Abstract
After infection of swine with porcine reproductive and respiratory syndrome virus (PRRSV), there is a rapid rise of PRRSV-specific nonneutralizing antibodies (NNA), while neutralizing antibodies (NA) are detectable not sooner than 3 weeks later. To characterize neutralizing epitopes, we selected phages from a 12-mer phage display library using anti-PRRSV neutralizing monoclonal antibody (MAb) ISU25-C1. In addition, phages carrying peptides recognized by swine antibodies with high seroneutralizing titer were isolated after subtracting from the library those clones binding to swine anti-PRRSV serum with no neutralizing activity. Two epitopes located in the ectodomain of PRRSV GP5 were identified. One of these epitopes, which we named epitope B, was recognized both by neutralizing MAb ISU25-C1 and swine neutralizing serum (NS) but not by swine nonneutralizing serum (NNS), indicating that it is a neutralizing epitope. Epitope B is sequential, conserved among isolates, and not immunodominant. Antibodies directed against it are detected in serum late after infection. In contrast, the other epitope, which we named epitope A, is hypervariable and immunodominant. Antibodies against it appear early after infection with PRRSV. This epitope is recognized by swine NNA but is not recognized by either neutralizing MAb ISU25-C1 or swine NA, indicating that it is not involved in PRRSV neutralization. During infection with PRRSV, epitope A may act as a decoy, eliciting most of the antibodies directed to GP5 and delaying the induction of NA against epitope B for at least 3 weeks. These results are relevant to the design of vaccines against PRRSV.
Porcine reproductive and respiratory syndrome (PRRS) is currently accepted as the most important infectious disease of swine (National Pork Producer Council, 1999/2000 Pork Issues Handbook, http://www.porkscience.org/documents/other/positionprrs.pdf), causing late-term reproductive failure and severe pneumonia in neonatal pigs. This disease is caused by PRRS virus (PRRSV) (15), a member of the Arteriviridae family, order Nidoviridales (5, 6, 8). This enveloped virus contains a 14.5-kb positive-strand RNA genome that encodes a replicase polyprotein (open reading frames [ORFs] 1a and 1b) and six structural proteins (ORFs 2 to 7). The products of ORFs 2 to 4 are minor membrane-associated glycoproteins (GP2, GP3, and GP4, respectively). The products of ORFs 5 to 7 are the three major structural proteins (GP5, N, and M proteins, respectively).
Pigs are generally infected with PRRSV following exposure of the mucosal surface of the respiratory tract to the virus. Initial replication in macrophages from lungs and regional lymph nodes is followed by viremia and systemic distribution of virus to other macrophage populations (32, 35). Large amounts of specific antibodies are first detectable in sera at around 9 days postinfection (p.i.) (18), while specific immunoglobulin G (IgG) antibodies peak at 3 or 4 weeks after infection. A hallmark of the swine antibody response against PRRSV is the abundant nonneutralizing antibodies (NNA) detected early in the infection followed by a low neutralizing antibody (NA) titer that appears not sooner than 3 weeks after infection (16, 41).
GP5 possesses a small putative ectodomain comprising approximately the first 40 residues of the mature protein. The ectodomain contains a variable number of N-glycosylation sites (33), and it has been proposed that linear neutralizing epitopes could be located in this region (28). Several murine monoclonal antibodies (MAbs) against GP5 have been elicited (28, 38). GP4 (20) and M protein (40) can also elicit neutralizing MAbs. However, those MAbs recognizing GP5 neutralize PRRSV more effectively than the others (38).
Antibodies from pigs also seem to recognize neutralizing epitopes in GP5, as suggested by the correlation between the titers of NAs and anti-GP5 antibodies in sera from convalescent swine that was established (11). Nevertheless, nonneutralizing epitopes are also present in PRRSV GP5 (31), and, unlike neutralizing epitopes, they are recognized during the early p.i. period (16). Immunodominant epitopes in PRRSV structural and nonstructural proteins have been characterized (20, 25, 26, 31). However, to date, there has been no molecular characterization of PRRSV neutralizing epitopes present in GP5.
In the last few years, experimental data showing the importance of NAs in protection against PRRSV infection has accumulated. For example, a protective role of NAs present in colostrum transferred to piglets was reported (12). Likewise, antibodies passively transferred to pigs (final titer, 8) cleared PRRSV viremia effectively (41). In addition, young pigs immunized with a DNA vaccine comprising the ORF 5 gene developed PRRSV-specific NAs and protective immunity (29). Finally, passively transferred NAs prevented transplacental infection and completely cleared PRRSV infection in pregnant sows (F. A. Osorio, J. A. Galeota, E. Nelson, B. Brodersen, A. Doster, R. Wills, F. Zuckermann, and W. W. Laegreid, submitted for publication). In all, these results clearly demonstrate the importance of NAs for protection against PRRSV.
Current vaccines against PRRSV have several drawbacks. Modified live vaccines protect against challenge with homologous isolates but generally have a limited effect against challenge with heterologous viruses (19, 37). Furthermore, live PRRSV vaccines generally provide at best partial protection against clinical disease but do not prevent infection (27). Additionally, live PRRSV vaccines can revert to virulence (3, 23). Since the attenuated vaccines induce an immune response resembling that induced by PRRSV natural infection, they do not induce high levels of NAs. Nevertheless, in those cases in which vaccines have been effective to some extent, protection could be correlated with an anamnestic NA response after challenge (27, 29). Killed-PRRSV vaccines, on the other hand, have proved to be less effective in prevention of both infection and disease (24). Thus, a better understanding of PRRSV neutralizing epitopes may contribute to the development of a new generation of vaccines that can elicit a protective response against PRRSV.
Herein, we show the presence of two independent epitopes located at the N-terminal region of GP5. One of these epitopes is located in a region recognized both by neutralizing MAb ISU25-C1 (40) and NAs from swine. The other epitope, which is immunodominant, is recognized by swine NNA.
MATERIALS AND METHODS
Viruses, sera, and MAbs. (i) Virus strains and cells.
PRRSV isolates PRRSV IA strain 97-7895 (1) (GenBank accession no. AF325691), PRRSV 16244B (2) (GenBank accession no. AF046869), NVSL (provided by National Veterinary Services Laboratory, U.S. Department of Agriculture, Animal and Plant Inspection Service, Ames, Iowa), American prototype PRRSV strain 2332 (American Type Culture Collection), PRRSV strain KY-35 (41), modified live vaccine virus (MLV) RespPRRS (NOBL Labs, Ames, Iowa), and modified live vaccine virus Prime PacPRRS (Schering Plough Animal Health, Elkorn, Nebr.) were propagated and titrated in MARC-145 cells. These cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. None of the strains were propagated more than twice in MARC-145 cells after last isolation from infected tissues (wild-type strains) or after retrieval from the commercial vaccine vial (MLV strains).
(ii) Preparation of HI sera with high titer of NAs.
Hyperimmune (HI) sera with high titer of NAs against PRRSV were prepared by hyperimmunization of swine. A total of 13 PRRSV-free female swine weighing 300 lb each were initially infected by oronasal inoculation with 106 50% tissue culture infective doses (TCID50) of the PRRSV IA strain 97-7895. At 5 weeks p.i., the animals were superinfected with a mixture of five additional PRRSV strains, which included 16244B, 2332 (American Type Culture Collection), NVSL, RespPRRS, and Prime PacPRRS strains. Each strain was given at a dose of 106 TCID50 through oronasal inoculation. Four weeks later each of the animals received a dose of 105 TCID50 of PRRSV IA strain 97-7895 emulsified in 2 ml of Freund's (complete) adjuvant via the intramuscular route. At intervals of 1.5 months thereafter, the animals received similar doses of PRRSV IA strain 97-7895 emulsified in 2 ml of Freund's (incomplete) adjuvant for a range of three to six applications. The seroneutralizing (SN) end point titer gradually increased in each of the hyperimmunized animals. Within a period of time that ranged from 7 (including three applications of virus plus incomplete Freund's adjuvant) to 14 months (including six applications of virus plus incomplete Freund's adjuvant), all of the 13 animals reached a final end point titer that ranged between 1:32 and 1:128, with most of the animals exhibiting an end point SN titer of ≥1:64. This point was considered to be the end of the hyperimmunization process; the animals were bled out, and all of their sera were individually collected and used to precipitate and concentrate the antibodies by ammonium sulfate treatment.
(iii) Standardization of the antibody stocks.
The total content of swine antibodies was measured by an indirect enzyme-linked immunosorbent assay (ELISA) specific for swine IgG (Bethyl Laboratories Inc., Montgomery, Tex.). The end point of PRRSV-neutralizing activity attained for the resulting master stock solution was 1:256 with strains IA 97-7895, 16244B, and 2332.
Anti-PRRSV nonneutralizing sera (NNS) were obtained at the beginning of the hyperimmunization protocol (day 20 after first infection), when nonneutralizing activity was detected (SN titer, <4) and the sample-to-positive (S/P) ratio in ELISA (IDEXX Laboratories Inc., Westbrook, Maine) was between 0.652 and 1.432 (the cutoff value for the S/P in this ELISA is 0.4). The S/P ratio is the difference in the optical density (OD) of the test serum between viral and control antigens divided by the difference in the OD of the positive reference between viral and control antigens. The concentrated antibodies had a SN titer of 1:256 against all PRRSV isolates used in the hyperimmunization and a titer of 2.232 by ELISA (IDEXX Laboratories Inc.).
Anti-PRRSV neutralizing sera (NS) were obtained at different times throughout the immunization protocol. The S/P ratios and SN titers for each serum are indicated in Tables 3 and 5.
TABLE 3.
Reactivity of MAb ISU25-C1 and swine anti-PRRSV hyperimmune sera to synthetic peptides bound to MBS-coated ELISA plates
| Peptide | Reactivitya with serum or primary antibody (SN titer, ELISA S/P ratio)b:
|
||||||
|---|---|---|---|---|---|---|---|
| 6 (64, 1.053) | 8 (128, 1.712) | 3 (64, 1.835) | 5 (64, 1.677) | 19 (<4, <0.4) | ISU19-A1 (<2, 0.441) | ISU25-C1 (8, 0.190) | |
| S1 (epitopes A and B) VLVNANNSSSSHFQSIYNC | 1.780 (± 0.045) | 0.503 (± 0.011) | 0.626 (± 0.006) | 0.720 (± 0.013) | 0.099 (± 0.008) | 0.020 (± 0.001) | −0.014 (± 0.014) |
| S2 (epitope A removed, epitope B present) SGSGA NNSSSSHFQSIYNC | 0.152 (± 0.033) | 0.189 (± 0.019) | 0.284 (± 0.002) | 0.051 (± 0.001) | 0.036 (± 0.006) | 0.013 (± 0.005) | −0.023 (± 0.000) |
| S3 (epitopes A and B removed) SGSGANNSSSSGS GSIYNC | 0.005 (± 0.021) | 0.092 (± 0.002) | 0.161 (± 0.014) | 0.019 (± 0.014) | 0.020 (± 0.011) | 0.017 (± 0.009) | −0.054 (± 0.399) |
| S4 (mimotope B) SHITSYHPAYFWC | −0.004 (± 0.014) | −0.185 (± 0.001) | −0.161 (± 0.001) | −0.09 (± 0.003) | −0.100 (± 0.001) | 0.037 (± 0.002) | 1.811 (± 0.014) |
| R4N (mimotope A) ALVNIPISNNLAC | 1.784 (± 0.022) | 0.599 (± 0.004) | 1.061 (± 0.003) | 1.205 (± 0.018) | 0.211 (± 0.002) | 0.041 (± 0.006) | −0.04 (± 0.031) |
| R1N (epitope A) VLVNSNNSSSSAC | 1.679 (± 0.085) | 0.653 (± 0.005) | 0.753 (± 0.016) | 0.716 (± 0.011) | 0.177 (± 0.001) | 0.094 (± 0.058) | −0.001 (± 0.006) |
| Δ4R1N (epitope A removed) SGSGSNNSSSSAC | −0.010 (± 0.021) | 0.123 (± 0.016) | 0.174 (± 0.001) | 0.010 (± 0.006) | −0.033 (± 0.018) | −0.007 (± 0.008) | −0.031 (± 0.002) |
Results are expressed as corrected OD at 405 nm (OD for selected phage − OD for unselected phage library) (± standard deviation).
SN titer was determined against the PRRSV Iowa strain. ELISA S/P ratio was determined as described in Materials and Methods. Sera 3, 5, 6, and 8 are from PRRSV-hyperimmunized pigs, serum 19 is a negative control (see Materials and Methods), ISU25-C1 and ISU19-A1 are anti-PRRSV neutralizing and nonneutralizing MAbs, respectively.
PRRSV-negative serum was obtained from another group of pigs of the same genetic background that were hyperimmunized against pseudorabies virus. These animals were negative for anti-PRRSV antibodies as detected by seroneutralization or ELISA (see Table 3).
(iv) MAbs.
ISU25-C1 is an IgG1(λ) anti-PRRSV KY-35 isolate MAb (40) directed against a sequential epitope on GP5, as demonstrated by the recognition of the protein electrophoresed under denaturing conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. It has neutralization activity against several PRRSV isolates (40), including PRRSV IA 97-7895 (this paper).
ISU19-A1 is a IgG1(κ) anti-PRRSV KY-35 isolate nonneutralizing MAb (40) directed against a conformational epitope on protein M of PRRSV. This MAb was used as a negative control in the present experiments.
Affinity purification of anti PRRSV antibodies. (i) Affinity purification of anti-PRRSV swine polyclonal antibodies.
Affinity purification of antibodies was performed in accordance with the Harlow and Lane protocol (13). Briefly, a 1.5-ml volume of NS (S/P ratio by ELISA, 1.053; SN titer, 1:64) was diluted 1:10 in 10 mM Tris, pH 7.5, and passed three times through a MARC-145 cell column in which 30 mg of uninfected MARC-145 cell lysates were coupled to CNBr-activated Sepharose (Sigma, St. Louis, Mo.). This antigenic extract was prepared by three cycles of freezing and thawing two 75-cm2 flasks of confluent uninfected MARC-145 cells. Nonbinding antibodies were recovered and passed three times through a cell-associated virus column in which 30 mg of MARC-145 PRRSV-infected cells (prepared by three cycles of freezing and thawing MARC-145 cells that had been previously infected with PRRSV for 48 h) was coupled to CNBr-Sepharose. After extensive washing with 10 mM Tris, pH 7.5, followed by 10 mM Tris, pH 7.5-0.5 M NaCl, specifically bound antibodies were eluted with 100 mM glycine, pH 2.5, and pH was adjusted with 1 M Tris, pH 8.
A further purification step was performed by passing the antibodies through a protein A/G-Sepharose resin (Sigma). After the purification, the swine antibody solution was highly enriched with anti-PRRSV NAs possessing an SN titer of 1:32. The absence of anti-MARC-145 cell antibodies in NAs was confirmed by immunofluorescence (data not shown).
(ii) Affinity purification of MAb ISU25-C1.
A 2-ml volume of hybridoma culture supernatant was passed three times through a cell-associated virus column. Washing and elution steps were performed as described before. Three fractions, containing a total of 68 μg of MAb ISU25-C1, were dialyzed against phosphate-buffered saline (PBS). The antibodies were then coupled to 50 mg of CNBr-activated Sepharose (Sigma) in accordance with the manufacturer's indications.
Seroneutralization.
The fluorescence focus neutralization assay was performed as described previously (39). Serial dilutions of test sera were incubated for 60 min at 37°C in the presence of 100 focus-forming units of the virus in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. The mixtures were added to 96-well microtitration plates containing confluent MARC-145 cells which had been seeded 72 h earlier. After incubation for 24 h at 37°C in a humidified atmosphere containing 5% CO2, the cells were fixed for 5 min with a solution of 50% methanol and 50% acetone. After extensive washing with PBS, the expression of N protein of PRRSV was detected with MAb SDOW17 (22), followed by incubation with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Sigma). Neutralization titers were expressed as the reciprocal of the highest dilution that inhibited 90% of the foci present in the control wells.
Immunoaffinity selection of phages from a 12-mer peptide library. (i) Biopanning using PRRSV affinity-purified swine antibodies.
Three rounds of biopanning were carried out. Briefly, 10 μl of the phage library (Ph.D.12; New England Biolabs, Watertown, Mass.), was incubated for 30 min at room temperature with 1 μg of anti-PRRSV affinity-purified antibodies with high SN titer. To capture the phage-antibody complexes, 50 μl of bovine serum albumin-blocked protein A/G-Sepharose was incubated for 15 min with the antibody-phage complexes.
The particles of protein A/G-Sepharose were then washed 10 times with Tris-buffered saline (TBS) containing increasing concentrations of Tween 20 in each panning (0.05, 0.1, and 0.5% in the first, second, and third pannings, respectively). Finally, phages were eluted by a 10-min incubation of the Sepharose in 100 mM glycine, pH 2.5. Sepharose particles were centrifuged for 30 s, and the eluate was transferred to another tube. The pH was adjusted with 1 M Tris, pH 8.5.
An aliquot of the eluted phages was titrated in Escherichia coli ER2738, and the rest of the eluate was amplified for 4.5 h in a mid-log-phase E. coli ER2738 culture. Phages were then precipitated with 3.33% polyethylene glycol (PEG) 8000-0.42 M NaCl for 16 h at 4°C and titrated.
The amplified enriched library was subjected to further cycles of selection. After the second panning with swine antibodies with high SN titer was performed, a third cycle of selection was performed with NNS to subtract those clones interacting with NNA. Those phages that did not bind to NNA were selected once more with the antibodies with high SN titer to further enrich for clones expressing peptides resembling neutralizing epitopes.
(ii) Biopanning using MAb ISU25-C1 as the ligand.
Two selections with different stringency conditions were performed with MAb ISU25-C1. In the first (low-stringency) selection, 2 μl of ISU25-C1 ascites fluid was diluted in 200 μl of 10 mM Tris, pH 7.5, and incubated for 30 min with the Ph.D.12 phage library. Then, 50 μl of blocked protein G-agarose was added to the phage-antibody mixture, and the mixture was incubated for 30 min with occasional mixing. The resin was washed with TBS containing 0.05% Tween 20 in the first panning, TBS containing 0.1% Tween 20 in the second panning, and TBS containing 0.5% Tween 20 in the third panning. Phages bound to ISU25-C1 were eluted with 100 mM glycine, pH 2.5.
In the second (high-stringency) selection, 2 μl of ISU25-C1 ascites fluid was diluted in 200 μl of 10 mM Tris, pH 7.5, and the mixture was incubated for 30 min with the Ph.D.12 phage library. Then, 50 μl of protein G-agarose (50% aqueous solution) was added to the phage-antibody mixture, and the mixture was incubated for 30 min with occasional mixing. In the second panning, a CNBr-activated Sepharose-coupled MAb was used. In the third panning, protein G was used again to capture the phage-antibody complexes from solution. The resin was thoroughly washed with 10 mM Tris, pH 7.5, in the first panning, with 10 mM Tris, pH 7.5, containing 0.5 M NaCl in the second panning, and with 10 mM Tris, pH 7.5, containing 0.05% Tween 20 in the third panning.
(iii) Biopanning with affinity-purifed anti-peptide S2 antibodies.
Anti-peptide S2 antibodies were affinity purified from anti-PRRSV pooled precipitated antibodies. Two micrograms of these antibodies was used for each round of selection. Three pannings with protein A/G capture were performed. For the first one, PBS with 0.1% Tween 20 was used for the binding and washing steps. For the second and third pannings, PBS with 0.5% Tween 20 was used.
Phage cloning and sequencing.
Nonamplified eluates were plated on Luria-Bertani (LB) top agar containing 50 μg of IPTG (isopropyl-β-d-thiogalactopyranoside)/ml and 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-μ-d-galactopyranoside), and, after 16 h of incubation, single phage plaques were amplified in 1 ml of mid-log-phase E. coli ER2537 in LB medium for 4.5 h at 37°C.
After one precipitation with PEG 8000 as described above, DNA from the phages was extracted and sequenced with a −96 gIII sequencing primer (CCC TCA TAG TTA GCG TAA CG) (New England Biolabs). Sequencing was performed manually by the dideoxy-mediated chain termination method (34) using T7 Sequenase, version 2.0, DNA polymerase (Amersham Life Sciences, Inc.) and α-35S-dATP. Samples were run in gels containing 50% urea and 6% polyacrylamide. Gels were dried onto a 3MM paper and exposed to film (BioMax MR; Kodak) for 36 h at room temperature for autoradiographic detection of the sequence.
Phage ELISA.
Individual clones were propagated for 4.5 h in E. coli ER2738 and then they were purified by two precipitations with PEG 8000. Phages (1010) were added to each well of an Immulon 2HB (Dynex Technologies Inc., Chantilly, Va.) in 0.1 M sodium bicarbonate, pH 8.6, and incubated for 16 h at room temperature in a humidified chamber. Plates were blocked with PBS containing 10% nonfat dry milk for 3 h at room temperature. Swine sera or MAbs from the tissue culture supernatant were preadsorbed in PBS containing 10% nonfat dry milk added with 100 μg of E. coli proteins/ml. In addition, 1010 PFU of nonselected phages/ml were added to the antibodies to reduce background caused by anti-M-13 activity found in animal sera. The plates were washed six times with PBS-0.05% Tween 20 (PBST), and the peroxidase-conjugated secondary antibody (1:500) was added for 30 min at 37°C. The plates were washed six times, and ABTS (2,2′-azinobis[3-ethylbenzthiazolinesulfonic acid]; KPL, Gaithersburg, Md.) substrate was added for 15 min. The results were read in a spectrophotometer at 405 nm.
The specific reactivity of the serum against the phage-displayed peptide was calculated as the difference between the reactivity against the tested phage clone and the reactivity against phages carrying an irrelevant peptide.
Peptide synthesis.
Peptides were synthesized by using solid-phase technology and 9-fluorenylmethoxy carbonyl chemistry at the Protein Core Facility of the Center for Biotechnology, University of Nebraska. The sequences of the synthetic peptides are shown in Table 1.
TABLE 1.
Sequences of the synthetic peptides useda
| Peptide | Sequence |
|---|---|
| S1 | VLVNANNSSSSHFQSIYNC |
| S2 | SGSGANNSSSSHFQSIYNC |
| S3 | SGSGANNSSSSGSGSIYNC |
| S4 | SHITSYPAYFWC |
| S6 | SGSGANSNSSSHLQLIYNLTLCELC |
| R4N | ALVNIPISNNLAC |
| R1N | VLVNSNNSSSSAC |
| Δ4R1N | SGSGSNNSSSSAC |
| P7 | QRAYLELPPWPPC |
Peptides were produced as described in Materials and Methods. All of them have a carboxy-terminal cysteine, which allows the coupling to MBS-activated proteins or ELISA plates.
Affinity purification of antipeptide antibodies.
Two milligrams of each peptide was coupled to 2 mg of maleimidobenzoyl-N-hydroxysuccimide ester (MBS)-activated keyhole limpet hemocyanin (KLH; Pierce, Rockford, Ill.) in accordance with the directions of the manufacturer. The KLH-peptide complexes were then coupled to 0.3 g of CNBr-activated Sepharose (Amersham Life Sciences, Inc.).
A 2-ml volume of anti-PRRSV-precipitated antibodies was passed through the column, and specifically bound antibodies were eluted with 100 mM glycine, pH 2.5. The antibodies were concentrated with Centricon 30 (Amicon, Beverly, Mass.) to a final concentration of 500 ng/μl. A 100-μl volume of a 20-μg/ml dilution of these antibodies was used in an MBS-activated ELISA plate coated with different synthetic peptides.
Peptide ELISA.
Reacti-Bind maleimide-activated plates (Pierce) were coated with synthetic peptides in accordance with the directions of the manufacturer. Briefly, synthetic peptides were diluted in PBS, pH 6.8, to a final concentration of 50 μg/ml and then were used to coat Reacti-Bind maleimide-activated plates for 5 h at room temperature. Remaining maleimide groups were blocked by a 1-h incubation with cysteine-HCl (10 μg/ml) diluted in PBS, pH 6.8. An extra blocking step was carried out by incubating the plate with PBS containing 10% nonfat dry milk. Sera, affinity-purified antibodies, or MAbs were added in different dilutions and incubated for 1 h at room temperature. The plates were washed six times with PBST, and the peroxidase-conjugated secondary antibody (1:500) was added for 30 min at 37°C. The plates were washed six times with PBST, and ABTS (KPL) substrate was added for 15 min. The results were read in a spectrophotometer at 405 nm.
MAb ISU25-C1 inhibition ELISA.
Several dilutions of peptide S4 and irrelevant peptide P7 were incubated with MAb ISU25-C1 for 1.5 h at room temperature. The antibody-peptide mixture was then added to ELISA strips coated with PRRSV-infected or uninfected MARC-145 cells (IDDEX Laboratories, Inc.) for 45 min. The plates were washed six times with PBST, and the peroxidase-conjugated antimouse antibody (1:500) was added for 30 min at 37°C. The plates were washed six times, and ABTS (KPL) substrate was added for 15 min. The results were read in a spectrophotometer at 405 nm and were expressed as the differences between values for infected and noninfected wells.
Anti-S2 affinity-purified antibody inhibition.
Several dilutions of peptide S2 or S3 or irrelevant peptide P7 were incubated with anti-S2 affinity-purified antibodies for 1.5 h at room temperature. Then, the antibody-peptide mixture was added to Immulon 2HB (Dynex Technologies Inc.) plates coated with 1010 S2-4 phages. The plates were washed six times with PBST, and the peroxidase-conjugated antiswine antibody (1:500) was added for 30 min at 37°C. The plates were washed six times, and ABTS (KPL) substrate was added for 15 min. The results were read in a spectrophotometer at 405 nm.
Sequencing of ORF 5 of PRRSV isolate KY-35.
PRRSV isolate KY-35 was grown in MARC-145 cells. When cytopathic effect reached 60%, the supernatant was discarded and cells were resuspended in TRIzol reagent (Gibco, Life Technologies, Grand Island, N.Y.). RNA was isolated and subjected to reverse transcription. Briefly, 1 μg of total RNA was added to a solution containing 50 mM Tris-HCl, pH 8.3, 50 mM KCl, 50 mM MgCl2, 50 mM dithiothreitol, 50 U of Moloney murine leukemia virus reverse transcriptase (Fermentas, MBI, Buffalo, N.Y.), and 20 pmol of primer (GCG GTC ACT ACT ATA TAC GG). This primer was developed by us, and it anneals with bases 14454 to 14473 of the PRRSV genome. The whole ORF 5 was amplified with primers CCATTCTGTTGGCAATTTGA and GCGGTCACTACTATATACGG; annealing was with nucleotides 13758 to 13777 and 14454 to 14473 of the PRRSV genome, respectively. The amplicon was cycle sequenced in both directions with the same primers used for amplification.
RESULTS
Affinity selection of PRRSV mimotopes.
The epitopes recognized by either swine antibodies with high SN titer or neutralizing MAb ISU25-C1 were selected from a 12-mer phage display library. The number of recovered phages after the third panning was at least 100-fold higher than that after the first panning in all of the selections performed.
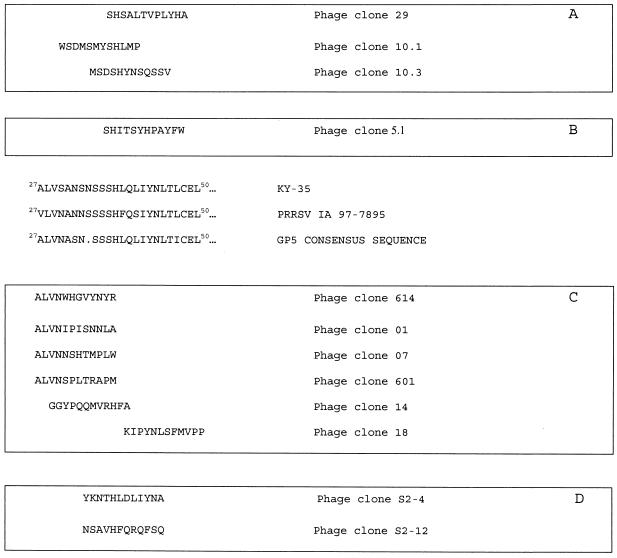
In Fig. 1A are shown the sequences of relevant peptides carried on clones recovered after the third panning of a low-stringency selection performed with MAb ISU25-C1. Three out of 15 clones with an SH motif were found. In another selection with MAb ISU25-C1 under high-stringency conditions, all phages in the third panning carried an identical peptide with a SHI motif (Fig. 1B). The SH(I/L) motif is found in residues 37 to 39 of GP5 of the PRRSV KY-35 isolate, which was the isolate used to immunize the mice for the production of MAb ISU25-C1.
FIG. 1.
Deduced amino acid sequences of relevant peptides carried on phage clones obtained in four different selections. Peptides expressed by phages selected with MAb ISU25-C1 under low- (A) or high-stringency (B) conditions, with swine antibodies possessing high SN titer (C), or with swine anti-peptide S2 affinity-purified antibodies (D) are shown aligned with PRRSV Iowa strain, PRRSV KY-35, and a consensus sequence deduced from the ORF 5 products of several North American strains (34). The sequences are given with the standard single-letter amino acid code.
Anti-PRRSV affinity-purified antibodies with high SN titer were used to select phages from the library, and several clones carrying amino-terminal motif ALVN were recovered (Fig. 1C). This is a consensus motif found in residues 27 to 30 of PRRSV GP5 (31). Another clone selected with the swine antibodies with high SN titer (clone 18) expressed a peptide with a YNL motif, which is also a consensus motif found in amino acids 43 to 45 of PRRSV GP5. Finally, clone 14 carried an HF motif, which is found in residues 38 to 39 of GP5 of a PRRSV IA 97-7895 isolate, which was the primary PRRSV strain used to immunize the pigs.
Anti-peptide S2 affinity-purified antibodies were used to select phages from the library, and 4 out of 12 selected clones corresponded to a single clone (clone S2-4), containing an HLXLIYN motif. This motif is also found between amino acids 38 and 44 of the consensus sequence of PRRSV GP5 (Fig. 1). Another clone selected with these antibodies (clone S2-12) contained an HFQ motif located between residues 38 and 40 of GP5 of the PRRSV Iowa strain.
Two independent epitopes are located in the ectodomain of PRRSV GP5.
Several phage clones were tested in a phage ELISA format. Table 2 shows that the peptide displayed by phage clone 5.1, which carries an SH(I/L) motif corresponding to a putative epitope that we named epitope B, was strongly recognized by MAb ISU25-C1. However, this peptide was not recognized by either swine anti-PRRSV NS or NNS. Conversely, phage clone 01, expressing a peptide with an ALVN amino-terminal motif corresponding to a putative recognition site that we named epitope A, was strongly recognized by anti-PRRSV NS and NNS but was not recognized by MAb ISU25-C1.
TABLE 2.
Reactivity of swine sera and MAbs to selected cloned phages
| Phagotope | Reactivitya with primary antibody:
|
||||
|---|---|---|---|---|---|
| ISU25-C1 | ISU19-A1 | NS 6b | NNS 5c | Swine negative serum | |
| Clone 01 (epitope A) ALVNIPISNNLA | 0.045 (± 0.008) | 0.087 (± 0.006) | 0.373 (± 0.016) | 0.855 (± 0.040) | −0.022 (± 0.002) |
| Clone 5.1 (epitope B) SHITSYHPAYFW | 0.500 (± 0.003) | 0.067 (± 0.005) | 0.027 (± 0.035) | 0.031 (± 0.018) | 0.037 (± 0.031) |
| Clone S2-4 (epitope B) YKNTHLDLIYNA | 0.008 (± 0.001) | 0.039 (± 0.014) | 0.371 (± 0.05) | 0.021 (± 0.020) | 0.045 (± 0.020) |
Results are expressed as corrected OD at 405 nm (OD for selected phage − OD for unselected phage library) (± standard deviation).
NS 6, NS from pig 6 with a neutralizing titer >64 and an ELISA S/P ratio of 1.053.
NNS 5, NNS from pig 5 with a neutralizing titer <4 and an ELISA S/P ratio of 0.652.
Phage clones 10.1, 29, 10.3, and S2-12 were tested in the phage ELISA, and all of them were negative with MAb ISU25-C1 and swine NS and NNS (data not shown).
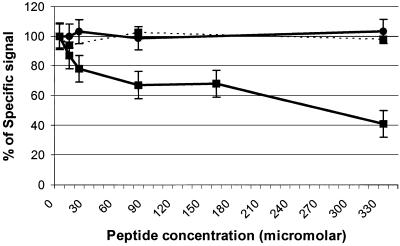
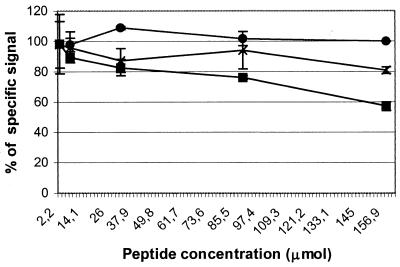
Based on the previous results, several peptides comprising one or both of these motifs in different amino acid contexts were chemically synthesized and tested using either swine anti-PRRSV sera or MAb ISU25-C1 (Table 3). MAb ISU25-C1 recognized peptide S4, which was deduced from the sequence of the peptide carried on phage clone 5.1. The specificity of MAb ISU25-C1 for peptide S4 was confirmed by an experiment involving inhibition of binding in an ELISA. Figure 2 shows that, at a concentration of 333 μM, peptide S4 inhibited 60% of the binding of MAb ISU25-C1 to PRRSV.
FIG. 2.
Inhibition by synthetic peptides of the binding of MAb ISU25-C1 (solid lines) and MAb ISU19-A1 (dotted lines) to PRRSV in ELISA. Serial fivefold dilutions of synthetic peptide S4 were used (▪). Unrelated peptide P7 was used with MAb ISU25-C1 as a negative control (•). Results are percentages of the anti-PRRSV-specific signal obtained with each MAb in wells to which no peptides were added. Values are averages of four readings.
Several different swine NS recognized peptides R4N and R1N. Peptide R4N corresponds to the sequence of the peptide expressed by phage clone 01, and peptide R1N represents amino acids 27 to 38 of the GP5 of the PRRSV IA 97-7895 isolate. Both peptides have an (A/V)LVN motif, which corresponds to amino acids 27 to 30 of GP5 of PRRSV. The signal was considerably reduced when these residues (27 to 30; VLVN in peptide R1N) were changed to irrelevant motif SGSG in peptide Δ4R1N (Table 3).
Peptide S1 represents residues 27 to 43 of GP5 of IA 97-7895 and contains motifs VLVN and SHF. This peptide was strongly recognized by swine NS. Peptide S2, which is identical to peptide S1 except that motif VLVN was replaced with an SGSG motif, was still recognized by swine NS, although with a considerable reduction in the ELISA signal. In addition, the signal was further reduced when motifs VLVN and SHF were replaced by amino acids SGSG and GSG, respectively (peptide S3), indicating that these motifs are key components of their respective epitopes.
Based on the previous results, anti-epitope A and anti-epitope B antibodies were affinity purified from a pool of precipitated antibodies with an SN titer of 512 by using KLH-conjugated peptides R4N and R1N and peptide S2, respectively. The peptide-KLH complexes were coupled to CNBr-activated Sepharose as the ligand, and antibodies eluted from these columns were used to detect synthetic peptides in ELISA (Table 4). Anti-peptide R4N and R1N affinity-purified antibodies recognized peptides R4N, R1N, and S1. All of these peptides contain an (A/V)LVN motif. However, these antibodies did not recognize peptides S2 or Δ4R1N in which N-terminal residues VLVN were changed to SGSG. Conversely, S2 affinity-purified antibodies recognized peptides S1 and S2, containing the SHF motif, but did not recognize peptides R4N, R1N, and S3, which lack this motif. Since peptide S3 only differs from peptide S2 in that in the former the SHF motif was replaced by GSG, these data confirm the importance of at least one of these three amino acids for antibody binding.
TABLE 4.
Reactivity of antipeptide affinity-purified antibodies to synthetic peptides bound to MBS-coated ELISA plates
| Peptide | Reactivitya for:
|
||||
|---|---|---|---|---|---|
| Eluted R4N-R1N column | Eluted S2 column | Eluted S2 + eluted R1N | Eluted S4 column | Eluted P7b column | |
| S1 (epitopes A and B) VLVNANNSSS SHFQSIYNC | 0.164 (± 0.012) | 0.269 (± 0.007) | 0.436 (± 0.006) | 0.007 (± 0.006) | −0.008 (± 0.003) |
| S2 (epitope A removed, epitope B present) SGSGANNSSSSHFQSIYNC | −0.004 (± 0.004) | 0.336 (± 0.005) | 0.351 (± 0.025) | 0.008 (± 0.000) | 0.007 (± 0.024) |
| S3 (epitopes A and B removed) SGSGANNSSSSGSGSIYNC | 0.001 (± 0.002) | 0.003 (± 0.004) | 0.000 (± 0.001) | 0.002 (± 0.001) | −0.009 (± 0.005) |
| S4 (mimotope B) SHITSYHPAYFWC | −0.007 (± 0.001) | 0.010 (± 0.021) | NDc | −0.005 (± 0.005) | −0.010 (± 0.003) |
| R4N (mimotope A) ALVNIPISNNLAC | 0.540 (± 0.027) | 0.002 (± 0.002) | ND | 0.010 (± 0.006) | 0.015 (± 0.007) |
| R1N (epitope A) VLVNSNNSSSSAC | 0.410 (± 0.024) | 0.000 (± 0.005) | ND | 0.009 (± 0.001) | 0.005 (± 0.019) |
| Δ4R1N (epitope A removed) SGSGSN NSSSSAC | 0.005 (± 0.008) | 0.001 (± 0.003) | ND | 0.004 (± 0.001) | 0.004 (± 0.009) |
Results are expressed as corrected OD at 405 nm (OD for tested peptide − OD for irrelevant peptide P7) (± standard deviation).
Irrelevant peptide P7 (QRAYLELPPWPP).
ND, not done.
The signal obtained by ELISA by using peptide S1 as the antigen and a mixture of affinity-purified anti-R4N and -R1N and anti-S2 antibodies as primary antibodies is higher than the signals obtained by using these antibodies separately (Table 4). This indicates that the binding of antibodies to one of the epitopes does not interfere with the binding of antibodies to the other epitope.
Altogether, these results indicate that in swine PRRSV NS there are antibodies directed to two putative epitopes located in the ectodomain of GP5. One of these epitopes (epitope A) comprises the stretch of amino acids including residues (A/V27)L28V29N30 of GP5. The other epitope (epitope B) comprises the stretch of amino acids including residues S37H38(F/L)39. The S37H38(L/I)39 motif recognized by neutralizing MAb ISU25-C1 is located in the epitope B area.
Further characterization of epitope B.
To further characterize epitope B, anti-peptide S2 affinity-purified antibodies were used to screen the phage display library. Four out of 12 selected phage clones carried a peptide with a YKNTHLDLIYNA sequence (Fig. 1D). Motif HXXXIYN, found in this phage, is also found in all of the five PRRSV isolates used to immunize the pigs. Moreover, motif HLQLIYN is present between residues 38 and 44 of the PRRSV consensus sequence encoded by ORF 5 (30). In addition, another phage clone (clone S2-12) selected with these antibodies expressed an HFQ motif, which is also located at amino acids 38 to 40 of the PRRSV Iowa strain. This may indicate that some of the anti-peptide S2 affinity-purified antibodies could also recognize the two amino acids located downstream from H39.
Phage clone S2-4 was assayed in a phage ELISA with MAb ISU25-C1 and swine anti-PRRSV sera (Table 2). While swine NS recognized this clone, swine NNS did not recognize it. In addition, MAb ISU25-C1 did not recognize this phage clone.
The deduced sequences of the peptides expressed by phages selected with MAb ISU25-C1, swine antibodies with high SN titer, and anti-peptide S2 affinity-purified swine antibodies (Fig. 1) suggested that epitope B is composed of two stretches of amino acids, located among positions S37H38(L/F)39 and among positions I42Y43N44L45 of PRRSV GP5. To confirm these results, inhibition experiments were performed with peptides S2 and S3 (Fig. 3). Although both peptides inhibited the binding of anti-peptide S2 affinity-purified antibodies to clone S2-4, peptide S2 inhibited it to a greater extent than peptide S3. This stresses the importance of motif SHF in the composition of epitope B.
FIG. 3.
Inhibition by synthetic peptides of the binding of anti-peptide S2 affinity-purified antibodies to phage clone S2-4. Serial fivefold dilutions of synthetic peptides S2 (▪) and S3 (×) and irrelevant peptide P7 (•) were used. Results are percentages of the signal obtained with anti-peptide S2 affinity-purified antibodies in wells to which no peptides were added. Values are the averages of four readings.
To determine if substitutions in the amino acids surrounding the core of epitope B could affect the recognition of this epitope, the binding of anti-peptide S2 affinity-purified antibodies was assessed with peptides S2 and S6. The latter corresponds to residues 27 to 51 of the GP5 of the PRRSV KY-35 isolate, except that residues 27 to 30 (epitope A) were replaced by irrelevant motif SGSG. Anti-peptide S2 affinity-purified antibodies recognized to a similar extent peptides S2 (OD, 0.336) and S6 (OD, 0.369), confirming that residues H38, Q40, I42, Y43, and N44 constitute the core of epitope B.
Difference in the kinetics of appearance of antibodies induced by epitopes A and B.
A characteristic feature of PRRSV infection is that anti-PRRSV NAs appear late after infection. Moreover, natural infection or vaccination with PRRSV induces low levels of NAs. Thus, to obtain SN titers over 16, it was necessary to subject the pigs to the complex hyperimmunization scheme described in Materials and Methods. In an attempt to determine if there was a correlation between time p.i. and epitope recognition by serum antibodies, sera from several pigs obtained at two different times of the hyperimmunization protocol were evaluated (Table 5).
TABLE 5.
Specific recognition, by two sequential serum samples from six HI pigs, of epitopes A and B
| Epitope | Recognitiona at indicated timeb by sample from pig:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
5
|
8
|
|||||||
| A (1.432, 0) | B (1.755, 32) | A (1.266, 4) | B (1.998, 64) | A (1.078, 0) | B (2.030, 512) | A (1.382, 4) | B (1.993, 128) | A (0.652, 0) | B (1.29, 64) | A (0.946, 0) | B (1.712, 128) | |
| A (clone S2-4) | 0.019 (± 0.020) | 0.072 (± 0.016) | 0.033 (± 0.004) | 0.170 (± 0.011) | 0.014 (± 0.011) | 0.155 (± 0.005) | 0.000 (± 0.027) | 0.493 (± 0.012) | 0.021 (± 0.004) | 0.062 (± 0.001) | 0.044 (± 0.015) | 0.249 (± 0.004) |
| B (clone 01) | 0.738 (± 0.018) | 0.289 (± 0.019) | 0.782 (± 0.039) | 0.237 (± 0.019) | 0.704 (± 0.069) | 0.594 (± 0.037) | 0.689 (± 0.019) | 0.755 (± 0.017) | 0.855 (± 0.04) | 0.431 (± 0.019) | 0.828 (± 0.008) | 0.231 (± 0.005) |
Recognition is expressed as corrected OD at 405 nm (OD for phage clone − OD for unselected phage library) (± standard deviation).
Times A and B correspond to early (20 days p.i.) and advanced (7 months p.i.) stages of the hyperimmunization protocol, respectively. Values in parentheses are ELISA S/P ratios, determined as indicated in Materials and Methods, followed by SN titers.
Sera obtained shortly after the beginning of the immunization, which were positive for PRRSV by ELISA but which had low or undetectable SN titers, reacted very strongly with epitope A (phage clone 01) and very weakly with epitope B (phage clone S2-4). These results confirm that epitope A is immunodominant and not associated with viral neutralization.
Sera from the same animals were tested after repeated immunizations, when SN titers had reached 32 to 128. Table 5 shows that, at this time, the response against epitope B is considerably increased. These results, together with our finding that epitope B is the target for neutralizing MAb ISU25-C1, demonstrate that this epitope is associated with PRRSV neutralization.
DISCUSSION
In this report, we identified and characterized two epitopes located in the ectodomain corresponding to the amino-terminal area of GP5 of PRRSV. The use of swine polyclonal antibodies in conjunction with murine MAbs allowed us to thoroughly characterize this antigenic region of GP5.
Residues 27A/(V)L28V29N30 of PRRSV GP5 are the main antibody recognition site for epitope A. The replacement of these four amino acids with an irrelevant SGSG motif considerably reduced the recognition of this epitope, which is present in several different synthetic peptides (Tables 3 and 4). Epitope A is strongly recognized by swine sera obtained early after infection, when neutralizing activity cannot be detected in these sera (Table 5), indicating that this epitope is not involved in viral neutralization. Nevertheless, epitope A is highly immunodominant all throughout the immune response. This observation could explain why, although a subtraction of nonneutralizing epitopes from the 12-mer phage display library with NNS was performed, several clones displaying peptides resembling epitope A were still selected with NS obtained from animals during late stages of infection (Fig. 1).
Epitope B is located in a region of GP5 that comprises residues 37 to 45. This region is recognized both by neutralizing MAb ISU25-C1 and swine sera with high neutralizing titers but not by swine NNS. A comparison of the sequences of several PRRSV isolates demonstrates that the area where epitope B is located is conserved. Moreover, anti-peptide S2 affinity-purified antibodies recognized to a similar extent the epitope variants present in PRRSV IA 97-7895 (HFQSIYN) and PRRSV KY-35 (HLQIYN), two highly divergent North American isolates. In addition, these antibodies recognized the mimotope carried by phage clone S2-4 (HLDLIYN). These observations could explain the broad neutralizing activity of MAb ISU25-C1 (40). Additionally, these results demonstrate that residues H38 and I43Y44N45 are the main recognition site in epitope B. Nevertheless, it seems that residues 39 to 41 contribute to the binding to antibodies, since the peptide carried on phage clone S2-4 contains leucines 40 and 42, which are also present in PRRSV isolate KY-35. This finding regarding the presence of a sequential neutralizing epitope is in agreement with previous reports indicating the ability of MAb ISU25-C1 to recognize the denatured GP5 (40). Furthermore, the presence of sequential neutralizing epitopes in GP5 of PRRSV has also been described by others (28).
The product of ORF 5 of lactate dehydrogenase-elevating virus (LDV), a murine arterivirus, has more than 60% similarity at the amino acid level with the GP5 of PRRSV (Fig. 4). A neutralizing epitope located between residues 37 and 60 of the LDV ORF 5 product was described (17). The key residues in this epitope were identified. Interestingly, they correspond to the residues identified by us as part of the main recognition site for NAs in epitope B. For example, threonine39 of the ORF 5 product of LDV, which corresponds to histidine38 in PRRSV GP5, seems to be key to recognition by anti-LDV neutralizing MAbs 159-12 and 159-18, as it was the only residue mutated (T→A) in an LDV neutralization escape mutant. Moreover, neutralizing MAb 159-19 (which reacts with the same epitope recognized by Mabs 159-12 and 159-18) recognized peptides with substitutions at I43 or Y44 to a lesser extent than the original peptide, indicating that both residues contribute to this epitope. It seems that residue H38 in PRRSV GP5 is also relevant for the binding of swine antibodies to epitope B, since the substitution of G for H completely eliminated the recognition (Tables 3 and 4 and Fig. 3). The use of anti-peptide S2 affinity-purified antibodies confirmed this assertion, since the use of these antibodies to select epitopes from the phage library resulted in a significant enrichment of phage clone S2-4. This phage clone carries a peptide with an HLDLIYN motif (Fig. 1). Thus, epitope B seems to be the counterpart of the LDV neutralizing epitope. The high degree of conservation among PRRSV isolates in the area where epitope B is located suggests that this region has an important functional or structural role for these viruses. Thus, the number of mutations in this area that do not adversely affect viability may be limited.
FIG. 4.
Comparison of the predicted amino acid sequences of GP5 of PRRSV and LDV. The predicted amino acid sequences of PRRSV isolate IA 79-7895 GP5 (1) and the product of LDV ORF 5 (9) are compared. Hyphens, identical amino acids; dots, gaps; single underlining (PRRSV), key residues in epitope A; double underlining (PRRSV and LDV), epitope B area; arrow, the only residue mutated in GP5 of an LDV neutralization escape mutant (20).
The titer of anti-epitope B antibodies in sera is low at the beginning of the hyperimmunization protocol (Table 5, time point A). At this time, NAs are not present in serum (SN titer, <4; ELISA S/P ratio, >0.652). Later, when the SN titer increases to >32, antibodies against epitope B dramatically rise. A similar scenario in humans infected with human immunodeficiency virus (HIV) was described. MAb I5e, recognizing a broadly neutralizing epitope on HIV gp120, was characterized (14). Sera obtained from patients less than 6 months after infection did not compete with this MAb for binding HIV in ELISA. However, sera obtained from patients six or more months after infection competed with neutralizing MAb I5e for the epitope in HIV. The recognition of this neutralizing epitope was associated with the development of a neutralizing activity in the sera (14). It seems that PRRSV also delays the induction of NAs, at least to epitope B, as described for the epitope of HIV recognized by MAb I5e.
Only a fraction of the repertoire of B-cell epitopes presented by a protein antigen are functionally capable of eliciting antibodies. It has been proposed that the chemical composition of a given epitope could influence at least one of the parameters that defines its immunodominance (21). Thus, it is possible that epitope A has evolved to behave as a decoy epitope, luring the immune response away from more-protective targets present in GP5 such as neutralizing epitope B. Indeed, epitope A exhibits some features expected of a decoy epitope (10), such as its high immunogenicity expressed rapidly after infection and its hypervariability (30). In other viral infections, decoy epitopes have been shown to act by suppressing the recognition of neutralizing epitopes. For example, the dampening of an immunodominant epitope on gp120 of HIV allowed the refocusing of the NA response toward a previously silent, qualitatively different, second-order neutralizing epitope (10). In addition, neutralizing epitope ERDRD on gp41 of HIV type 1 was demonstrated to be immunogenic in rabbits. However, this response is completely eliminated when epitope ERDRD is presented on a peptide juxtaposed to immunodominant epitope IEEE, located two amino acids away in the native sequence of gp41 (7). Thus, decoy epitope IEEE decreases the antibody response to the ERDRD neutralizing epitope. Epitope A of PRRSV, which is located seven amino acids apart from neutralizing epitope B, could be acting similarly to the ERDRD epitope of HIV.
Although MAb ISU25-C1 did not bind to any of the peptides representing fractions of the PRRSV IA 97-7095 ectodomain (Table 3), the consensus motif which it recognizes (SH[L/I]) aligns with that of GP5 of PRRSV Iowa strain in this region. In contrast, swine HI sera did not react with the mimotope recognized by MAb ISU25-C1 but instead reacted with the native sequence deduced from PRRSV GP5. Differences in recognition by mouse and pig antibodies have been reported before (25). It is not surprising since swine antibodies are limited to the usage of the products of only one family of variable heavy genes which are homologous to the 7183 VH family of mouse antibodies (4) whereas MAb ISU25-C1 uses the most distant J558 VH family of antibodies (unpublished results), confirming that epitopes can only be defined in terms of their complementary paratopes (36). Alternatively, the nonrecognition of synthetic peptides by MAb ISU25-C1 could be a consequence of the absence of glycosylation. The epitope B region is highly glycosylated, and MAb ISU25-C1 does not recognize an E. coli-expressed GP5 (R. Bastos, personal communication). Thus, besides the amino acids described here, sugars could be involved in this epitope.
Despite being separated by only seven amino acids, both antigenic regions identified here are independent epitopes. Both epitopes can be recognized simultaneously by different populations of antibodies in serum without any interference between them (Table 4). Moreover, the increase in the titer of antibodies against epitope B correlates with a decrease in the level of antibodies against epitope A (Table 5).
In summary, herein we demonstrate that two epitopes exist in the ectodomain of GP5 of PRRSV. Epitope A is immunodominant and induces a rapid rise of antibodies without neutralizing activity. NAs targeting epitope B appear later in the immune response, when PRRSV is being cleared from the animal. Epitope B is conserved among isolates, making it a suitable target for a more efficient vaccine for PRRSV.
Recent results from different laboratories have stressed the importance of NAs in protection against infection. The findings described herein could help in the understanding of PRRSV pathogenesis and the development of a protective immune response against it.
Acknowledgments
This work was supported by United States Department of Agriculture grant SBIR-USDA 33610-10337 and grant USDA NRICGP 99-35204-8041.
We thank Gautham Sarath and Nancy Caceres from the Protein Core Facility of the Center for Biotechnology of the University of Nebraska.
REFERENCES
- 1.Allende, R., G. F. Kutish, W. Laegreid, Z. Lu, T. L. Lewis, D. L. Rock, J. Friesen, J. A. Galeota, A. R. Doster, and F. A. Osorio. 2000. Mutations in the genome of porcine reproductive and respiratory syndrome virus responsible for the attenuation phenotype. Arch. Virol. 145:1149-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80:307-315. [DOI] [PubMed] [Google Scholar]
- 3.Botner, A., B. Strandbygaard, K. J. Sorensen, P. Have, K. G. Madsen, E. S. Madsen, and S. Alexandersen. 1997. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet. Rec. 141:497-499. [DOI] [PubMed] [Google Scholar]
- 4.Butler, J. E., J. Sun, P. Weber, P. Navarro, and D. Francis. 2000. Antibody repertoire development in fetal and newborn piglets. III. Colonization of the gastrointestinal tract selectively diversifies the preimmune repertoire in mucosal lymphoid tissues. Immunology 100:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 6.Chen, Z. Y., and P. G. W. Plagemann. 1995. Detection of related positive-strand RNA virus genomes by reverse transcription polymerase chain reaction using degenerate primers for common replicase sequences. Virus Res. 39:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleveland, S. M., E. Buratti, T. D. Jones, P. North, F. Baralle, L. McLain, T. McInerney, Z. Durrani, and N. J. Dimmock. 2000. Immunogenic and antigenic dominance of a nonneutralizing epitope over a highly conserved neutralizing epitope in the gp41 envelope glycoprotein of human immunodeficiency virus type 1: its deletion leads to a strong neutralizing response. Virology 266:66-78. [DOI] [PubMed] [Google Scholar]
- 8.Conzelmann, K. K., N. Visser, P. van Woensel, and H. J. Thiel. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:659-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W. P. Tsai, G. Lin, J. J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 11.Gonin, P., B. Pirzadeh, C. A. Gagnon, and S. Dea. 1999. Seroneutralization of porcine reproductive and respiratory syndrome virus correlates with antibody response to the GP5 major envelope glycoprotein. J. Vet. Diagn. Investig. 11:20-26. [DOI] [PubMed] [Google Scholar]
- 12.Gorcyca, D. E. 1996. The effect of maternal immunity on respiratory disease caused by PRRS virus, p. 61. In Proceedings 14th IPVS Congress. Bologna, Italy.
- 13.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keffaber, K. K. 1989. Reproductive failure of unknown etiology. AASP Newsl. 2:1-10. [Google Scholar]
- 16.Labarque, G. G., H. J. Nauwynck, K. Van Reeth, and M. B. Pensaert. 2000. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J. Gen. Virol. 81:1327-1334. [DOI] [PubMed] [Google Scholar]
- 17.Li, K., Z. Chen, and P. Plagemann. 1998. The neutralization epitope of lactate dehydrogenase-elevating virus is located on the short ectodomain of the primary envelope glycoprotein. Virology 242:239-245. [DOI] [PubMed] [Google Scholar]
- 18.Loemba, H. D., S. Mounir, H. Mardassi, D. Archambault, and S. Dea. 1996. Kinetics of humoral immune response to the major structural proteins of the porcine reproductive and respiratory syndrome virus. Arch. Virol. 141:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X. J. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 74:309-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meulenberg, J. J. M., A. P. van Nieuwstadt, A. van Essen-Zandbergen, and J. P. M. Langeveld. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 71:6061-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakra, P., V. Manivel, R. A. Vishwakarma, and K. V. Rao. 2000. B cell responses to a peptide epitope. X. Epitope selection in a primary response is thermodynamically regulated. J. Immunol. 164:5615-5625. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, H. S., M. B. Oleksiewicz, R. Forsberg, T. Stadejek, A. Botner, and T. Storgaard. 2001. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 82:1263-1272. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, T. L., J. Nielsen, P. Have, P. Baekbo, R. Hoff-Jorgensen, and A. Botner. 1997. Examination of virus shedding in semen from vaccinated and from previously infected boars after experimental challenge with porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 54:101-112. [DOI] [PubMed] [Google Scholar]
- 25.Oleksiewicz, M. B., A. Botner, P. Toft, P. Normann, and T. Storgaard. 2001. Epitope mapping of porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 75:3277-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oleksiewicz, M. B., A. Botner, P. Toft, T. Grubbe, J. Nielsen, S. Kamstrup, and T. Storgaard. 2000. Emergence of porcine reproductive and respiratory syndrome virus deletion mutants: correlation with the porcine antibody response to a hypervariable site in the ORF 3 structural glycoprotein. Virology 15:135-140. [DOI] [PubMed] [Google Scholar]
- 27.Osorio, F. A., F. Zuckermannn, R. Wills, W. Meier, S. Christian, J. Galeota, and A. Doster. 1998. PRRSV: comparison of commercial vaccines in their ability to induce protection against current PRRSV strains of high virulence. Allen D. Lennan Swine Conf. 25:179-182. [Google Scholar]
- 28.Pirzadeh, B., and S. Dea. 1997. Monoclonal antibodies to the ORF5 product of porcine reproductive and respiratory syndrome virus define linear neutralizing determinants. J. Gen. Virol. 78:1867-1873. [DOI] [PubMed] [Google Scholar]
- 29.Pirzadeh, B., and S. Dea. 1998. Immune response in pigs vaccinated with plasmid DNA encoding ORF5 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 79:989-999. [DOI] [PubMed] [Google Scholar]
- 30.Pirzadeh, B., C. A. Gagnon, and S. Dea. 1998. Genomic and antigenic variations of porcine reproductive and respiratory syndrome virus major envelope GP5 glycoprotein. Can. J. Vet. Res. 62:170-177. [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez, M. J., J. Sarraseca, J. Fominaya, E. Cortes, A. Sanz, and J. L. Casal. 2001. Identification of an immunodominant epitope in the C terminus of glycoprotein 5 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 82:995-999. [DOI] [PubMed] [Google Scholar]
- 32.Rossow, K. D., D. A. Benfield, S. M. Goyal, E. A. Nelson, J. Christopher-Hennings, and J. E. Collins. 1996. Chronological immunohistochemical detection and localization of porcine reproductive and respiratory syndrome virus in gnotobiotic pigs. Vet. Pathol. 33:551-556. [DOI] [PubMed] [Google Scholar]
- 33.Rowland, R. R., M. Steffen, T. Ackerman, and D. A. Benfield. 1999. The evolution of porcine reproductive and respiratory syndrome virus: quasispecies and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology 259:262-266. [DOI] [PubMed] [Google Scholar]
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sur, J. H., A. R. Doster, J. S. Christian, J. A. Galeota, R. W. Wills, J. J. Zimmerman, and F. A. Osorio. 1997. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J. Virol. 71:9170-9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Regenmortel, M. H. 1998. From absolute to exquisite specificity. Reflections on the fuzzy nature of species, specificity and antigenic sites. J. Immunol. Methods 216:37-48. [DOI] [PubMed] [Google Scholar]
- 37.van Woensel, P. A., K. Liefkens, and S. Demaret. 1998. European serotype PRRSV vaccine protects against European serotype challenge whereas an American serotype vaccine does not. Adv. Exp. Med. Biol. 440:713-718. [DOI] [PubMed] [Google Scholar]
- 38.Weiland, E., M. Wieczorek-Krohmer, D. Kohl, K. K. Conzelmann, and F. Weiland. 1999. Monoclonal antibodies to the GP5 of porcine reproductive and respiratory syndrome virus are more effective in virus neutralization than monoclonal antibodies to the GP4. Vet. Microbiol. 66:171-186. [DOI] [PubMed] [Google Scholar]
- 39.Wu, W.-H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine respiratory and reproductive syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]
- 40.Yang, L., M. L. Frey, K. J. Yoon, J. J. Zimmerman, and K. B. Platt. 2000. Categorization of North American porcine reproductive and respiratory syndrome viruses: epitopic profiles of the N, M, GP5 and GP3 proteins and susceptibility to neutralization. Arch. Virol. 145:1599-1619. [DOI] [PubMed] [Google Scholar]
- 41.Yoon, I. J., H. S. Joo, S. M. Goyal, and T. W. Molitor. 1994. A modified serum neutralization test for the detection of antibody to porcine reproductive and respiratory syndrome virus in swine sera. J. Vet. Diagn. Investig. 6:289-292. [DOI] [PubMed] [Google Scholar]