-kb TRL4 RNA was detected by hybridization to a complementary fluorescently labeled “antisense” riboprobe. The specificity of the 2.7-kb TRL4 RNA staining was controlled using a similarly generated riboprobe specific for the herpes simplex virus type I thymidine kinase mRNA and a “sense” riboprobe, with sequence identity to the TRL4 RNA. No specific staining was observed using either control riboprobe, validating the specificity of the 2.7-kb TRL4 RNA fluorescence (data not shown). The specificity of the 2.7-kb TRL4 RNA fluorescence for HCMV-infected cells was further controlled by costaining HFFs infected with CRQ208 at a multiplicity of infection of 1.0 PFU/cell for ppUL44 DE protein (antibody BS510). Fluorescent cell staining was visualized by confocal microscopy (Fig. 4g). Only cells showing specific ppUL44 nuclear staining (red) also showed specific cytoplasmic 2.7-kb TRL4 RNA staining (green). Uninfected cells visible by Hoechst nuclear staining demonstrated no DE protein or RNA expression (Fig. 4g). The 2.7-kb TRL4 RNA was detected exclusively in the cytoplasm of infected cells.
FIG. 4.
Photomicrographs of fluorescence in situ hybridization analysis of viral DE RNA expressed at 48 h following infection with wild-type CRQ208 and CR208 at various multiplicities. Panels a, b, d, and e are fluorescence in situ hybridization of the 2.7-kb TRL4 DE RNA (green) overlaid with corresponding Hoechst nuclear counterstain (blue); panels c and f are cells immunostained for IE2 protein using SMX antibody diluted 1:2 (green) overlaid with corresponding Hoechst nuclear counterstain (blue); panel g is fluorescence in situ hybridization of the 2.7-kb TRL4 DE RNA (green) overlaid with corresponding immunostaining for pUL44 DE protein (red) and Hoechst nuclear counterstain (blue). HFFs were infected with CRQ208 at a multiplicity of 5 PFU/cell (panel a), 0.5 PFU/cell (panels b and c), or 1.0 PFU/cell (panel g) or with CR208 at a multiplicity of 5 PFU/cell (panel d) or 0.5 PFU/cell (panels e and f). All images were obtained using a Leica TCS-SP confocal microscope with Leica TCS software. All images were created from a projected stack of horizontal cross-sections, representing the entire depth of the cell monolayer. Overlaid images in panels a, b, d, e, and g were generated using Leica TCS software. Overlaid images in panels c and f were generated using Adobe Photoshop software. All corresponding images were treated identically. Panels a to f represent fields 700 by 700 μm square. Panel g represents a field 130 by 130 μm square.
Although the staining pattern has some granularity, we have been unable to convincingly demonstrate the localization of this RNA to any cellular organelles (data not shown). Our fluorescence results differ from the nuclear and cytoplasmic staining observed in permissive HFs by Wu et al. (81), in that we observed no evidence of nuclear accumulation of the 2.7-kb TRL4 RNA in thin nuclear cross-sections of productively infected cells.
Monolayers of HFF cells were infected with CRQ208 or CR208 at various multiplicities. At 48 h p.i., cells were stained for either IE2 protein expression (antibody SMX) or for the 2.7-kb TRL4 RNA using a fluorescent riboprobe (Fig. 4). At an input multiplicity of 5 PFU/cell, high-level accumulation of the IE2 protein (data not shown) and the 2.7-kb TRL4 RNA was observed in both CRQ208 and CR208 infections (Fig. 4a and d, respectively). At an input multiplicity of 0.5 PFU/cell, however, whereas similar proportions of cells expressed IE2 and 2.7-kb TRL4 RNA during wild-type infections (Fig. 4b and c, respectively), a profound reduction in the number of cells expressing the 2.7-kb TRL4 RNA was observed in CR208 infection compared to the proportion of cells expressing IE2 (Fig. 2e versus f).
The results shown in Fig. 4a to f were quantified as before. The numbers of nuclei specifically stained for IE2 in representative fields of infected cells were counted and expressed as a percentage of the total number of cells, determined by counting Hoechst-stained nuclei. Likewise, the numbers of cells exhibiting specific cytoplasmic 2.7-kb TRL4 RNA staining were counted and expressed as a percentage of the total number of cells determined by counting Hoechst-stained nuclei. The results, shown in Table 3, show that at a high multiplicity of infection of 5 PFU/cell, a high proportion of cells expressed 2.7-kb TRL4 RNA in both CRQ208 and CR208 infection. A higher proportion of cells expressed 2.7-kb TRL4 RNA after CR208 infection than after CRQ208 infection (85% versus 75%, respectively), and this was also reflected in the proportions of cells expressing IE2 protein at a multiplicity of infection of 0.5 PFU/cell (28% versus 9.7%, respectively). This indicates that the input multiplicities were not exactly equivalent and that the actual input of CR208 was higher than that of CRQ208.
TABLE 3.
Frequencies of nuclear IE2 and cytoplasmic TRL4 DE RNA staining at 48 h p.i.
| Protein | CRQ208
|
CR208
|
||||||
|---|---|---|---|---|---|---|---|---|
| 5 PFU/cell
|
0.5 PFU/cell
|
5 PFU/cell
|
0.5 PFU/cell
|
|||||
| No. of cells stained/total | % Positive cells | No. of cells stained/total | % Positive cells | No. of cells stained/total | % Positive cells | No. of cells stained/total | % Positive cells | |
| IE2 | NDa | ND | 20/207 | 9.7 | ND | ND | 31/110 | 28 |
| DE RNA | 117/156 | 75 | 21/220 | 9.5 | 117/137 | 85 | 1/160 | 0.6 |
ND, not determined.
At the calculated input multiplicity of 0.5 PFU/cell, the proportion of cells expressing 2.7-kb TRL4 RNA was greatly reduced after CR208 infection compared to CRQ208 infection (0.6% versus 9.5%, respectively). Thus, during a low-multiplicity infection by wild-type virus, there was approximate parity between numbers of IE2 protein-expressing and 2.7-kb TRL4 RNA-expressing cells, but during a low-multiplicity CR208 infection, the majority of cells accumulating IE2 failed to accumulate 2.7-kb TRL4 RNA. These DE RNA data reflect the data obtained for DE proteins, suggesting that the failure to accumulate DE proteins is likely to be due to the failure to accumulate DE RNA. This indicates that IE1 p72 is required for DE RNA accumulation and may therefore be required for DE promoter transactivation during low-multiplicity infections.
Defective delayed-early promoter activity in the absence of IE1 p72.
We wished to determine if lower DE promoter activity was responsible for the lower proportion of cells accumulating DE RNAs during low-multiplicity CR208 infection. Conventional methods for the direct measurement of promoter activity (for example, nuclear run-on techniques) lack sensitivity in the low-multiplicity scenario in which IE1 p72 function is most significant. We therefore followed an indirect measurement of DE promoter activity, transfection of DE promoter-reporter constructs followed by infection with wild-type and ie1 mutant viruses. Plasmid constructs in which the luciferase reporter gene was placed under the control of the UL44, UL54, UL57, UL69, UL98, and UL112-113 promoters were obtained or generated.
Basal levels of luciferase activity expressed from the UL57 promoter were not distinguishable from background levels. Otherwise, small but significant basal activity was measured: 2.3-fold background (UL44-luc); 1.8-fold background (UL54-luc); 3.4-fold background (UL69-luc); 3.8-fold background (UL98-luc); and 11.6-fold background (UL112-luc). Each DE promoter reporter construct was trans-activated 500- to 3,000-fold by HCMV (Towne) infection of transfected cells at a multiplicity of infection of 10 PFU/cell. Similar results were obtained using either primary fibroblasts or the human papillomavirus 16 E6/E7-immortalized fibroblast line ihf-2. The high ratios of activation obtained suggested the feasibility of using this system to measure DE promoter activity during low-multiplicity HCMV infections.
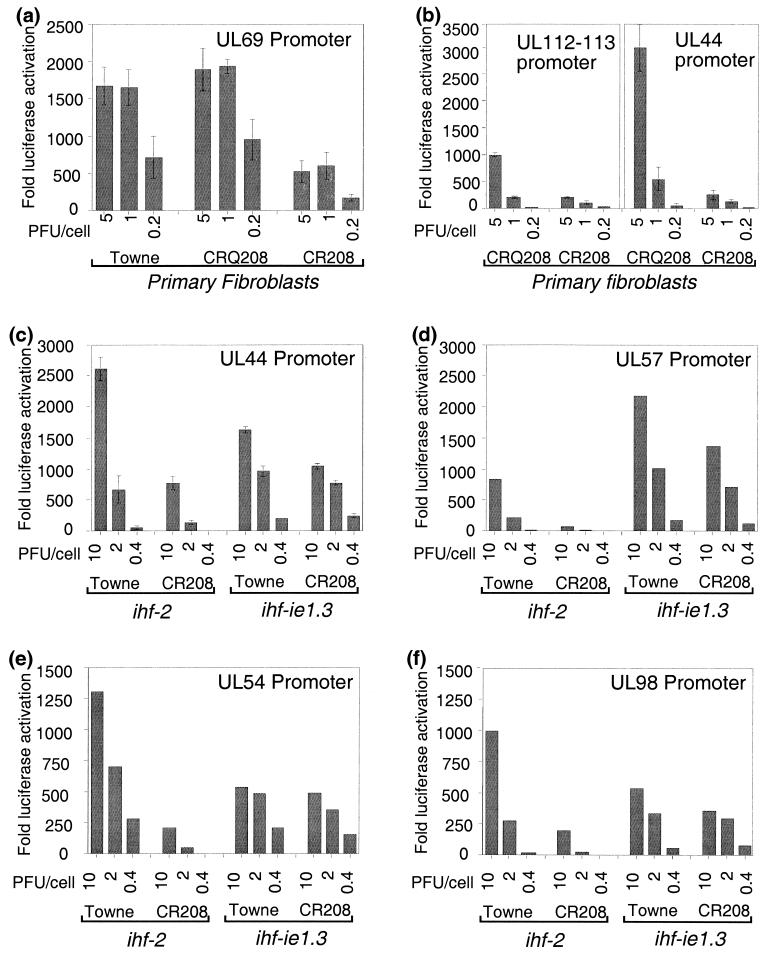
An experiment using the UL69 promoter reporter construct is shown in Fig. 5a. Multiple cultures of primary fibroblasts were transfected with a fixed quantity of reporter plasmid and infected at 24 h posttransfection with various multiplicities of wild-type Towne virus, ie1 mutant CR208, or the rescued CRQ208 virus. Luciferase activity was measured at 24 h p.i. Although the wild-type and rescued viruses produced similar levels of promoter activation, activation by CR208 infection was defective at all multiplicities tested. UL54, UL57, and UL98 reporters gave very similar results in primary fibroblasts; activation by CR208 infection was defective at all multiplicities tested (data not shown).
FIG. 5.
Luciferase reporter gene analysis of DE promoter activity during infection by Towne, CR208, and CRQ208 at various multiplicities of infection. HFF, ihf-2, or ihf-ie1.3 cells were transfected with luciferase reporter constructs, infected as indicated, and assayed as described in Materials and Methods. Results from representative experiments with the UL69 promoter (a), UL112-113 and UL44 promoters (b), UL44 promoter (c), UL57 promoter (d), UL54 promoter (e), and UL98 promoter (f) are shown. Data points in panels a, b, and c are the mean ratio of luciferase activation over basal, derived from triplicate data points. Error bars denote a 95% confidence interval for the estimated mean. Data points in panels d, e, and f are the ratio of luciferase activation over basal, obtained in representative single-data point experiments.
A similar analysis was performed using a UL112-113 promoter reporter construct in an attempt to ascertain whether higher promoter activity could account for the significant accumulation of UL112-113 RNAs and proteins observed at low multiplicity of infection in the absence of IE1 p72. A comparative experiment together with the UL44 reporter is shown in Fig. 5b, and UL112-113 reporter activity surprisingly followed a pattern very similar to that of the other DE reporters, showing a deficiency in the absence of IE1 p72 at all multiplicities tested. We were therefore unable to correlate the increased accumulation of UL112-113 RNAs and proteins relative to other DE proteins with any reduced dependence of the UL112-113 promoter on IE1 p72 function during low-multiplicity CR208 infection.
To determine that the activation defects observed after CR208 infection resulted from the absence of IE1 p72, similar experiments were undertaken comparing the E6/E7-immortalized, IE1 p72-expressing human fibroblast line ihf-ie1.3 with a control E6/E7-immortalized human fibroblast line, ihf-2. We have previously demonstrated that ihf-ie1.3 cells are permissive to CR208 infection after low-multiplicity input, whereas ihf-2 cells are not (23). Multiple cultures of the two cell lines were transfected with a fixed quantity of a UL44 promoter-luciferase construct and infected after 24 h at various multiplicities with either Towne or CR208 virus. Luciferase activity was determined at 24 h p.i. and is presented in Fig. 5c as the induction ratio above the basal level.
In noncomplementing ihf-2 cells, activity of the UL44 promoter was clearly deficient at all multiplicities of CR208 compared to Towne infection. This defect was most apparent at low multiplicity of infection (0.4 PFU/cell), where 55-fold induction was seen after Towne infection, compared to 9-fold induction after CR208 infection. However, when these experiments were repeated in cells expressing the IE1 p72 protein, this deficiency was corrected. Activation of the luciferase reporter gene was comparable at all multiplicities, most notably at 0.4 PFU/cell, where high-level activation of the UL44 promoter was observed during infection by both CR208 (248-fold induction) and Towne (206-fold induction). Slightly reduced induction ratios for CR208 versus Towne at higher multiplicities may possibly be accounted for by incomplete expression of IE1 p72 in the ihf-ie1.3 cell culture; at any one time during the course of these experiments, between 70 and 90% of cells in the ihf-ie1.3 culture stained positive with a monoclonal antibody to IE1 p72 (data not shown).
Similar data are shown for the UL57, UL54, and UL98 DE reporters in Fig. 5d to f. These data show that IE1 p72 is vital for the proper activation of DE promoter reporter constructs in this system and that the activation defect observed during infection by CR208 was corrected by the provision of IE1 p72 in trans and therefore attributable to the absence of IE1 p72 protein.
DISCUSSION
The IE1 p72 protein is essential during low-multiplicity HCMV infections of primary fibroblasts for full progression into the DE phase of the lytic life cycle. We have previously demonstrated that low-multiplicity infection by the ie1 mutant CR208 is frequently abortive, with a failure to accumulate a DE protein and establish replication compartments (23). This was characterized during in situ analysis of viral antigens by a disproportionately low number of cells which accumulated the ppUL44 protein relative to those which expressed IE2 p86. A strongly nonlinear relationship was observed between CR208 inoculum volume and either the resulting plaque number or the proportion of cells which expressed ppUL44. This observation suggested that the rare cells in which ppUL44 accumulated after low-multiplicity infection represented productive multiple-hit infections.
Here, we have carried out a broader analysis to show that three further viral DE proteins (pUL57, pUL98, and pUL69) exhibited a very similar low-multiplicity infection accumulation defect when analysis was carried out at the level of individual cells—a disproportionately small number of cells accumulated these DE proteins relative to those which expressed IE2 p86. The small proportion of cells accumulating these DE proteins was furthermore observed not to increase over time. These results imply a broad block to viral DE protein accumulation during low-multiplicity infection in the absence of IE1 p72.
To address the nature of this defect to DE gene expression, we first investigated steady-state levels of DE RNAs by Northern blotting. These experiments revealed defective accumulation of several DE RNAs (UL44, UL69, and TRL4) during low-multiplicity infection by the ie1 mutant. To assess whether DE RNA accumulation patterns at the single-cell level were similar to those observed for DE proteins, we conducted in situ hybridization analysis of the abundant 2.7-kb DE transcript originating from the TRL4 open reading frame. Again, a disproportionately small number of cells infected with CR208 at low multiplicity accumulated the TRL4 transcript relative to those expressing IE2 p86. This defect was not observed in wild-type or rescue viruses. These results imply that the block to DE gene expression lies at the level of RNA accumulation.
As IE1 p72 is known to transactivate DE promoters in concert with IE2 p86, defective transcription from DE promoters is a likely explanation for the observed block. We therefore assessed DE promoter activity during infection by mutant and wild-type viruses at different multiplicities, using plasmid reporter constructs. Defective activity was observed from transfected DE reporter constructs during low-multiplicity infection by ie1 mutant CR208, which could be corrected by the provision of IE1 p72 in trans. When considered together with the DE RNA accumulation data, these transient reporter studies argue that a transcriptional defect was probably responsible for the impasse observed at the onset of the DE phase during low-multiplicity infections in the absence of functional IE1 p72.
It should be noted that strong (10- to 100-fold) nonspecific activation of heterologous reporter constructs upon infection by human and simian CMVs has been documented (52, 54) and may include a posttranscriptional component (52). However, the much greater levels of activation that we obtained using HMCV DE reporter constructs (500- to 3,000-fold) strongly support a specific component to the activation observed. Nevertheless, we acknowledge that there may be a significant nonspecific contribution to activation and that our transient experiments do not fully rule out posttranscriptional effects. In addition, there was lack of agreement between the defective behavior of the UL112-113 promoter as measured by these experiments and the normal accumulation of UL112-113 RNAs observed during low-multiplicity CR208 infection. The analysis of DE reporters in the context of the ie1 mutant viral genome will in the longer term provide the most direct and reliable assessment of transcription during infection in the absence of IE1 p72.
The precise mechanism by which IE1 p72 is involved in the transcription of DE genes remains elusive. Although IE1 p72 has been shown to transactivate some cellular genes, it has not been shown to significantly activate viral DE gene promoters when acting alone. It is generally thought that IE2 p86 is the major factor directing transcription at many DE promoters and that IE1 p72 acts to augment its effects (50), although the relative magnitudes of these effects appear to be cell type specific. Of particular relevance to this study, experiments using permissive primary fibroblasts showed that both IE1 p72 and IE2 p86 together were required to significantly activate the DE viral DNA polymerase promoter (74). The mechanism for this cooperation is unknown; no direct interaction between IE1 p72 and IE1p86 has been shown, although both proteins can bind concomitantly to the hTAFII130 component of the basal transcriptional machinery (43). This suggests that IE1 p72 may facilitate the recruitment of basal transcription factors to IE2 p86-responsive promoters and/or stabilize the transcriptional preinitiation complex.
Our results demonstrate that IE1 p72 function is vital during low-multiplicity infections and that an essential role in DE gene transcription is the probable reason. Even at high multiplicity, transcriptional activity from DE reporter constructs in the absence of IE1 p72 was impaired compared to wild-type HCMV infection. We also observed a slight delay in the accumulation of DE RNAs at high multiplicity. It is therefore likely that IE1 p72 is necessary under all conditions for the full and efficient transcription of DE genes. Despite this impairment, significant DE gene transcription must ensue under high-multiplicity conditions to allow completion of the viral lytic cycle, since all the DE proteins and RNAs studied accumulated in the majority of infected cells to high levels. Indeed, productive infection was observed at high input multiplicities (23).
The absolute requirement for IE1 for the accumulation of DE RNAs and proteins is therefore circumvented at high multiplicity. This may be because the ancillary functions of IE1 p72 are performed by other viral or cellular factors. Viral factors which may have the ability to influence DE gene expression include the IE proteins from the US3, UL36-38 (16), and TRS1/IRS1 loci (30, 73) and the virion proteins pp71 (8, 14), pUL69 (80), and pTRS1/IRS1 (64). Each of these factors may be present in CR208-infected cells prior to the onset of DE phase. However, as these proteins influence transcription and DE gene transcription appears still to be compromised during high-multiplicity CR208 infection, it is not clear how these factors might act to augment the accumulation of DE proteins. Alternative explanations for productive infection by CR208 after high-multiplicity input are that IE2 p86 is produced in sufficient quantity from multiple templates to negate the requirement for IE1 p72 or that multiple DE gene templates may allow sufficient DE expression in the presence of IE2 p86 alone.
The use of DE reporter constructs in transient-transfection assays has shown that additional viral proteins can enhance IE2 p86-mediated transactivity. Since some of these factors are not expressed with IE kinetics, they may not be expressed at low multiplicity in the absence of IE1 p72 and may thus be unable to influence the progression into DE phase. One of the DE loci shown to act in this way is UL112-113 (30). Our results show that, although accumulation of these proteins was defective at low multiplicities in the absence of IE1 p72, they accumulated in a greater proportion of cells than did other DE proteins studied. First, this discordant expression demonstrates that ppUL112-113 proteins are unable to complement the lack of IE1 p72 during IE2 p86-mediated expression at other DE loci. Second, this indicates that expression from this DE locus is less dependent on IE1 p72 than are other DE genes. Indeed, when steady-state RNA levels were assessed, UL112-113 transcripts accumulated to apparently normal levels during low-multiplicity infection in the absence of IE1 p72.
Accordingly, IE2 p86 activation of the UL112-113 promoter has been observed to be virtually independent of IE1 p72's effects in transfected permissive U373 cells (37). IE2 p86 activation of transcription from this promoter has been shown to be mediated by DNA binding sites for both IE2 p86 and the cellular CREB factor (6, 40, 68). In the context of the viral genome, both promoter elements are important for efficient DE expression from an ectopically placed UL112-113 promoter (63). Despite the higher steady-state levels of UL112-113 mRNAs relative to other DE RNAs during CR208 infection, we were unable to demonstrate higher sensitivity of a transfected UL112-113 promoter to CR208 infection at low multiplicity of infection. This suggests either that our promoter activation assay lacks the refinement to distinguish subtle transcriptional differences or that significant UL112-113 RNA and protein accumulation can occur in the face of reduced transcription, even at lower multiplicities.
The role of IE1 p72 in the expression of DE genes may not solely involve interaction with IE2 p86 at DE gene promoters. IE1 p72 has been shown to activate cellular genes independently, including two genes directly involved in cellular DNA replication (DHFR and polymerase α) (27, 76). IE1 p72 may function similarly to activate cellular genes whose products are involved in viral DE gene activation. Alternatively, IE1 p72 may directly activate cellular phosphorylation cascades and transcription factor pathways that are required for HCMV DE gene transcription. IE1 p72 physically interacts with the cellular pocket protein p107 (60), and kinase activity has been attributed to IE1 p72, so that it is able to phosphorylate cellular E2F and pocket proteins (55), affecting the transcription of E2F-responsive genes. IE1 p72 is also capable of inducing cellular transition into S-phase, where p53 function is inhibited (12, 60).
Transition through G1 may not be a vital role for IE1 p72 in the expression of DE genes per se, since IE2 p86 can also mediate this function (12, 53). However, IE1 p72 clearly has broad activities within the host cell which favor an environment conducive to HCMV lytic replication, and these may include the generation or regulation of cellular factors required for DE gene transcription. If cell cycle stimulation by IE1 p72 constitutes by these means an essential low-multiplicity function, this must be replaced at higher multiplicities by other viral functions, possibly by IE2 p86, possibly by other IE or virion proteins, perhaps as a result of signaling deriving from the initial virus-cell interaction. Intense research interest in this field should produce more definitive candidates.
Another known function of IE1 p72 that may be important in regulating the transition into DE phase is its ability to disrupt nuclear polymorphonuclear leukocyte (PML) bodies, also known as ND10s and PODs (3, 78). These bodies represent the primary deposition site of viral genomes, are directly adjacent to sites of initial viral gene transcription, and are thought to direct the formation of viral replication compartments (4, 29). PML bodies are not disrupted and replication compartments do not form during abortive low-multiplicity CR208 infection; however, the disruption of PML bodies is not essential for lytic cycle progression per se, since they remain intact during productive high-multiplicity CR208 infections (2; R. F. Greaves and G. W. Wilkinson, unpublished data). It is interesting, however, that retardation of PML body disruption by overexpression of the PML oncoprotein has growth-inhibitory effects on wild-type HCMV infection somewhat similar to those seen in low-multiplicity CR208 infection—lytic cycle progression is delayed and DE gene expression is impaired (2). It has therefore been suggested that PML body disruption is required for the efficient transcription of DE genes at low multiplicity.
Like IE1 p72, IE2 p86 localizes to PML bodies prior to their disruption by IE1 p72 (3, 29). Although IE gene transcription can take place from genomes deposited at PML bodies (29), IE2 p86 protein may be functionally impeded while localized at PML bodies, requiring their disruption by IE1 p72 to mediate efficient DE gene transcription. In support of this hypothesis, accumulation of IE2 p86 at PML bodies appears accentuated during CR208 infection (2). During high-multiplicity infections by CR208, it is conceivable that enough IE2 p86 is generated that sufficient free and functional protein exists to direct DE gene transcription. Alternatively, other IE or virion proteins might functionally substitute for IE1 p72 at high multiplicities, modifying PML bodies without so obviously disrupting them.
IE1 p72 has been shown to positively autoregulate the MIEP, probably via NF-κB transcription factor binding sites, by a mechanism that is likely to involve cellular factors and not to entail direct binding of DNA (65). Although initial observations indicated an important role for this function in viral replication (51), our previous experiments (23) and those presented here suggest equivalent early accumulation of IE2 p86 during permissive infections by wild-type virus and during abortive infections by CR208. Infection of permissive U373 cells with Towne and CR208 viruses at low multiplicity has led to similar observations (2). We therefore argue that it is unlikely that autoregulation by IE1 p72 plays an essential role in lytic infection.
An essential role for IE2 during infection is supported by the antiviral properties of phosphorothioate oligonucleotides antisense to IE2 mRNAs (7). Furthermore, transfected IE2-null recombinant viral genomes lacking exon 5 of the major IE gene failed to express DE RNAs (47). To date, ie2 mutant viruses have not been successfully cultured in complementing cell lines, and therefore the requirement for IE2 during high-multiplicity infections has yet to be tested. However, a murine CMV mutant unable to express IE3 (the homolog of HCMV IE2) was defective at all multiplicities tested and failed to express DE genes (5). We therefore consider it likely that IE2 proteins will prove essential at all multiplicities of infection.
In contrast, IE1 p72 is only essential for the infection of fibroblasts at low multiplicity. Under such low-multiplicity conditions, we have shown that IE1 p72 is required for the proper accumulation of a broad sample of DE proteins and DE RNAs. We have also shown data which imply that, even at high multiplicity, DE gene transcription (although sufficient for the completion of the lytic cycle) is impaired in the absence of IE1 p72.
Whether the stimulatory function of IE1 p72 involves direct interaction of IE1 p72 with DE gene promoters or the modulation of intracellular events which come to bear on viral DE gene transcription remains to be determined. Nevertheless, it is increasingly apparent that IE1 p72 is a highly important factor in the normal and efficient progression of the HCMV lytic cycle.
Acknowledgments
This work was supported by an MRC Senior (Nonclinical) Fellowship awarded to R.F.G., preceded by a Wellcome Career Development Fellowship.
We thank John Sinclair and Patrick Sissons for sponsorship of this work at Cambridge and Martin Allday and Paul Farrell for continuing support of the work at Imperial College.
REFERENCES
- 1.Adam, B. L., T. Y. Jervey, C. P. Kohler, G. L. Wright, Jr., J. A. Nelson, and R. M. Stenberg. 1995. The human cytomegalovirus UL98 gene transcription unit overlaps with the pp28 true late gene (UL99) and encodes a 58-kilodalton early protein. J. Virol. 69:5304-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azad, R. F., V. Brown-Driver, K. Tanaka, R. M. Crooke, and K. P. Anderson. 1993. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob. Agents Chemother. 37:1945-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldick, C. J., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075-22082. [DOI] [PubMed] [Google Scholar]
- 10.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 11.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 12.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caswell, R., C. Hagemeier, C. J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 14.Chau, N. H., C. D. Vanson, and J. A. Kerry. 1999. Transcriptional regulation of the human cytomegalovirus US11 early gene. J. Virol. 73:863-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. I. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-170. [DOI] [PubMed] [Google Scholar]
- 16.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein, B., I. Muller, and J. Collins. 1982. Cloning of the complete human cytomegalovirus genome in cosmids. Gene 18:39-46. [DOI] [PubMed] [Google Scholar]
- 20.Geballe, A. P., F. S. Leach, and E. S. Mocarski. 1986. Regulation of cytomegalovirus late gene expression: γ genes are controlled by posttranscriptional events. J. Virol. 57:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. Han, R. J. Lutz, S. Watanabe, E. D. McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. [DOI] [PubMed] [Google Scholar]
- 23.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenaway, P. J., and G. W. Wilkinson. 1987. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 7:17-31. [DOI] [PubMed] [Google Scholar]
- 25.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, E. S. 1975. Human cytomegalovirus. III. Virus-induced DNA polymerase. J. Virol. 16:298-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunninghake, G. W., B. G. Monks, L. J. Geist, M. M. Monick, M. A. Monroy, M. F. Stinski, A. C. Webb, J. M. Dayer, P. E. Auron, and M. J. Fenton. 1992. The functional importance of a cap site-proximal region of the human prointerleukin 1β gene is defined by viral protein trans-activation. Mol. Cell. Biol. 12:3439-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwayama, S., T. Yamamoto, T. Furuya, R. Kobayashi, K. Ikuta, and K. Hirai. 1994. Intracellular localization and DNA-binding activity of a class of viral early phosphoproteins in human fibroblasts infected with human cytomegalovirus (Towne strain). J. Gen. Virol. 75:3309-3318. [DOI] [PubMed] [Google Scholar]
- 32.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal, and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemble, G. W., A. L. McCormick, L. Pereira, and E. S. Mocarski. 1987. A cytomegalovirus protein with properties of herpes simplex virus ICP8: partial purification of the polypeptide and map position of the gene. J. Virol. 61:3143-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerry, J. A., M. A. Priddy, and R. M. Stenberg. 1994. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J. Virol. 68:4167-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klucher, K. M., D. K. Rabert, and D. H. Spector. 1989. Sequences in the human cytomegalovirus 2.7-kilobase RNA promoter which mediate its regulation as an early gene. J. Virol. 63:5334-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klucher, K. M., M. Sommer, J. T. Kadonaga, and D. H. Spector. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 39.LaFemina, R. L., M. C. Pizzorno, J. D. Mosca, and G. S. Hayward. 1989. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology 172:584-600. [DOI] [PubMed] [Google Scholar]
- 40.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukac, D. M., J. R. Mannupello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonough, S. H., S. I. Staprans, and D. H. Spector. 1985. Analysis of the major transcripts encoded by the long repeat of human cytomegalovirus strain AD169. J. Virol. 53:711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields (ed.), Fields virology, 3rd ed., vol. 2. Lipincott-Raven, Philadelphia, Pa. [Google Scholar]
- 51.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosca, J. D., D. P. Bednarik, N. B. Raj, C. A. Rosen, J. G. Sodroski, W. A. Haseltine, G. S. Hayward, and P. M. Pitha. 1987. Activation of human immunodeficiency virus by herpesvirus infection: identification of a region within the long terminal repeat that responds to a trans-acting factor encoded by herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 84:7408-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Hare, P., and G. S. Hayward. 1984. Expression of recombinant genes containing herpes simplex virus delayed-early and immediate-early regulatory regions and trans activation by herpesvirus infection. J. Virol. 52:522-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pajovic, S., E. L. Wong, A. R. Black, and J. C. Azizkhan. 1997. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol. Cell. Biol. 17:6459-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 58.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 60.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Post, L. E., A. J. Conley, E. S. Mocarski, and B. Roizman. 1980. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc. Natl. Acad. Sci. USA 77:4201-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puchtler, E., and T. Stamminger. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodems, S. M., C. L. Clark, and D. H. Spector. 1998. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection. J. Virol. 72:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romanowski, M. J., E. GarridoGuerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambucetti, L. C., J. M. Cherrington, G. W. G. Wilkinson, and E. S. Mocarski. 1989. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheaffer, A. K., S. P. Weinheimer, and D. J. Tenney. 1997. The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J. Gen. Virol. 78:2953-2961. [DOI] [PubMed] [Google Scholar]
- 72.Staprans, S. I., and D. H. Spector. 1986. 2.2-kilobase class of early transcripts encoded by cell-related sequences in human cytomegalovirus strain AD169. J. Virol. 57:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the α gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stenberg, R. M., D. R. Thomsen, and M. F. Stinski. 1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 49:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wade, M., T. F. Kowalik, M. Mudryj, E. S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walker, S., C. Hagemeier, J. G. Sissons, and J. H. Sinclair. 1992. A 10-base-pair element of the human immunodeficiency virus type 1 long terminal repeat (LTR) is an absolute requirement for transactivation by the human cytomegalovirus 72-kilodalton IE1 protein but can be compensated for by other LTR regions in transactivation by the 80-kilodalton IE2 protein. J. Virol. 66:1543-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 79.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, T. C., W. A. Lee, M. C. Pizzorno, W. C. Au, Y. J. Chan, R. H. Hruban, G. M. Hutchins, and G. S. Hayward. 1992. Localization of the human cytomegalovirus 2.7-kb major early beta-gene transcripts by RNA in situ hybridization in permissive and nonpermissive infections. Am. J. Pathol. 141:1247-1254. [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]