Abstract
Cells infected with mammalian reoviruses often contain large perinuclear inclusion bodies, or “factories,” where viral replication and assembly are thought to occur. Here, we report a viral strain difference in the morphology of these inclusions: filamentous inclusions formed in cells infected with reovirus type 1 Lang (T1L), whereas globular inclusions formed in cells infected with our laboratory's isolate of reovirus type 3 Dearing (T3D). Examination by immunofluorescence microscopy revealed the filamentous inclusions to be colinear with microtubules (MTs). The filamentous distribution was dependent on an intact MT network, as depolymerization of MTs early after infection caused globular inclusions to form. The inclusion phenotypes of T1L × T3D reassortant viruses identified the viral M1 genome segment as the primary genetic determinant of the strain difference in inclusion morphology. Filamentous inclusions were seen with 21 of 22 other reovirus strains, including an isolate of T3D obtained from another laboratory. When the μ2 proteins derived from T1L and the other laboratory's T3D isolate were expressed after transfection of their cloned M1 genes, they associated with filamentous structures that colocalized with MTs, whereas the μ2 protein derived from our laboratory's T3D isolate did not. MTs were stabilized in cells infected with the viruses that induced filamentous inclusions and after transfection with the M1 genes derived from those viruses. Evidence for MT stabilization included bundling and hyperacetylation of α-tubulin, changes characteristically seen when MT-associated proteins (MAPs) are overexpressed. Sequencing of the M1 segments from the different T1L and T3D isolates revealed that a single-amino-acid difference at position 208 correlated with the inclusion morphology. Two mutant forms of μ2 with the changes Pro-208 to Ser in a background of T1L μ2 and Ser-208 to Pro in a background of T3D μ2 had MT association phenotypes opposite to those of the respective wild-type proteins. We conclude that the μ2 protein of most reovirus strains is a viral MAP and that it plays a key role in the formation and structural organization of reovirus inclusion bodies.
Many animal viruses induce the formation of inclusion bodies in the nuclei and/or cytoplasm of infected cells. These inclusions, which may also be termed viral factories, viroplasms, or viral replication complexes, are generally believed to be the sites of active viral genome replication and particle assembly within infected cells (34). One model for the function of viral inclusion bodies is that they act to concentrate and sequester proteins, nucleic acids, and other small molecules essential for those viral processes. A recent study of cells infected with African swine fever virus (ASFV) showed that its perinuclear inclusion bodies have many of the same properties as cellular aggresomes (28) and may thus arise in part from a programmed cellular response to overexpressed or misfolded proteins (35).
Various cellular elements have been proposed to participate in forming viral inclusion bodies, including membranous organelles and cytoskeletal elements (10, 14, 38). For example, the maturation of ASFV inclusions requires an intact microtubular network, and the inclusions are surrounded by a cage of vimentin (intermediate) filaments, two features they share with cellular aggresomes in addition to their perinuclear localization (28). Members of the Reoviridae family of double-stranded RNA viruses, including orbiviruses, reoviruses, and rotaviruses, form large perinuclear inclusion bodies that are believed to interact with the cytoskeletons of infected cells (13, 18, 46, 52, 55, 56). Early electron and fluorescence microscopy studies by Dales and by Spendlove and Lennette (13, 55) described a filamentous reticulum of inclusion material within reovirus-infected cells. These filamentous inclusions were shown to be associated with microtubules (MTs) and formed an electron-dense coat surrounding the MTs, in which the viral particles were embedded. The material coating the MTs consisted of thin, coiled filaments and granular material. Subsequently, this coat was shown to be at least partly of viral origin (14). Treatment of cells with colchicine early in infection caused the inclusions to lose their filamentous characteristics, consistent with a role for MTs in inclusion formation and morphology (55, 56). An in vitro study of the association between MTs and purified virions found that the capacity of virions to associate with MTs mapped to the viral S1 genome segment, which encodes the structural protein σ1 and the nonstructural protein σ1s (1). A later study found that the viral nonstructural protein μNS could be coprecipitated from infected-cell cytoskeletal extracts by using monoclonal antibodies against tubulin or vimentin (43). Sharpe et al. (52) additionally described a reorganization of the vimentin filament network in reovirus-infected CV-1 cells without a discernible effect on the organization of MTs or microfilaments. From these findings, associations between reovirus inclusions and two different cytoskeletal elements, MTs and intermediate filaments, seem likely.
Despite the preceding observations, many questions remain about the viral and host cell factors required for morphogenesis of the reovirus inclusion bodies. Two more recent studies have added the core protein μ2 and the nonstructural protein σNS to the list of reovirus proteins that may play key roles in inclusion formation (3, 41). The role of μ2 appears especially interesting in that its encoding M1 genome segment was shown to be the primary genetic determinant of a difference in the rate of inclusion formation by either of two reovirus strains: type 1 Lang (T1L) and type 3 Dearing (T3D) (41). In the present study, we used immunofluorescence (IF) microscopy to identify and analyze a previously unreported difference between the inclusion morphologies of these two strains: inclusions induced by reovirus T1L are filamentous, whereas those induced by our laboratory's isolate of reovirus T3D are globular. Our studies revealed that the filamentous distribution involves association of the viral inclusion material with MTs and that the reovirus M1 genome segment, which encodes the μ2 protein, is the primary determinant of this association and the associated difference in inclusion morphology between reovirus strains T1L and T3D. Filamentous inclusions were seen with nearly all (21 of 22) of the other reovirus strains we tested. Other results showed that the μ2 proteins from reoviruses that induce filamentous inclusions bind to and stabilize MTs in the absence of the other viral proteins. We conclude that the μ2 protein of most reovirus strains is a viral MT-associated protein (MAP) and that it plays a key role in the formation and structural organization of reovirus inclusion bodies.
MATERIALS AND METHODS
Cells and viruses.
L cells adapted to spinner culture were maintained in Joklik's modified Eagle's minimal essential medium (Irvine Scientific) containing 2% fetal bovine serum (HyClone), 2% bovine calf serum (HyClone), 2 mM l-glutamine (Irvine Scientific), 100 U of penicillin G (Irvine Scientific) per ml, and 100 μg of streptomycin (Irvine Scientific) per ml. HeLa, CHO, CV-1, and Mv1Lu cells were maintained in Dulbecco's modified Eagle's medium (Gibco BRL) containing 10% fetal bovine serum (HyClone) and 10 μg of gentamicin solution (Gibco BRL) per ml. The reovirus strains T1L and T3D were laboratory stocks. The isolate of T3D in general use in our laboratory is designated T3DN. An independent isolate of T3D obtained from the laboratory of L. W. Cashdollar (Medical College of Wisconsin) is designated T3DC. The differences between T3DN and T3DC may reflect their independent culture in different laboratories, possibly dating back to the preceding generation of investigators (B. N. Fields and W. K. Joklik) from whose work the current clonal isolates were derived. We tested the inclusion phenotypes of 22 other reovirus strains obtained from either the Fields laboratory or the T. S. Dermody laboratory (see the supplemental table at http://micro.med.harvard.edu/nibert/suppl/parker02a/suppl2.html for details). Although originally reported as type 2 (32), the clone 12 isolate in use in our laboratory is type 3 based on its capacity to agglutinate bovine erythrocytes, a type 3-specific property (44) (data not shown). We also used previously described T1L × T3D reassortant viruses (reviewed in reference 44). Third-passage L-cell lysate stocks of twice-plaque-purified reovirus clones were used for cell infections.
Antibodies and reagents.
Monoclonal antibodies to vimentin (clone V9), α-tubulin (B-5-1-2), Cy3-conjugated β-tubulin (TUB 2.1), and acetylated α-tubulin (clone 6-11B-1) were obtained from Sigma. Alexa 594-conjugated phalloidin was obtained from Molecular Probes. Goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG conjugated to Alexa 488 or Alexa 594 were obtained from Molecular Probes. The antibodies against viral antigens have been described previously (7, 23, 59). Antisera against the λ1 and μ2 proteins were generated in rabbits by the polyclonal antibody service in the animal care unit of the University of Wisconsin Medical School (Madison) by immunization with proteins purified from Escherichia coli expression systems (J. Kim and M. L. Nibert, unpublished data). All antibodies were titrated to optimize the signal-to-noise ratios. All enzymes were from New England Biolabs unless otherwise stated. For some experiments, we used Oregon Green or Texas Red conjugates of μNS that were prepared from protein A-purified rabbit anti-μNS IgG conjugated to Oregon Green or Texas Red using kits obtained from Molecular Probes. To detect expressed μ2, we used a rabbit anti-μ2 polyclonal serum. During titration of the anti-μ2 serum, we noted that the serum cross-reacted with a nuclear protein in uninfected cells fixed with paraformaldehyde. However, the cross-reactivity of the anti-μ2 serum with nuclear proteins was minimal when cells were fixed with 100% methanol at −20°C for 3 min. We therefore fixed the infected and M1-transfected cells in 100% methanol for these experiments.
Amplification and DNA sequencing of M1 genome segments.
Purified cores of reoviruses T1L, T3DC, and T3DN were used to generate plus-strand transcripts. Full-length cDNA copies of each M1 genome segment were synthesized using plus-strand transcripts and a primer corresponding to the 18 5′-most nucleotides of the minus strand of the previously published reovirus M1(T3D) sequence (60) with an EcoRI restriction site added to the 5′ end of the primer. PCR amplification was performed using cDNA, the original primer, and a second primer corresponding to the 18 5′-most nucleotides of the plus strand of the published M1(T3D) sequence (60) with an EcoRI restriction site added to the 5′ end of the primer. For each isolate, the PCR products from two reactions were gel purified and combined before DNA was sequenced. cDNA synthesis and PCR amplification were performed as described previously (27). Amplification with these primers fixed only the first 5 nucleotides of the M1 coding region.
DNA sequencing was performed at the Dana-Farber/Harvard Cancer Center High-Throughput DNA Sequencing Facility using cycle-sequencing protocols with fluorescent dideoxy terminators (Applied Biosystems-Perkin-Elmer). The first two sequencing reactions were performed with the primers used for PCR amplification. The primers for subsequent reactions were designed from the published sequences of T1L (62) and T3D (60) to provide a complete sequence for each strand of the M1 PCR products. After the complete sequence of each strand was independently determined, the sequences from the two complementary strands were compared. All discrepancies between the sequences from the two strands were resolved by reevaluating the original sequence data.
Cloning the M1(T1L) gene into pCI-neo.
We originally obtained the M1(T1L) gene as a gift from E. G. Brown (63) and cloned this gene into pBluescript II KS(+) (pBS) (Stratagene). However, we found that this clone contained two nucleotide changes at position 919 (C to A) and position 1151 (T to C) that caused amino acid changes in the encoded protein sequence compared to our determination of the viral M1(T1L) sequence. Therefore, we used restriction enzyme fragments of our viral M1(T1L) PCR products to correct the above-mentioned nucleotide changes and to generate clones pBS-M1(T1L) and pCI-M1(T1L). These clones were subsequently sequenced to ensure that the M1(T1L) gene sequence matched the sequence of our M1(T1L) PCR product.
Cloning T3D M1 genes.
The PCR product generated for M1(T3DC) as described above was cut with EcoRI and ligated to pBS cut with EcoRI. The resulting pBS-M1(T3DC) was sequenced to ensure that the encoded protein sequence of the cloned M1(T3DC) gene matched the sequence of the PCR product. The M1(T3DN) gene was similarly cloned into pBS to generate pBS-M1(T3DN) and was sequenced. The M1 gene was removed from pBS-M1(T3DN) and pBS-M1(T3DC) at the EcoRI sites flanking the gene and ligated to EcoRI-cut pCI-neo to generate pCI-M1(T3DN) and pCI-M1(T3DC).
Generation of M1 mutants at amino acid 208.
To change the amino acids at position 208 of different M1 genes, pBS-M1(T3DN), pBS-M1(T3DC), and pBS-M1(T1L) were cut with BsrGI and MfeI to remove nucleotides 475 to 639. The small fragment (475 to 639) from pBS-M1(T3DN) was placed into the pBS-M1(T1L) background to generate pBS-M1(T1L)-P208S. The small fragment (475 to 639) from pBS-M1(T3DC) was placed into the pBS-M1(T3DN) background to generate pBS-M1(T3DN)-S208P. All clones were sequenced from nucleotides 300 to 1300 and confirmed to contain no additional changes. The mutated M1 genes were subcloned into pCI-neo as described above for the wild-type genes to generate pCI-M1(T1L)-P208S and pCI-M1(T3DN)-S208P.
Transfections and infections.
Cells were seeded the day before transfection or infection at a density of 1.5 × 104 per cm2 in six-well plates (9.6 cm2 per well) containing round glass coverslips (18-mm diameter). The cells were transfected with 2 μg of DNA using 6 μl of Lipofectamine (Life Technologies, Inc.) as suggested by the manufacturer. For some experiments, acid-washed coverslips were pretreated with poly-l-lysine to improve cell adherence. The cells on coverslips were inoculated with T1L or T3D virus at a multiplicity of infection (MOI) of 5 in phosphate-buffered saline (PBS) containing 2 mM MgCl2, and the virus was adsorbed for 1 h at room temperature before fresh medium was added. The cells were further incubated for 12 to 48 h at 37°C before being processed for IF microscopy.
IF and IF microscopy.
Cells to be processed for IF microscopy were fixed for 10 min at room temperature in 2% paraformaldehyde or for 3 min at −20°C in 100% methanol unless otherwise stated. In some cases, the cytoplasm was extracted for 20 s with 0.1% Triton X-100 in 80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.8)-5 mM MgCl2-1 mM EGTA prior to fixation with methanol. Cells fixed in paraformaldehyde were washed with PBS and permeabilized and blocked in PBS containing 1% bovine serum albumin and 0.1% Triton X-100 (PBSA-0.1%Tx100). After methanol fixation, the cells were incubated in PBSA-0.1%Tx100 three times for 5 min at room temperature (RT), prior to incubation in primary antibody. Primary antibodies were diluted in PBSA-0.1%Tx100 and incubated with the cells for 25 to 40 min at RT. After three washes in PBS, secondary antibodies diluted in PBSA-0.1%Tx100 were added and incubated with the cells for 25 min at RT. Coverslips were incubated with 300 nM DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes) in PBS for 5 min to counterstain the cell nuclei, briefly washed in PBS, and then mounted on glass slides with Prolong (Molecular Probes). Samples were examined with either a 63×, 1.4 NA or a 100×, 1.3 NA objective using a Nikon TE-300 inverted microscope equipped with phase and fluorescence optics. Images were collected digitally using a progressive-scan, interline-cooled, charge-coupled device camera (Hamamatsu Corp.) and analyzed with Metamorph 5.1 (Universal Imaging Corp.). Confocal images were collected using a 63×, 1.4 NA oil immersion objective attached to a Zeiss LSM 410 confocal microscope. Sections (0.2 μM thick) were collected in each channel, and appropriate controls for fluorescence bleed-through were used. All images were processed and prepared for presentation using Photoshop 5.5 (Adobe Systems, Inc.).
Nocodazole treatment.
In preliminary experiments, we found that treatment of CV-1 cells with concentrations of nocodazole as low as 500 nM for 1 h completely depolymerized MTs detected by α-tubulin staining. We consequently used a concentration of 10 μM nocodazole for all subsequent experiments.
Immunoblot analysis of acetylated α-tubulin.
To quantify the amounts of acetylated α-tubulin in infected and transfected cells, we collected cell lysates 18 to 24 h after infection or transfection. The cells were washed briefly in MT stabilization buffer (MTSB) (80 mM PIPES [pH 6.8], 1 mM MgCl2, 1 mM EGTA) and then scraped into 1 ml of MTSB and pelleted. The pelleted cells were resuspended in a small volume of MTSB and then lysed in sample buffer, boiled, and subjected to electrophoresis on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels. Proteins in the gels were electroblotted to nitrocellulose in 25 mM Tris-192 mM glycine, pH 8.3. Binding of monoclonal antibodies to acetylated α-tubulin or to α-tubulin was detected with alkaline phosphatase-coupled goat anti-mouse IgG (Bio-Rad) and the colorimetric reagents p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (Bio-Rad). Immunoblots were quantified by densitometry.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new sequences in this study are AF461682, AF461683, and AF461684.
RESULTS
Viral inclusions formed in reovirus T1L- versus T3DN-infected cells have distinct morphologies.
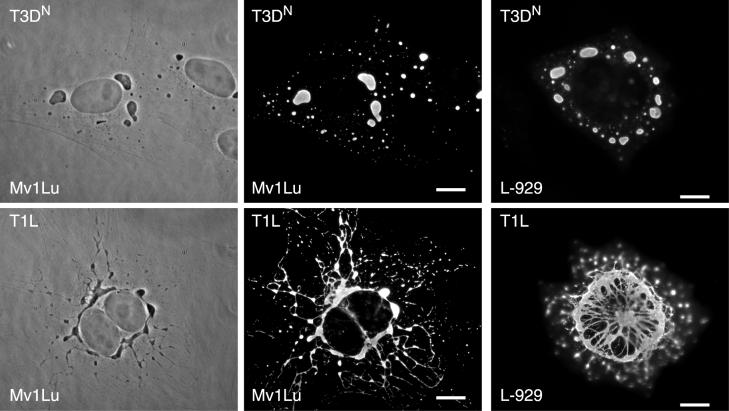
While examining the subcellular distribution of the reovirus nonstructural protein μNS in infected cells by IF microscopy, we noted a striking difference between the morphologies of inclusions induced by reoviruses T1L and T3DN. The μNS-containing inclusions in T1L-infected cells at 18 h postinoculation (p.i.) formed an irregular meshwork that was concentrated around the nucleus, often in a ringlike structure, with radiating filaments of inclusion material connecting more peripheral nodes. In contrast, the μNS-containing inclusions in T3DN-infected cells lacked filamentous extensions and were discrete and globular in shape (Fig. 1). The strain difference in inclusion morphology was detected in all of the cell lines we tested (CHO, CV-1, HeLa, L929, and Mv1Lu) (Fig. 1 and supplemental data at http://micro.med.harvard.edu/nibert/suppl/parker02a/suppl1.html), although we saw minor differences in the appearance of the T1L-induced filamentous inclusions in some lines (Fig. 1, compare L929 to Mv1Lu). The distinct inclusion morphologies of T1L and T3DN were also apparent in phase-contrast images of infected cells (Fig. 1). Larger inclusions were found closer to the nucleus, and smaller inclusions were found mainly in the periphery of cells infected with either strain. The difference between T1L and T3DN inclusions became apparent by 12 h p.i. and could be easily distinguished at 18 h p.i. At 48 h p.i., the difference was still clearly discernible (data not shown). In addition, the MOI did not affect the inclusion phenotype, as the strain-specific morphology of inclusions was clear at multiplicities of 0.5, 5, and 50 PFU per cell (data not shown).
FIG. 1.
Morphologies of viral inclusions formed in reovirus T1L- or T3DN-infected cells. Mv1Lu cells (left two columns) or L-929 cells (right column) were infected with virus strain T3DN or T1L at an MOI of 5 and then fixed at 18 h p.i. After being processed, the cells were visualized by phase-contrast microscopy (left column) or IF microscopy (right two columns). The inclusions were localized by immunostaining them with rabbit anti-μNS serum followed by Alexa 488-conjugated goat anti-rabbit IgG. Scale bars, 10 μm.
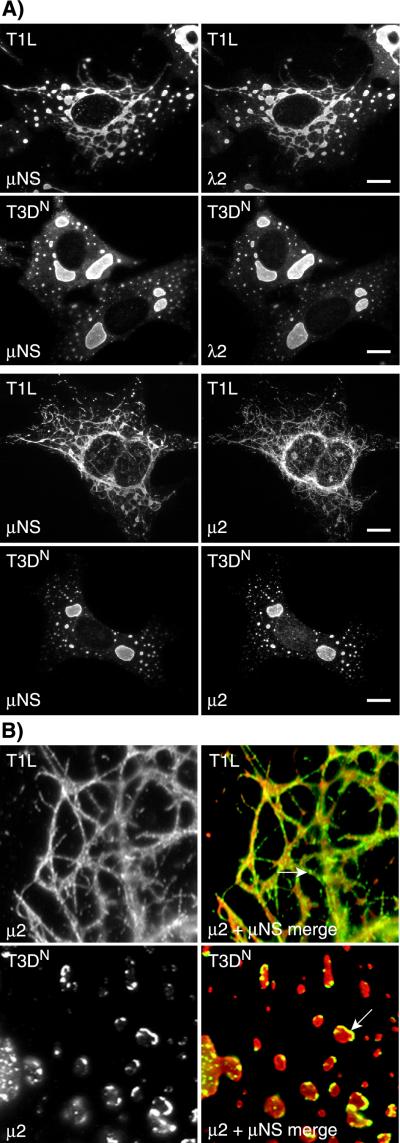
As we had initially detected the difference between T1L and T3DN inclusions using an anti-μNS serum, we investigated whether their different inclusion phenotypes could be detected with antibodies against other viral proteins as well. We dually labeled T1L- and T3DN-infected cells with the anti-μNS serum and antisera or antibodies against the reovirus protein σNS, μ2, λ1, λ2, or λ3 and examined them by IF microscopy. All of these proteins clearly colocalized with μNS in both T1L- and T3DN-induced inclusions (Fig. 2A and data not shown) and thus reflected the same difference between T1L and T3DN inclusion morphologies first detected by staining for μNS.
FIG.2.
Morphologies of viral inclusions in T1L- and T3DN-infected cells detected by immunostaining for other viral proteins. T1L- or T3DN-infected CV-1 cells were fixed and immunostained at 18 h p.i. (A) The subcellular localizations of μNS (left column) and λ2 (upper right column) were detected by immunostaining with rabbit anti-μNS serum and a mouse monoclonal antibody to λ2 (57), followed by Alexa 594-conjugated goat anti-rabbit IgG and Alexa 488-conjugated goat anti-mouse IgG. The subcellular localizations of μNS (left column) and μ2 (lower right column) were detected by immunostaining first with rabbit anti-μ2 serum followed by Alexa 488-conjugated goat anti-rabbit IgG (green) and then with rabbit IgG that was purified from the anti-μNS serum and then conjugated with Texas Red (red). Scale bars, 10 μM. (B) Higher magnification of the codistribution of μNS and μ2 in T1L- and T3DN-infected cells. Merged images are shown in the right column: μ2 (green) and μNS (red). Arrows indicate localized accumulations of μ2.
To visualize more clearly the relative distributions of μ2 and μNS within the viral inclusions, infected cells were extracted prior to fixation (see Materials and Methods). Following this procedure, the μ2 protein was seen concentrated in granules on the margins of the inclusions (Fig. 2B). These μ2-containing granules were present in the inclusions induced by both T1L and T3DN but were more obvious with the latter. The presence of μ2 granules on the margins of the inclusions suggests that the inclusions contain significant substructure, which may reflect the fact that different steps in viral replication and assembly take place in different regions of the inclusions (3).
The M1 genome segment determines the phenotypic difference in inclusion morphology.
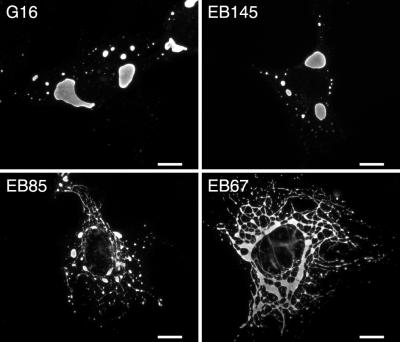
To determine the genetic basis for the difference in inclusion morphology between reoviruses T1L and T3DN, we examined the inclusions induced by viruses from an available panel of T1L × T3D reassortants (9, 16, 44). The μNS-containing inclusions were classified as either filamentous or globular after examination by IF microscopy at 18 h p.i. in Mv1Lu cells. We were easily able to distinguish globular inclusions based on the absence of filamentous extensions connecting more localized accumulations of inclusion material. We found that the morphologies of inclusions formed by the reassortants segregated into two groups that closely resembled the filamentous and globular phenotypes of the two parents (representative examples are shown in Fig. 3). Analysis of the reassortant genomes revealed that the parental origin of the M1 genome segment segregated with the inclusion phenotype (Table 1): all reassortants with a T1L M1 formed filamentous inclusions, and all reassortants with a T3D M1 formed globular inclusions. M1 encodes protein μ2, which is a structurally minor component of the reovirus core and was localized to the inclusions as discussed above.
FIG. 3.
Morphologies of viral inclusions formed in Mv1Lu cells infected with T1L × T3D reassortant viruses. Mv1Lu cells were inoculated with the reassortant viruses G16, EB145, EB85, and EB67 and then fixed at 18 h p.i. The inclusions were localized by immunostaining for viral protein μNS, as described for Fig. 1. The upper images show reassortants with globular inclusions; the lower images show reassortants with filamentous inclusions. Scale bars, 10 μm.
TABLE 1.
Association of inclusion phenotype with reassortant genotype
| Reassortant | Parental origina
|
Inclusion phenotype | Acetylated/total α-tubulin ratiob | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L3 | M1 | M2 | M3 | S1 | S2 | S3 | S4 | |||
| EB39 | L | D | D | L | D | D | D | D | D | D | Filamentous | 32.7 |
| T1L | L | L | L | L | L | L | L | L | L | L | Filamentous | 10.1 |
| H5 | D | D | L | L | L | D | L | D | L | D | Filamentous | 9.1 |
| H41 | D | D | L | L | L | D | L | L | D | L | Filamentous | 5.1 |
| EB118 | D | D | L | L | D | D | D | D | L | L | Filamentous | 3.3 |
| EB67 | L | D | L | L | L | D | L | L | L | L | Filamentous | 3.1 |
| H14 | L | L | D | L | L | L | L | D | D | L | Filamentous | 2.1 |
| EB85 | L | L | L | L | L | D | L | D | L | L | Filamentous | 1.3 |
| EB136 | D | D | D | L | D | L | D | D | D | D | Filamentous | NDc |
| H60 | D | D | L | L | D | D | D | D | D | L | Filamentous | ND |
| EB124 | D | D | D | D | L | D | D | L | L | L | Globular | ND |
| EB146 | L | L | L | D | L | L | L | L | L | D | Globular | 1.1 |
| EB145 | D | D | D | D | D | L | L | D | D | D | Globular | 1.1 |
| EB31 | L | L | L | D | L | L | L | D | D | L | Globular | 1 |
| T3D | D | D | D | D | D | D | D | D | D | D | Globular | 0.3 |
| G16 | L | L | L | D | L | L | L | D | L | L | Globular | 0.2 |
| EB113 | L | L | L | D | L | L | L | L | D | L | Globular | 0.1 |
L, T1L; D, T3D. The origins in boldface correlate with the inclusion phenotype.
The ratio of acetylated α-tubulin to total α-tubulin was quantified from Western blots of lysates from infected cells at 18 h p.i.
ND, not done.
The globular inclusion phenotype is uncommon among reovirus strains.
Given the dramatic difference in the appearance of the viral inclusions induced by reoviruses T1L and T3DN, we examined the prevalence of the filamentous versus globular inclusion phenotypes among other reovirus strains. Of 22 additional strains (supplemental data are at http://micro.med.harvard.edu/nibert/suppl/parker02a/suppl2.html), only one other, T3/Human/Wash., D.C./Clone 12/1957 (32) (see Materials and Methods), induced globular inclusions. All 21 other strains, representing all three established serotypes of the nonfusogenic mammalian reoviruses, showed a filamentous phenotype. Interestingly, we found that a different laboratory isolate of T3D obtained from L.W. Cashdollar (T3DC) showed a filamentous inclusion phenotype similar to those of most other strains and thus dramatically different from the globular phenotype of our own laboratory's T3D isolate (T3DN).
Sequence comparisons of M1 genome segments.
To identify the sequences within the M1 genome segment that are responsible for the difference in inclusion morphology, we sequenced cDNA copies of the M1 plus-strand transcripts generated by cores of reoviruses T1L, T3DN, and T3DC. The deduced amino acid differences between these sequences and the previously published M1 sequences are shown in Table 2. We found two nucleotide differences between our M1(T1L) sequence and the published M1(T1L) sequence (62). One of these nucleotide changes causes a Leu-to-Phe change in the deduced amino acid sequence of μ2 at position 302. The published μ2(T3D) sequence also has a Phe at position 302; thus, this change reduces the number of amino acid differences between μ2(T1L) and μ2(T3D) to 9 from the previously reported 10 (60).
TABLE 2.
Amino acid differences in the T1L and T3D μ2 proteins
| Reovirus strain | Amino acid in μ2 at position:
|
Inclusion phenotypeb | μ2 distributionc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93 | 150 | 208a | 300 | 302 | 347 | 372 | 434 | 458 | 652 | 726 | |||
| T1Ld | V | Q | P | V | L | F | I | V | H | M | N | ND | Filamentous |
| T1Le | V | Q | P | V | F | F | I | V | H | M | N | Filamentous | Filamentous |
| T3DC f | A | R | P | M | F | L | M | I | Q | I | S | Filamentous | Filamentous |
| T3DN | A | Q | S | M | F | L | I | I | Q | I | S | Globular | Diffuse |
Amino acids in boldface correlate with differences in inclusion phenotype.
Morphology of inclusions during reovirus infection as determined by IF with anti-μNS serum. ND, not determined.
Distribution of μ2 in cells transfected with expression vector containing the M1 gene as determined by IF.
Previously published sequence of T1L from another laboratory (62).
Clonal isolate of T1L from our laboratory.
Sequence identical to previously published M1(T3D) (60).
The sequence determined for M1(T3DC) was identical to the published M1(T3D) sequence (60). In contrast, the sequence determined for M1(T3DN) had six nucleotide differences from that of M1(T3DC). Three of these nucleotide differences cause changes in the deduced amino acid sequence of μ2 at positions 150, 208, and 372 (Table 2). The deduced amino acid residues of μ2(T3DN) at positions 150 and 372 are identical to those of μ2(T1L). However, μ2(T3DN) differs at position 208 from both μ2(T1L) and μ2(T3DC). Analysis of these differences in the amino acid sequences of the μ2(T1L), μ2(T3DN), and μ2(T3DC) proteins revealed that T1L and T3DC, viruses that induced filamentous inclusions, had a Pro at position 208 whereas T3DN, a virus that induced globular inclusions, had a Ser at position 208. None of the other deduced amino acid differences between μ2 proteins derived from T1L, T3DN, and T3DC correlated with the different inclusion phenotypes (Table 2). A single-amino-acid difference in μ2 may thus be capable of determining the difference in inclusion morphology (see below for further analysis).
The filamentous extensions of inclusions formed by reoviruses T1L and T3DC are colinear with MTs.
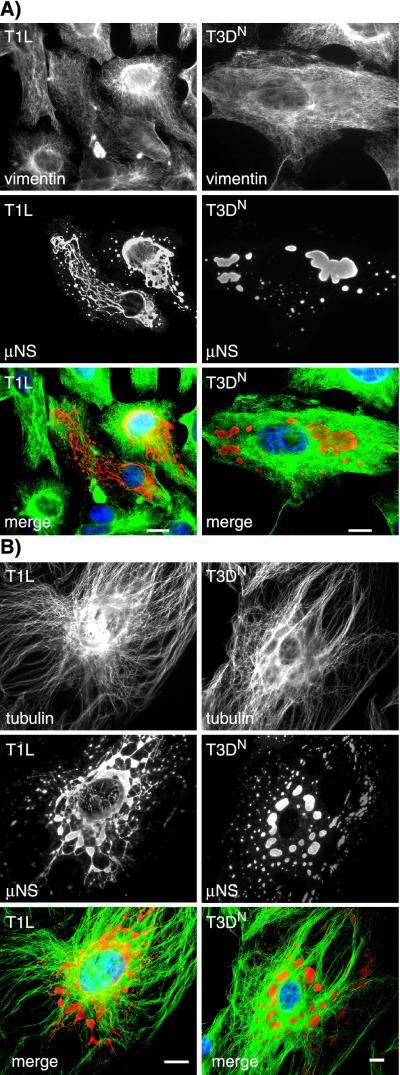
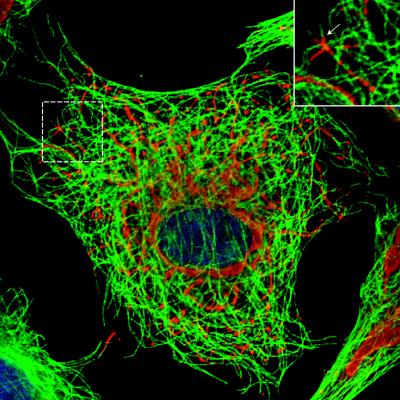
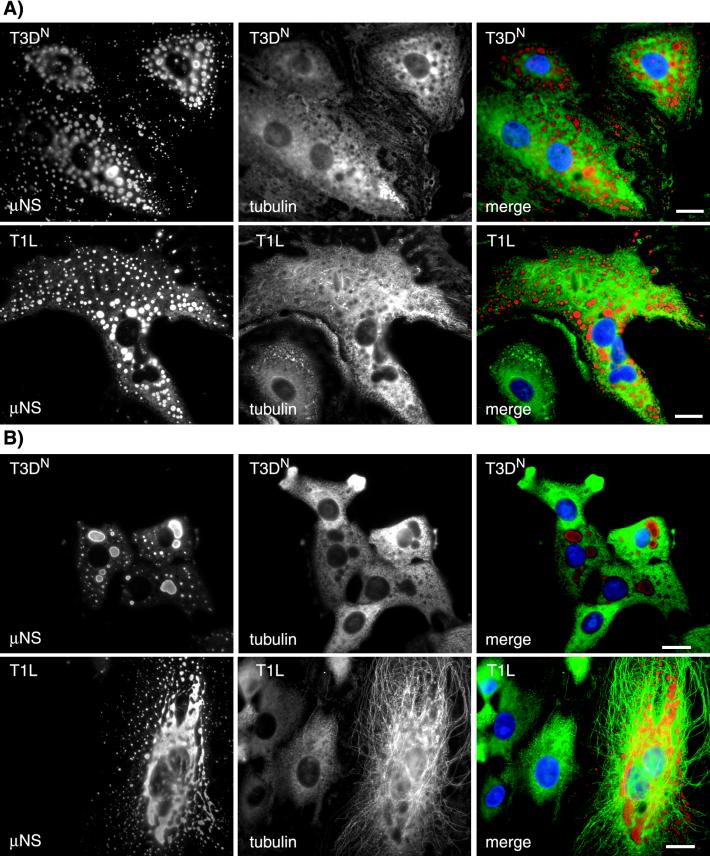
Given the filamentous nature of the inclusions formed by reoviruses T1L and T3DC, we explored a possible association with cytoskeletal elements. In initial experiments, we used IF microscopy to examine the distributions of μNS together with vimentin, actin, and α-tubulin in reovirus-infected CV-1 and Mv1Lu cells. A previous study by Sharpe et al. (52) described an association between reovirus inclusions and the intermediate filament protein vimentin, accompanied by significant changes in the vimentin filament network in CV-1 cells at 48 h p.i. At the earlier time point of 18 h p.i. in our experiments, however, we did not detect any significant colocalization between μNS-containing inclusions and vimentin in either T1L- or T3DN-infected CV-1 or Mv1Lu cells (Fig. 4A and data not shown). In addition, at 18 h p.i., the vimentin filament network was only mildly displaced by the inclusions of either strain in CV-1 or Mv1Lu cells (Fig. 4A and data not shown). In agreement with the results of Sharpe et al. (52), we found no association between the viral inclusions and the actin cytoskeleton (data not shown). There was no obvious disruption of the MTs in T1L-, or T3DN-infected CV-1 and Mv1Lu cells compared to noninfected cells. However, we found that the filamentous inclusions formed in T1L- and T3DC-infected cells were colinear with MTs in images collected by IF microscopy (Fig. 4B and data not shown). We used confocal microscopy to confirm the colinearity of MTs and the μNS-containing inclusions in T1L-infected cells (Fig. 5). Although extensive colocalization between α-tubulin and μNS was not apparent, we noted gaps in the α-tubulin-containing MT filaments that were bridged by μNS-containing inclusion material (Fig. 5). We interpret these findings to indicate that the μNS-containing inclusion material was coating the MTs and thus occluding the underlying α-tubulin in those sections of colinear filaments in which tubulin staining was not observed. The inclusions formed in T3DN-infected cells, although not colinear with the MTs, appeared to be distributed along them (Fig. 4B).
FIG.4.
Relationships between viral inclusions and the intermedi-ate filament and MT cytoskeleton. CV-1 cells were infected with T1L or T3DN at an MOI of 5 and then fixed in 100% methanol at 18 h p.i. The cells were immunostained with rabbit anti-μNS serum and then with Alexa 594-conjugated goat anti-rabbit IgG (red) together with mouse monoclonal antibodies to either vimentin (A) or α-tubulin (B), followed by Alexa 488-conjugated goat anti-mouse IgG (green). The nuclei were counterstained with DAPI (blue). Scale bars, 10 μm.
FIG. 5.
Colinearity between the inclusions formed in T1L-infected cells and MT filaments. Mv1Lu cells were infected with T1L at an MOI of 5 and then fixed in 100% methanol at 18 h p.i. The cells were immunostained for μNS (red) and α-tubulin (green) as described for Fig. 4. The nuclei were counterstained with DAPI (blue). Confocal images were collected, and the image shown is a pseudocolored, merged image of three independently collected channels. The inset shows the colinearity between tubulin and μNS at higher magnification.
Effects of nocodazole on reovirus inclusions.
To investigate the apparent relationship between reovirus inclusions and MTs, we examined the effect of treating reovirus-infected cells with the MT-depolymerizing drug nocodazole. In T1L-infected cells treated with nocodazole for 12 h beginning at 6 h p.i., the μNS-containing inclusions at 18 h p.i. were no longer filamentous but were instead round, discrete, smaller, and more numerous than those seen in untreated T1L-infected cells (Fig. 6A; compare with Fig. 4B). In addition, we noted that these small round inclusions were distributed throughout the cytosol rather than concentrated in the perinuclear region. When T3DN-infected cells were similarly treated with nocodazole for 12 h, the μNS-containing inclusions at 18 h p.i. remained globular but were smaller and more numerous than those in untreated cells and were also dispersed throughout the cytosol (Fig. 6A; compare with Fig. 4B). These findings suggest that intact MTs are required for the formation of filamentous inclusions in T1L-infected cells as well as for the formation of larger perinuclear inclusions in T3DN-infected cells.
FIG. 6.
Effects of the MT-depolymerizing drug nocodazole on the morphologies and distributions of viral inclusions in T3DN- and T1L-infected CV-1 cells. Nocodazole (10 μM) was added to cells infected with T3DN or T1L at 6 (A) or 17 (B) h p.i. The drug was left on the cells until they were fixed in 100% methanol at 18 h p.i. The cells were immunostained for μNS (red) and α-tubulin (green) as described for Fig. 4. The nuclei were counterstained with DAPI (blue). Scale bars, 10 μm.
The filamentous nature of the μNS-containing inclusions in T1L-infected CV-1 cells was minimally affected when the cells were treated with nocodazole for 1 h beginning at 17 h p.i. and then processed for microscopy (Fig. 6B). Similarly, the globular appearance and perinuclear distribution of μNS-containing inclusions in T3DN-infected CV-1 cells was unaffected by treatment with nocodazole for 1 h at 17 h p.i. (Fig. 6B). We noted, however, that unlike MTs in uninfected or T3DN-infected cells, which were completely depolymerized by nocodazole treatment for 1 h at 17 h p.i., the MTs in T1L-infected cells were largely resistant to depolymerization (Fig. 6B and data not shown). We conclude that MTs are stabilized in T1L-infected cells but not in T3DN-infected cells.
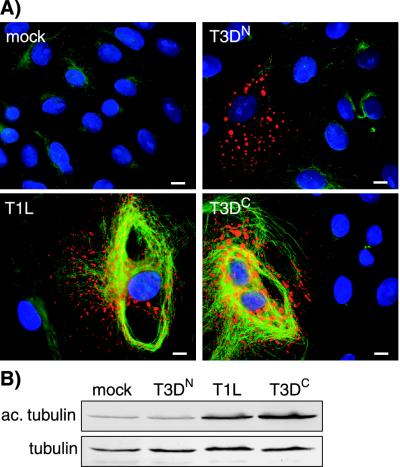
Infection with reoviruses T1L and T3DC, but not T3DN, induces hyperacetylation of MTs.
One commonly used marker for MT stabilization is acetylation of α-tubulin (49). We examined the distribution of acetylated α-tubulin in reovirus-infected CV-1 cells by IF microscopy using monoclonal antibody 6-11B-1 (48). The amounts of acetylated α-tubulin were increased in T1L- and T3DC-infected CV-1 cells compared to that in mock- or T3DN-infected cells (Fig. 7A). The amount of acetylated α-tubulin in T3DN-infected cells appeared comparable to that in mock-infected cells. We found similar results in Mv1Lu and CHO cells (data not shown). To quantify the difference in the levels of acetylated α-tubulin in T1L-, T3DC-, T3DN-, and mock-infected cells, we collected lysates from infected Mv1Lu cells at 18 h p.i. and assessed the amount of acetylated α-tubulin relative to total α-tubulin by immunoblotting. Lysates from T1L- and T3DC-infected cells contained three- and fourfold more acetylated α-tubulin, respectively, than mock- or T3DN-infected cells (Fig. 7B). Similar amounts of total tubulin were present in all lysates (Fig. 7B). We conclude that cells infected with reovirus T1L or T3DC have hyperacetylated MTs, indicating that their MTs are stabilized. In contrast, the MTs in T3DN-infected cells have levels of acetylated α-tubulin comparable to that in mock-infected cells, indicating that their MTs are not significantly stabilized.
FIG. 7.
Hyperacetylation of MTs in T1L- and T3DC-infected, but not T3DN-infected, CV-1 cells. (A) CV-1 cells were infected with T1L, T3DC, or T3DN at an MOI of 5 and then fixed 18 h p.i. in 100% methanol. The cells were immunostained with rabbit anti-μNS serum and then with Alexa 594-conjugated goat anti-rabbit IgG (red) together with a mouse monoclonal antibody to acetylated α-tubulin (47, 48), followed by Alexa 488-conjugated goat anti-mouse IgG (green). The nuclei were counterstained with DAPI (blue). Scale bars, 10 μm. (B) Cells in 60-mm-diameter dishes were mock infected or infected with T1L, T3DC, or T3DN at an MOI of 5, and lysates were collected 18 h later. The lysates were subjected to electrophoresis on 10% polyacrylamide gels and electroblotting to nitrocellulose for immunodetection of acetylated α-tubulin (ac. tubulin) and α-tubulin (tubulin).
The M1 genome segment determines the level of α-tubulin acetylation in infected cells.
Given the correlation between the inclusion phenotype and the degree of MT stabilization in T1L- and T3D-infected cells, we hypothesized that the M1 genome segment may also determine MT stabilization. To test this hypothesis, we used immunoblotting to measure the relative amounts of acetylated and total α-tubulin in Mv1Lu cells infected with a subset of the T1L × T3D reassortants used for genetic analysis of the inclusion phenotype described above. After ranking the reassortants by the ratio of acetylated to total α-tubulin as measured by densitometry, we found that reassortants possessing an M1 genome segment derived from T1L had higher relative amounts of acetylated α-tubulin than reassortants with an M1 segment derived from T3D. Statistical analysis of the reassortant data using the Mann-Whitney U test revealed that the parental origin of only the M1 genome segment segregated with this ratio to a significant degree (P = 0.0006) (Table 1). The μ2-encoding M1 segment is thus the primary genetic determinant of the differences in α-tubulin hyperacetylation and, by inference, of the differences in MT stabilization observed in Mv1Lu cells infected with these different viruses.
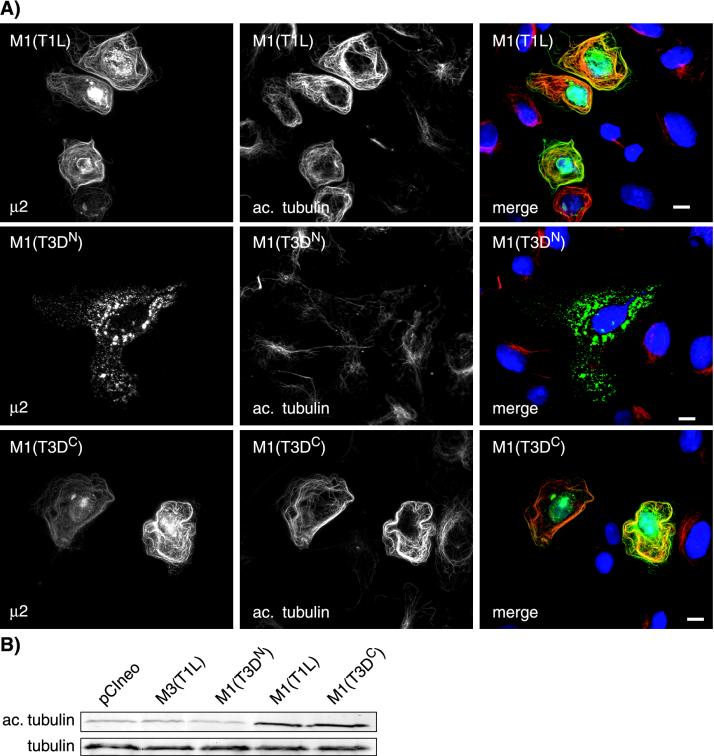
The μ2 protein colocalizes with and causes bundling of MTs in cells transfected with the M1 gene of reoviruses T1L and T3DC but not T3DN.
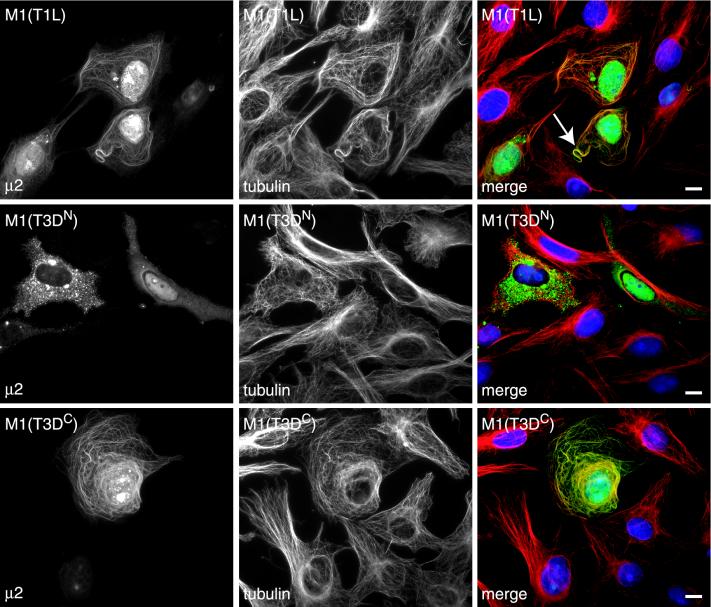
Having shown that the M1 genome segment determines both the inclusion phenotype and the degree of MT stabilization in T1L- and T3DN-infected cells, we hypothesized that its encoded protein, μ2, may colocalize with MTs in cells transfected with cDNA clones of M1(T1L) and M1(T3DC) but not M1(T3DN). The M1 genes of reoviruses T1L, T3DC, and T3DN were each cloned into the mammalian expression vector pCI-neo, generating plasmids designated pCI-M1(T1L), pCI-M1(T3DC), and pCI-M1(T3DN). We transfected CV-1 cells with the M1-containing plasmids and examined the subcellular distribution of the μ2 protein. In cells transfected with pCI-M1(T1L) or pCI-M1(T3DC), the μ2 protein was seen in the cytoplasm and nucleus either diffuse or in small aggregates and was also prominently associated with filamentous structures in the cytoplasm (Fig. 8). The filamentous structures labeled with the anti-μ2 serum clearly colocalized with MTs (Fig. 8), and in many transfected cells, MT bundling was seen (Fig. 8). In cells transfected with pCI-M1(T3DN), μ2 was similarly distributed, but there was no association of μ2 with filamentous structures (Fig. 8). In all three transfections, μ2 was concentrated in the nuclei of some cells (data not shown). While it appears that there may be multiple forms of μ2 given the different localizations, it is clear that μ2(T1L) and μ2(T3DC) colocalize with MTs whereas μ2(T3DN) does not (Fig. 8).
FIG. 8.
Colocalization of MTs with T1L and T3DC μ2, but not T3DN μ2, in M1-transfected CV-1 cells. The cells were transfected with 2 μg of plasmid pCI-M1(T1L), pCI-M1(T3DC), or pCI-M1(T3DN) and then fixed 18 h later. The cells were immunostained for μ2 (green) and α-tubulin (red) as described for Fig. 4. The nuclei were counterstained with DAPI (blue). The arrow indicates MT bundling. Scale bars, 10 μm.
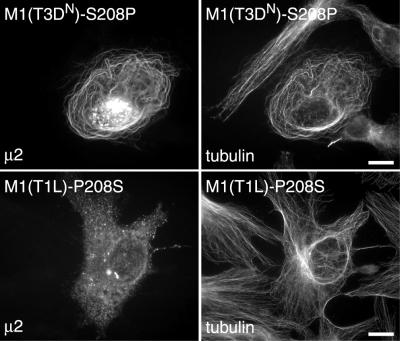
Exchanging the amino acid at position 208 in μ2 changes the association with MTs.
Above, we identified variation at amino acid position 208 in μ2 as responsible for the differences in inclusion morphology between the T1L, T3DC, and T3DN reoviruses (Table 2). To test whether this variation determined the different capacities of these μ2 proteins to associate with MTs, we constructed plasmids pCI-M1(T1L)-P208S and pCI-M1(T3DN)-S208P such that residue 208 was changed from Pro to Ser in the encoded μ2(T1L) protein and from Ser to Pro in the encoded μ2(T3DN) protein. We then transfected CV-1 cells with each of these plasmids and examined the subcellular distributions of μ2 and tubulin as described above. Cells transfected with pCI-M1(T3DN)-S208P contained μ2 with a filamentous distribution and clear colocalization with MTs (Fig. 9), similar to the distribution of μ2(T1L) in transfected cells. In contrast, cells transfected with pCI-M1(T1L)-P208S had a μ2 distribution pattern similar to that of μ2(T3DN) in transfected cells and thus did not display any filamentous distribution of μ2 (Fig. 9). These results indicate that the difference in distribution of μ2 is due to the variation at amino acid 208.
FIG. 9.
Subcellular distributions of μ2(T3DN)-S208P and μ2(T1L)-P208S in transfected cells. CV-1 cells were transfected with plasmid pCI-M1(T3DN)-S208P or pCI-M1(T1L)-P208S and then fixed 18 h later. The cells were immunostained with rabbit anti-μ2 serum followed by Alexa 488-conjugated goat anti-rabbit IgG and a Cy3-conjugated mouse monoclonal antibody to β-tubulin (tubulin). Scale bars, 10 μm.
MTs are hyperacetylated in cells expressing μ2(T1L) and μ2(T3DC) but not μ2(T3DN).
As we had mapped the phenotype of hyperacetylated α-tubulin in infected cells to the M1 genome segment, we examined directly whether expression of μ2(T1L) and μ2(T3DC) increased the amount of acetylated α-tubulin in transfected cells. CV-1 cells were transfected with pCI-M1(T1L), pCI-M1(T3DC), and pCI-M1(T3DN) and then immunostained for μ2 and acetylated α-tubulin 18 h later. As seen in Fig. 10A, those cells expressing μ2(T1L) and μ2(T3DC) had greatly increased amounts of acetylated α-tubulin compared to adjacent nontransfected cells, whereas cells expressing μ2(T3DN) did not show any increase in the level of acetylated α-tubulin in the μ2-positive cells. The μ2(T1L) and μ2(T3DC) proteins also clearly colocalized with acetylated α-tubulin in transfected cells (Fig. 10A). Additionally, we found that the amount of acetylated α-tubulin detected by immunoblotting of cell lysates was greater in cells transfected with pCI-M1(T1L) and pCI-M1(T3DC) than in cells transfected with pCI-M1(T3DN) or control transfections (Fig. 10B). We also analyzed the levels of acetylated α-tubulin in immunostained cells expressing the mutant forms of μ2 in which residue 208 was alternated between Pro and Ser. In those experiments, increased levels of acetylated α-tubulin were seen in cells transfected with pCI-M1(T3DN)-S208P, whereas control levels of acetylated α-tubulin were seen in cells transfected with pCI-M1(T1L)-P208S (supplemental data are at http://micro.med.harvard.edu/nibert/suppl/parker02a/suppl3.html), consistent with the filamentous distribution of the former but not the latter (Fig. 9).
FIG. 10.
Hyperacetylation of MTs in CV-1 cells expressing μ2 derived from T1L and T3DC but not T3DN. (A) Cells were transfected with 2 μg of plasmid pCI-M1(T1L), pCI-M1(T3DC), or pCI-M1(T3DN) and then fixed 18 h later. The cells were immunostained with rabbit anti-μ2 serum and then with Alexa 488-conjugated goat anti-rabbit IgG (green) together with a mouse monoclonal antibody to acetylated α-tubulin (ac. tubulin) (47, 48), followed by Alexa 594-conjugated goat anti-mouse IgG (red). The nuclei were counterstained with DAPI (blue). Scale bars, 10 μm. (B) Cells in 60-mm-diameter dishes were transfected with plasmid pCI-neo, pCI-M3(T1L), pCI-M1(T1L), pCI-M1(T3DC), or pCI-M1(T3DN), and lysates were collected 18 h later. The lysates were subjected to electrophoresis on SDS-10% polyacrylamide gel electrophoresis gels and electroblotting to nitrocellulose for immunodetection of acetylated α-tubulin (ac. tubulin) and α-tubulin (tubulin).
DISCUSSION
The replication complexes of viruses that reproduce their genomes and assemble their particles in the cytoplasm of host cells are commonly associated with either membranous organelles (36, 51, 58) or elements of the cytoskeleton (24, 26, 29). Cells infected with reoviruses develop large cytoplasmic inclusion bodies in which viral replication and assembly are thought to occur (45). In this report, we demonstrated two strain-dependent reovirus inclusion morphologies, filamentous and globular, the former reflecting an association with MTs. Our data indicate that MTs are stabilized following infection with reovirus strains that form filamentous inclusions and that the μ2-encoding M1 genome segment genetically determines this phenotype. In support of this finding, MTs formed bundles and were stabilized in cells expressing the μ2 protein derived from reovirus strains that formed filamentous inclusions. These properties of MT bundling and stabilization are classically associated with cellular MAPs.
Other cellular and viral MAPs.
Cellular MAPs modulate the dynamic instability of MTs. Some MAPs, such as tau, stabilize MTs by binding and reducing the rate of switching from growth to rapid disassembly (30, 37). Other functions ascribed to MAPs include nucleation of MT polymerization, regulation of organelle transport, and anchoring of proteins involved in signal transduction (37). The association of MAPs with MTs is regulated by phosphorylation and dephosphorylation, which are performed by specific MAP kinases and phosphatases (31, 33, 54). Given the similar properties of μ2 and MAPs, it will be important to determine whether μ2 shares other of their characteristics.
A number of other viral proteins associate with MTs and possess properties similar to those of MAPs. These include the plant tobamovirus movement protein (5), the aphid transmission factor of cauliflower mosaic virus (4), the vaccinia virus A10L and L4R proteins (50), and the herpes simplex virus type 1 tegument protein VP22 (20). What roles do these viral MAPs play in the viral life cycles? The plant movement proteins, through their interaction with MTs, are able to transport viral RNA to the plasmodesmata, where the RNA is transported between adjacent cells (2, 15). The vaccinia proteins A10L and L4R bind to viral cores and interact with the MT cytoskeleton, leading to loss of centrosomal proteins and function; however, it is not known what roles these proteins play in the vaccinia life cycle (50). The herpes simplex virus type 1 VP22 protein binds and stabilizes MTs (20) and in addition has the property of trafficking intercellularly (21). VP22 is also found in the nuclei of cells and has been shown to associate with chromatin (19). We found that μ2 is similarly sometimes found in the nuclei of transfected cells, but we found no evidence that it can traffic between cells (J. S. L. Parker, unpublished data).
The role of μ2 and MTs in reovirus inclusion formation and morphology.
Previous ultrastructural studies of reovirus-infected cells demonstrated the presence of MTs within the viral inclusions (13). Immunoelectron microscopy using ferritin-conjugated anti-reovirus antibodies showed that many MTs in infected cells are coated with an unidentified viral protein(s) (14). Subsequent work demonstrated that μNS could be coprecipitated from infected-cell extracts with an anti-tubulin antibody, suggesting that μNS might interact with MTs (43). We have found, however, that μNS does not associate with MTs when expressed in cells in the absence of other viral proteins (T. J. Broering and J. S. L. Parker, unpublished data). Our findings in this study indicate that MTs in T1L-infected cells are coated with inclusion material containing multiple different viral proteins, including μ2, μNS, λ1, λ2, λ3, and σNS (Fig. 2 and data not shown), and that this inclusion material occludes the accessibility of anti-tubulin antibodies to MTs buried within the inclusions. Moreover, we showed that μ2(T1L) and μ2(T3DC) colocalize with and stabilize MTs when expressed in cells in the absence of other viral proteins and that the μ2-encoding M1 genome segment is the sole genetic determinant of the MT-dependent filamentous inclusion morphology and MT stabilization phenotype of reovirus T1L. Thus, although the filamentous inclusion material seen coating MTs in this and previous studies contains multiple reovirus proteins, the association of that material with MTs is likely mediated by μ2.
We believe that in cells infected with most reovirus strains, including T1L and T3DC, μ2 firmly anchors the inclusion material to MTs, allowing it to remain spread out along the MTs. We propose that in addition to binding to MTs, μ2 must bind to one or more other reovirus proteins in order to anchor the inclusion material to the MTs. In contrast, in T3DN-infected cells, μ2 does not firmly anchor the inclusion material to MTs and hence the inclusions develop into globular structures. Our experiments with nocodazole indicate that MTs are nonetheless required for the coalescence of smaller, more peripherally located inclusions into larger perinuclear inclusions in both T1L- and T3DN-infected cells. Thus, a less stable, possibly μ2-independent, interaction between reovirus inclusions and MTs likely also occurs, allowing smaller inclusions to be moved from the periphery of the cytoplasm to the perinuclear region as infection proceeds. The viral and cellular mediators of this additional interaction remain to be identified.
Mbisa et al. (41) recently reported that inclusion bodies are seen in T3D-infected L-929 cells by 18 h p.i., whereas inclusions are not seen in T1L-infected cells until 39 h p.i. In both cases, the inclusions were described as globular. This difference in the kinetics of inclusion formation was found to be primarily determined by the M1 genome segment, with the S3 segment playing a secondary role (41). In contrast, we were easily able to detect inclusions induced by either strain by 18 h p.i. (Fig. 1). This discrepancy may be due to differences in the fixation procedures used for IF microscopy. We found that the acetone fixation procedure used by Mbisa et al. (41) causes disruption of the filamentous inclusions induced by T1L, making them appear diffuse at earlier times postinfection, whereas the globular inclusions induced by T3DN are relatively unaffected (data not shown). We found no strain difference in the kinetics of inclusion formation when L cells were fixed with 2% paraformaldehyde, with inclusions being detectable 6 to 8 h after infection with either T1L or T3DN (data not shown). Different fixation and permeabilization methods are known to change the subcellular distribution of proteins. Solvent fixation with acetone or methanol causes protein coagulation and disruption of the tertiary structure of proteins, whereas aldehyde fixation causes protein cross-linking and is generally thought to preserve cellular architecture and organelles (42). We believe that the fixation method chosen by Mbisa et al. (41) prevented those authors from detecting the M1-based strain difference in inclusion morphology and contributed to their conclusion that there is an M1-based strain difference in the kinetics of inclusion formation.
Possible significance of the different inclusion morphologies and similarities of reovirus inclusions to cellular aggresomes.
We found that most reovirus strains, including another laboratory's isolate of T3D (T3DC), induce filamentous inclusions. In some early reports, the inclusions induced by T3D infections were in fact described as filamentous (13). The rarity of the globular phenotype among our test group (only 2 of 24 strains) suggests that it may confer a selective disadvantage during natural infections or, reciprocally, that the filamentous phenotype may confer a selective advantage. One consequence of a filamentous versus a globular inclusion morphology is that the surface area/volume ratio is much larger for any given volume of inclusion material. The larger surface area of filamentous inclusions may allow for more efficient viral replication through better access to small-molecule substrates or newly synthesized proteins or RNA from the surrounding cytosol. It may also allow more efficient release of newly assembled viral particles from the inclusions, either for later steps in assembly or for infection of other cells. In infected cells, μ2 formed granular substructures at the margins of the inclusions, whether the inclusions were filamentous or globular (Fig. 2B). We suspect that these granules represent sites of active viral genome replication and core assembly. Studies of the in vivo particle morphogenesis of bluetongue virus have shown that the maturation of virions occurs at the periphery of the viral inclusion bodies (8). Thus, if active genome replication and core assembly by reoviruses occurs at the margins of the inclusions, then filamentous inclusions would provide a larger surface area for assembly of the outer-capsid proteins onto newly forming core particles to generate mature virions.
A similarity to cellular aggresomes was recently noted for the cytoplasmic inclusion bodies of the iridovirus ASFV (28). The formation of aggresomes is thought to represent a programmed cellular response to protein overexpression or misfolding, and it occurs to excess in a number of neurodegenerative illnesses, including Alzheimer's and Parkinson's diseases (35). As we now recognize from findings in this and previous studies, reovirus inclusion bodies share features with aggresomes as well. These features include their perinuclear localization and dependence on MTs for formation and morphology. We have also recently shown that a number of other protein markers of cellular aggresomes are associated with the reovirus inclusions (Parker, unpublished data). The large globular inclusions of our laboratory's isolate of reovirus T3D rapidly develop the appearance of aggresomes as they coalesce in the perinuclear region. The inclusions of reovirus T1L and the other strains with a filamentous phenotype develop more of this appearance at later times in infection, but the continuing filamentous distribution of much of the inclusion material remains a distinctive feature. One possibility is that the role of the reovirus μ2 protein in anchoring the inclusions to MTs is to resist the usual process of aggresome coalescence. Further studies are needed to confirm the link between reovirus inclusion bodies and aggresomes and to address the significance of this phenomenon for reovirus replication and its effects on host cells.
Other reported roles of MTs during reovirus infection.
In addition to a role for MTs in the morphogenesis of viral inclusion bodies, MTs have been shown to be involved in virus spread to the central nervous system via peripheral nerves in vivo and in the movement of virus from the periphery of cells to the perinuclear region during virus uptake in vitro. MTs are involved in the spread of reoviruses from peripheral inoculation sites along nerves to the central nervous system (57). This spread along nerves is believed to occur by fast axonal transport along MTs and can be blocked by pretreatment of mice with the MT-depolymerizing agent colchicine (57). It is not known if fast axonal transport of reovirus occurs within vesicles or if viral particles themselves are moved along MTs by attaching to MT motors. The perinuclear accumulation of fluorescently tagged parent virions following infection of L cells is also prevented by treatment of the cells with either colchicine or nocodazole (22). No link between the roles of MTs in reovirus particle movement and inclusion formation has yet been demonstrated.
Other properties of μ2.
The μ2 protein is found within viral cores in low copy numbers (most likely 12 to 24), close to the icosahedral fivefold vertices (12, 17), where it may modulate transcription of the viral genome (61). The μ2 protein binds both double-stranded and single-stranded RNAs (6) and is a determinant of the NTPase activity of cores (47). Although a direct enzymatic function for μ2 has not been demonstrated, μ2 contains two regions of sequence with limited resemblance to the A and B motifs of known NTP-binding proteins (47). The multiple locations of μ2 in transfected cells (e.g., Fig. 8) suggest that μ2 may assume multiple forms within cells. Posttranslational modifications, such as phosphorylation, are known to modify the intracellular location of proteins in many cases (11) and remain possible for μ2. Complex phenotypes associated with reovirus infection are also genetically influenced by the μ2-encoding M1 genome segment, including the capacity of reovirus T1L to replicate more efficiently than T3D in mouse primary myocardial cells (40); the capacity of T1L, but not T3D, to replicate efficiently in human and bovine endothelial cells (39); the capacity of the reovirus 8B variant to cause myocarditis in mice (53); the capacity of T1L, but not T3D, to replicate in the endothelium and liver in SCID mice (25); and the greater virulence of T1L than T3D in SCID mice (25). These M1-influenced differences in replication efficiency and pathogenesis suggest that μ2 may interact with specific host factors that determine the outcome of infection. Our finding that μ2 interacts with MTs suggests several possibilities for how these differences may be determined at the molecular level.
Acknowledgments
We are grateful to Earl Brown for supplying his laboratory's reovirus T1L M1 clone and to Bill Cashdollar for supplying stocks of his laboratory's reovirus T3D isolate. Many thanks also to Caroline Piggott, who measured the titers of many of the antisera used in this study; Laura Breun and Elaine Freimont for technical assistance; and the other members of our laboratory for helpful discussions.
This work was supported in part by NIH grant R29 AI-39533 (to M.L.N.) and by a USDA Hatch grant through the University of Wisconsin Extension (to M.L.N.). J.S.L.P. is the recipient of an individual NRSA fellowship (F32 AI-10134). T.J.B. acknowledges previous support provided by predoctoral fellowships from the Wisconsin Alumni Research Foundation and NIH research training grant T32 GM0712 to the Molecular Biosciences Program at the University of Wisconsin, Madison.
REFERENCES
- 1.Babiss, L. E., R. B. Luftig, J. A. Weatherbee, R. R. Weihing, U. R. Ray, and B. N. Fields. 1979. Reovirus serotypes 1 and 3 differ in their in vitro association with microtubules. J. Virol. 30:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beachy, R. N., and M. Heinlein. 2000. Role of P30 in replication and spread of TMV. Traffic 1:540-544. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc, S., I. Schmidt, M. Vantard, H. B. Scholthof, G. Kuhl, P. Esperandieu, M. Cerutti, and C. Louis. 1996. The aphid transmission factor of cauliflower mosaic virus forms a stable complex with microtubules in both insect and plant cells. Proc. Natl. Acad. Sci. USA 93:15158-15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyko, V., J. Ferralli, J. Ashby, P. Schellenbaum, and M. Heinlein. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2:826-832. [DOI] [PubMed] [Google Scholar]
- 6.Brentano, L., D. L. Noah, E. G. Brown, and B. Sherry. 1998. The reovirus protein μ2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 72:8354-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brookes, S. M., A. D. Hyatt, and B. T. Eaton. 1993. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J. Gen. Virol. 74:525-530. [DOI] [PubMed] [Google Scholar]
- 9.Brown, E. G., M. L. Nibert, and B. N. Fields. 1983. The L2 gene of reovirus serotype 3 controls the capacity to interfere, accumulate deletions and establish persistent infection, p. 275-288. In R. W. Compans and D. H. L. Bishop (ed.), Double-stranded RNA viruses. Elsevier Biomedical, New York, N.Y.
- 10.Caliguiri, L. A., and I. Tamm. 1969. Membranous structures associated with translation and transcription of poliovirus RNA. Science 166:885-886. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, P. 2000. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem. Sci. 25:596-601. [DOI] [PubMed] [Google Scholar]
- 12.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218-228. [DOI] [PubMed] [Google Scholar]
- 13.Dales, S. 1963. Association between the spindle apparatus and reovirus. Proc. Natl. Acad. Sci. USA 50:268-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dales, S., P. J. Gomatos, and K. C. Hsu. 1965. The uptake and development of reovirus in strain L cells followed with labeled viral ribonucleic acid and ferritin-antibody complexes. Virology 25:193-211. [DOI] [PubMed] [Google Scholar]
- 15.Deom, C. M., M. Lapidot, and R. N. Beachy. 1992. Plant virus movement proteins. Cell 69:221-224. [DOI] [PubMed] [Google Scholar]
- 16.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dryden, K. A., D. L. Farsetta, G.-J. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 225:33-46. [DOI] [PubMed] [Google Scholar]
- 18.Eaton, B. T., A. D. Hyatt, and J. R. White. 1987. Association of bluetongue virus with the cytoskeleton. Virology 157:107-116. [DOI] [PubMed] [Google Scholar]
- 19.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 22.Georgi, A., C. Mottola-Hartshorn, A. Warner, B. Fields, and L. B. Chen. 1990. Detection of individual fluorescently labeled reovirions in living cells. Proc. Natl. Acad. Sci. USA 87:6579-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillian, A. L., S. C. Schmechel, J. Livny, L. A. Schiff, and M. L. Nibert. 2000. Reovirus protein σNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J. Virol. 74:5939-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, S., B. P. De, J. A. Drazba, and A. K. Banerjee. 1998. Involvement of actin microfilaments in the replication of human parainfluenza virus type 3. J. Virol. 72:2655-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller, B. L., M. L. Barkon, G. P. Vogler, and H. W. Virgin IV. 1995. Genetic mapping of reovirus virulence and organ tropism in severe combined immunodeficient mice: organ-specific virulence genes. J. Virol. 69:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamaguchi, M., K. Nishikawa, T. Toyoda, T. Yoshida, T. Hanaichi, and Y. Nagai. 1985. Transcriptive complex of Newcastle disease virus. II. Structural and functional assembly associated with the cytoskeletal framework. Virology 147:295-308. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, S. J., D. L. Farsetta, J. Kim, S. Noble, T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus L3 gene sequences and evidence for a distinct amino-terminal region of the λ1 protein. Virology 258:54-64. [DOI] [PubMed] [Google Scholar]
- 28.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill, V. M., S. A. Harmon, and D. F. Summers. 1986. Stimulation of vesicular stomatitis virus in vitro RNA synthesis by microtubule-associated proteins. Proc. Natl. Acad. Sci. USA 83:5410-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa, N. 1994. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 6:74-81. [DOI] [PubMed] [Google Scholar]
- 31.Hoshi, M., K. Ohta, Y. Gotoh, A. Mori, H. Murofushi, H. Sakai, and E. Nishida. 1992. Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induces an alteration in their function. Eur. J. Biochem. 203:43-52. [DOI] [PubMed] [Google Scholar]
- 32.Hrdy, D. B., L. Rosen, and B. N. Fields. 1979. Polymorphism of the migration of double-stranded RNA genome segments of reovirus isolates from humans, cattle, and mice. J. Virol. 31:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson, L., and M. Caplow. 1981. Modification of microtubule steady-state dynamics by phosphorylation of the microtubule-associated proteins. Proc. Natl. Acad. Sci. USA 78:3413-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knipe, D., C. Samuel, and P. Palese. 2001. Virus-host cell interactions, p. 133-170. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 35.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 36.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maccioni, R. B., and V. Cambiazo. 1995. Role of microtubule-associated proteins in the control of microtubule assembly. Physiol. Rev. 75:835-864. [DOI] [PubMed] [Google Scholar]
- 38.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matoba, Y., W. S. Colucci, B. N. Fields, and T. W. Smith. 1993. The reovirus M1 gene determines the relative capacity of growth of reovirus in cultured bovine aortic endothelial cells. J. Clin. Investig. 92:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matoba, Y., B. Sherry, B. N. Fields, and T. W. Smith. 1991. Identification of the viral genes responsible for growth of strains of reovirus in cultured mouse heart cells. J. Clin. Investig. 87:1628-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16-26. [DOI] [PubMed] [Google Scholar]
- 42.Melan, M. 1999. Overview of cell fixatives and cell membrane permeants. Methods Mol. Biol. 115:45-55. [DOI] [PubMed] [Google Scholar]
- 43.Mora, M., K. Partin, M. Bhatia, J. Partin, and C. Carter. 1987. Association of reovirus proteins with the structural matrix of infected cells. Virology 159:265-277. [DOI] [PubMed] [Google Scholar]
- 44.Nibert, M. L., R. L. Margraf, and K. M. Coombs. 1996. Nonrandom segregation of parental alleles in reovirus reassortants. J. Virol. 70:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott Willams & Wilkins, Philadelphia, Pa.
- 46.Nilsson, M., C. H. von Bonsdorff, K. Weclewicz, J. Cohen, and L. Svensson. 1998. Assembly of viroplasm and virus-like particles of rotavirus by a Semliki Forest virus replicon. Virology 242:255-265. [DOI] [PubMed] [Google Scholar]
- 47.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71:7728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piperno, G., and M. T. Fuller. 1985. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101:2085-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piperno, G., M. LeDizet, and X. J. Chang. 1987. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 104:289-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. Gonzalez, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 53.Sherry, B., and B. N. Fields. 1989. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J. Virol. 63:4850-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheterline, P., and J. G. Schofield. 1975. Endogenous phosphorylation and dephosphorylation of microtubule-associated proteins isolated from bovine anterior pituitary. FEBS Lett. 56:297-302. [DOI] [PubMed] [Google Scholar]
- 55.Spendlove, R. S., and E. H. Lennette. 1963. The role of the mitotic apparatus in the intracellular location of reovirus antigen. J. Immunol. 90:554-560. [PubMed] [Google Scholar]
- 56.Spendlove, R. S., E. H. Lennette, J. N. Chin, and C. O. Knight. 1964. Effect of antimitotic agents on intracellular reovirus antigen. Cancer Res. 24:1826-1833. [PubMed] [Google Scholar]
- 57.Tyler, K. L., D. A. McPhee, and B. N. Fields. 1986. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science 233:770-774. [DOI] [PubMed] [Google Scholar]
- 58.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiener, J. R., J. A. Bartlett, and W. K. Joklik. 1989. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein μ2 and the major nonstructural protein μNS, respectively. Virology 169:293-304. [DOI] [PubMed] [Google Scholar]
- 61.Yin, P., M. Cheang, and K. M. Coombs. 1996. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 70:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou, S., and E. G. Brown. 1992. Nucleotide sequence comparison of the M1 genome segment of reovirus type 1 Lang and type 3 Dearing. Virus Res. 22:159-164. [DOI] [PubMed] [Google Scholar]
- 63.Zou, S., and E. G. Brown. 1996. Stable expression of the reovirus μ2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology 217:42-48. [DOI] [PubMed] [Google Scholar]