Abstract
Rabies virus nucleoprotein (N) plays vital roles in regulation of viral RNA transcription and replication by encapsidation of the nascent genomic RNA. Rabies virus N is phosphorylated, and previous studies demonstrated that mutation of the phosphorylated serine at position 389 to alanine resulted in reduction of viral transcription and/or replication of a rabies virus minigenome. In the present study, we mutated the serine (S) at position 389 to alanine (A), glycine (G), aspartic acid (D), asparagine (N), glutamic acid (E), and glutamine (Q) and examined the effects of these mutations on rabies virus transcription and replication in the minigenome. Furthermore, mutations from S to A, S to D, and S to E were also incorporated into the full-length infectious virus. Mutation of the serine to each of the other amino acids resulted in the synthesis of an unphosphorylated N and reduction of viral transcription and replication in the minigenome. Mutations from S to A and S to D also resulted in reduction of both viral transcription and replication in full-length infectious viruses. Growth curve studies indicated that production of the mutant virus with the S-to-A mutation (L16A) was as much as 10,000-fold less than that of the wild-type virus (L16). Northern blot hybridization with rabies virus gene probes revealed that the rates of viral transcription and replication were reduced by as much as 10-fold in the mutant viruses when the N was not phosphorylated. Interpretation of the data from the minigenome system and the full-length infectious virus indicates that phosphorylation of rabies virus N is necessary for replication. Further studies involving cycloheximide treatment of infected cells revealed that viral transcription was also reduced when the N was not phosphorylated. Taken together, these results provide definitive evidence that N phosphorylation plays an important role in the processes of rabies virus transcription and replication.
Within the Rhabdoviridae family, rabies virus is the prototype of the Lyssavirus genus and Vesicular stomatitis virus (VSV) is the prototype of the Vesiculovirus genus (23). Rhabdovirus genomic RNA is encapsidated with nucleoprotein (N), and this N-RNA complex, together with the phosphoprotein (P, also termed NS) and RNA-dependent RNA polymerase (L), forms the RNP complex. The N proteins of the rhabdoviruses, like the N proteins from other members of the order Mononegavirales, play vital roles in regulating viral RNA transcription and replication by encapsidating de novo-synthesized viral genomic RNA (for a review, see reference 23). Although rabies virus N and VSV N do not have a high degree of homology in primary nucleotide and protein sequences, they have conserved regions and similar protein characteristics. For example, the N protein of rabies virus has four conserved amino acid stretches homologous with those of VSV (22). In addition, similar helical structures exist in both rabies virus and VSV N proteins. Both N proteins have an α-helix continuing from the N terminus through most of the protein with a β-turn toward the C terminus (2).
However, there is one major structural difference between rabies virus N and VSV N. Rabies virus N is phosphorylated, whereas VSV N is not (20). The phosphorylation has been mapped to the serine residue at position 389 of the rabies virus N (7). Phosphorylation of rabies virus N, but not VSV N, has raised questions as to how the phosphorylation of rabies virus N is involved in the regulation of rabies virus RNA transcription and replication (27). We demonstrated that dephosphorylation of rabies virus N or mutation of serine 389 to alanine (S389A) resulted in an increased binding to in vitro-synthesized leader RNA (28). Furthermore, mutation of the phosphorylated serine to alanine resulted in the reduction of viral transcription and replication of a rabies virus minigenomic RNA (28). However, in the minigenome system, viral proteins necessary for viral transcription and replication were synthesized by T7 polymerase, and thus their synthesis was not under the control of rabies virus regulatory machinery. In the present study, we further mutated the phosphorylated serine (S) to alanine (A), glycine (G), aspartic acid (D), asparagine (N), glutamic acid (E), and glutamine (Q) and investigated the effects of these N mutants on viral transcription and replication in the minigenome as well as the rescued infectious virus. The results from the studies revealed that both viral transcription and replication were reduced when the N is not phosphorylated, suggesting that N phosphorylation plays an important role in modulating both transcription and replication of rabies virus. Furthermore, the results from this study indicate that the effects of N phosphorylation on viral transcription and replication are due to a combination of the net negative charge of the phosphate moiety and the structure of the serine residue.
MATERIALS AND METHODS
Cells, viruses, plasmids, and antibodies.
BSR (a clone of BHK) and BSR T7/5 (BSR cells stably expressing T7 polymerases) (4) were grown in Dulbecco's minimal essential medium and transfected with plasmids as described previously (28). Recombinant vaccinia virus expressing bacterial T7 RNA polymerase (vTF7-3) was prepared as described previously (14). Plasmids used for expression of the full-length infectious virus (pSAD-L16), rabies virus minigenome (pSDI-CAT), and rabies virus L (pT7T-L) (6, 18) were obtained from K. Conzelmann. Plasmids for expression of N (pRN) and P (pRP) were constructed previously in our laboratory (13). Polyclonal antiserum against rabies virus N was prepared in rabbits as described previously (10).
Site-directed mutagenesis.
Mutation of the serine 389 of the rabies virus N to A, G, D, N, E, or Q was carried out by site-directed mutagenesis, using the method of Weiner et al. (24). Six pairs of primers, as summarized in Table 1, were synthesized and were designed to contain one or two nucleotide changes that resulted in the mutation of the serine codon. PCR was performed with each of the six primer pairs using pRN (13) as a template. The PCR products were subjected to digestion with DpnI, which digests methylated and hemimethylated DNA at the GmeATC site, thereby digesting the pRN DNA template. The undigested PCR products (not methylated) were used to transform competent XL-1 Blue cells. The mutations in the plasmids were confirmed by nucleotide sequencing.
TABLE 1.
Primers used to make mutations of serine at position 389 to alanine, glycine, aspartic acid, asparagine, glutamic acid, and glutamine on rabies virus N
| Primer | Primer sequencea (5′-3′) | Amino acid resulting from change |
|---|---|---|
| SA5 | GATGATGGAACTGTCAACGCTGACGACGAGG | Alanine |
| SA3 | GTAGTCCTCGTCGTCAGCGTTGACAGTTCC | |
| SG5 | GATGATGGAACTGTCAACGGTGACGACGAGG | Glycine |
| SG3 | GTAGTCCTCGTCGTCACCGTTGACAGTTCC | |
| SD5 | GATGATGGAACTGTCAAGATGACGACGAGG | Aspartic acid |
| SD3 | GTAGTCCTCGTCGTCATCGTTGACAGTTCC | |
| SN5 | GATGATGGAACTGTCAACAATGACGACGAGG | Asparagine |
| SN3 | GTAGTCCTCGTCGTCATTGTTGACAGTTCC | |
| SE5 | GATGATGGAACTGTCAACGAAGACGACGAGG | Glutamic acid |
| SE3 | GTAGTCCTCGTCGTCTTCGTTGACAGTTCC | |
| SQ5 | GATGATGGAACTGTCAACCAAGACGACGAGG | Glutamine |
| SQ3 | GTAGTCCTCGTCGTCTTGGTTGACAGTTCC |
Mutated codon is in boldface.
To introduce the mutations of rabies virus N into the infectious clone, an SphI fragment containing the serine codon of the N from the full-length infectious clone (pSAD-L16) (nucleotides 482 to 4041 of the rabies virus genome [5]) was cloned into the SphI site of pGEM-3Z. The resulting plasmid was used as the template for construction of the mutants as described above for pRN. The serine at position 389 was mutated to A, D, or E with the surrogate vector. After confirmation by sequence analysis, each of the mutated SphI fragments was cloned back to pSAD-L16. Three mutated clones with the expected mutation and correct orientation were obtained and were designated pSAD-L16A, pSAD-L16D, and pSAD-L16E, respectively.
Transfection.
Transfection of BSR cells with plasmids was performed as described previously (28). Briefly, BSR cells were infected with recombinant vaccinia virus (vTF7-3) at a multiplicity of infection (MOI) of 5 PFU per cell. One hour after infection, cells were transfected with different combinations of mixed plasmids by using Lipofectamine (Life Technologies, Rockville, Md.). Transfected cells were harvested at various time points for further analysis.
CAT assay.
Chloramphenicol acetyltransferase (CAT) activities were measured with the Quan-T-CAT assay (Amersham Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's protocol. Transfected cells were lysed, and the supernatants were incubated with biotinylated chloramphenicol and [3H]acetyl coenzyme A. Then streptavidin-coated beads were added to the reaction mixture. After the free radioactive materials were removed, the pellets were resuspended in scintillation fluid for quantitation by scintillation spectrometry. The CAT activities were expressed as counts per minute. The relative CAT activities in cells transfected with each of the mutated N proteins were calculated by using the CAT activity in cells transfected with wild-type (wt) N as 100%.
Radiolabeling and immunoprecipitation of proteins.
Transfected or infected BSR cells were labeled with either [35S]methionine or [32P]phosphoric acid (Amersham Pharmacia Biotech) as described previously (28). Cells were harvested and subjected to immunoprecipitation with anti-N antibodies followed by electrophoresis on 12% polyacrylamide-10% sodium dodecyl sulfate (SDS) gel and autoradiography.
Northern and Western blotting.
Transfected or infected BSR cells and purified viruses were subjected to Northern and/or Western blotting. For Northern blotting, BSR cells were lysed with Trizon reagent (Life Technologies) and total RNA was prepared according to the manufacturer's specifications. Poly(A)+ mRNA was purified from total RNA by using the mRNA isolation kit (Roche, Indianapolis, Ind.). RNA preparations were denatured with a 10 mM sodium phosphate buffer (pH 7.4) containing 50% (vol/vol) formamide at 65°C for 15 min and electrophoresed on a 1.1% agarose gel containing 1.1 M formaldehyde and 10 mM sodium phosphate. The RNA then was transferred and covalently fixed onto a nylon membrane for hybridization with CAT, rabies virus gene, or β actin probes. Quantitation of RNA bands was done by densitometry. For Western blots, BSR cells were lysed with radioimmunoprecipitation assay (RIPA) buffer, and proteins were directly separated by SDS-polyacrylamide gel electrophoresis (PAGE). After transfer to a nitrocellulose membrane, rabies virus N was detected by rabbit anti-N polyclonal antibodies as described previously (10).
Selection of mutant rabies viruses.
Selection of mutant viruses was performed either in BSR cells infected with vTF7-3 (14) or in BSR T7/5 cells (4, 17). Briefly, BSR cells were infected with vTF7-3 at a MOI of 1. One hour later, the cells were transfected with 10 μg of pRN, 2.5 μg of pRP, 1.5 μg of pT7T-L, and 10 μg of pSAD-L16, pSAD-L16A, pSAD-L16D, or pSAD-L16E. After incubation for 48 h, cells were resuspended with the medium and subjected to three cycles of freezing and thawing to release cell-associated virus. Vaccinia virus was eliminated by centrifugation and then filtration through a 0.2-μm-pore-size filter unit (Millipore) as described previously (18). Alternatively, BSR T7/5 cells were transfected with 10 μg of pSAD-16 or pSAD-L16A, together with 10 μg of pTIT-N, 2.5 μg of pTIT-P, and 2.5 μg of pTIT-L as described previously (17). To confirm that the mutant viruses contain the desired mutations, total RNA was extracted from BSR cells infected with each of these viruses and subjected to PCR amplification for the N gene using primers 10g (5′CTACAATGGATGCCGAC3′) and 304 (5′TTGACGAAGATCTTGCTCAT3′) as described previously (19). These primers can amplify the complete N coding sequence from the genomic RNA. The amplified fragment was directly sequenced with primer 113 (5′GTAGGATGCTATATGGG3′) (19), which immediately precedes the area of the mutations on the N gene. The mutant viruses bearing S to A, S to D, and S to E mutations were designated L16A, L16D, and L16E, respectively.
Virus growth curve.
BSR cells growing in six-well plates were infected with wt or mutant rabies viruses at a MOI of 1 focus-forming unit (FFU)/cell. After incubation at 37°C for 1 h, virus inocula were removed and cells were washed with phosphate-buffered saline to remove any unabsorbed virus. The cells were replenished with fresh medium, and 100 μl of culture supernatant was removed at 6, 12, 24, 36, 48, 60, and 72 h after infection. Virus aliquots were titrated in duplicate in BSR cells as described previously (12).
Treatment of infected cells with CHX.
Cycloheximide (CHX) was purchased from Sigma (St. Louis, Mo.) and was added into cells infected with rabies virus at a final concentration of 150 μg/ml 1 h after infection as described previously (3). At 6, 12, and 24 h after infection, cells were harvested and RNA was extracted for Northern blot hybridization.
RESULTS
The effects of N phosphorylation on viral transcription and replication are due to a combination of the structure of the S and the net negative charge of the phosphate moiety.
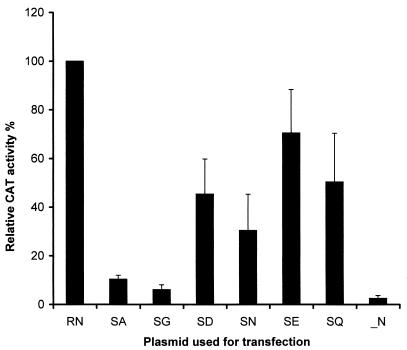
Previously, we demonstrated that phosphorylation of rabies virus N plays important roles in the process of viral transcription and replication (28). Mutation of the phosphorylated S to A results in reduced transcription and replication of a rabies virus minigenome. To determine whether the effects of rabies virus N phosphorylation on viral transcription and replication are caused by the net negative charge of the phosphate moiety or the structure of S or both, the phosphorylated serine at position 389 of the N was mutated to A, G, D, N, E, and Q. BSR cells were infected with recombinant vaccinia virus vTF7-3, followed by transfection with plasmids expressing the rabies virus minigenome (pSDI-CAT), rabies virus L (pT7T-L), and rabies virus P (pRP), together with plasmids expressing rabies virus N or mutant N (pRN, pRN-SA, pRN-SG, pRN-SD, pRN-SN, pRN-SE, or pRN-SQ), as described previously (6, 28). After incubation for 48 h, BSR cells were harvested, and CAT activities were measured by using the Quan-T-CAT assay. The relative CAT activities, which are a measure of minigenome transcription in cells transfected with each of the mutated N plasmids, were calculated by using the CAT activity in cells transfected with wt N as 100%. The experiment was repeated three times, and the average relative CAT activities are summarized in Fig. 1. The CAT activities relative to that for wt N were 6, 9, 28, 40, 46, and 62% in cells transfected with pRN-SG, pRN-SA, pRN-SN, pRN-SD, pRN-SQ, and pRN-SE, respectively. In cells transfected with all other plasmids but lacking an N-expressing plasmid, the relative CAT activity was less than 2%. These results suggest that the effects of N phosphorylation on viral transcription and replication are caused by a combination of the net negative charge of the phosphate moiety and the structure of the serine residue.
FIG. 1.
N phosphorylation affects viral RNA transcription in the minigenome. BSR cells were infected with recombinant vaccinia virus vTF7-3 and then transfected with plasmid pRP, pT7T-L, pSDI-CAT together with pRN (RN), pRN-SA (SA), pRN-SG (SG), pRN-SD (SD), pRN-SN (SN), pRN-SE (SE), or pRN-SQ (SQ). Cells were harvested for measurement of CAT activity by the Quan-T-CAT assay. Error bars, standard deviations.
Increasing or decreasing the concentrations of N-expressing plasmid cannot compensate for the effects of N phosphorylation on viral transcription and replication.
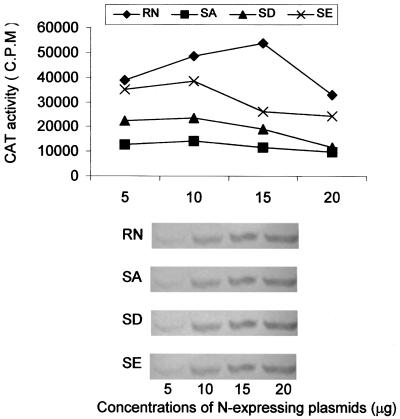
In studying the effects of phosphorylation of VSV P protein, Spadafora et al. (21) observed that unphosphorylated P was much less active in supporting viral transcription at low concentrations. To determine if N concentration has any effect on the ability of the mutant N proteins to achieve optimal transcription and replication of the rabies virus minigenome, various amounts (5, 10, 15, and 20 μg) of the plasmids expressing N (pRN) or mutant N (pRN-SA, pRN-SD, or pRN-SE) were used for transfection, and the cells were harvested for CAT assay. As shown in Fig. 2, 10 μg of each mutant N-expressing plasmid and 15 μg of the wt N-expressing plasmid (pRN) resulted in optimal CAT activities. A larger or smaller amount led to slightly reduced CAT activities (20%), indicating that increasing or decreasing the concentration of N does not have a major impact on viral transcription for either the wt N or the mutant N. To ensure that the levels of N expressed corresponded to the amount of N-expressing plasmid added, transfected cells were lysed with RIPA buffer and the cell lysates were subjected to analysis by SDS-PAGE, followed by Western blotting using the polyclonal anti-N antibodies (13). As shown at the bottom of Fig. 2, for wt N and each of the mutant N proteins, the amount of N expressed was proportional to the amount of N-expressing plasmid added.
FIG. 2.
Effects of N concentrations on viral transcription. BSR cells were infected with recombinant vaccinia virus vTF7-3 and then transfected with plasmid pRP, pT7T-L, pSDI-CAT together with pRN, pRN-SA (SA), pRN-SD (SD), or pRN-SE (SE). Various amounts (5, 10, 15, and 20 μg) of N-expressing plasmids were used. Cells were harvested for CAT assay (top) and Western blotting with anti-N antibodies (bottom).
Phosphorylation of rabies virus N affects both transcription and replication of the rabies virus minigenome.
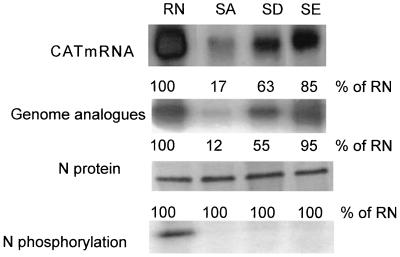
In our previous study (28) and the study described above, only CAT activities were assayed to measure viral transcription and replication. To further determine whether N phosphorylation affects viral transcription or replication or both, Northern blot hybridization was performed to measure the transcripts and the genomic analogues in the transfected cells. To measure viral transcription, total RNA was prepared from transfected cells and poly(A)+ mRNA was purified from the total RNA. To measure viral replication, transfected cells were extracted with distilled H2O and the RNP complex was immunoprecipitated with polyclonal anti-N antibodies. The complex was subjected to treatment with Trizon reagent to obtain genomic analogues. The RNA preparations were hybridized with a CAT probe labeled with [α-32P]dCTP by nick translation from CAT cDNA. As shown in Fig. 3, the amounts of CAT transcripts and the genomic RNA analogues were reduced when the N was unphosphorylated. The transcription and replication activities ranged from high to low in the following order: wt N and N with mutations from S to E, S to D, and S to A. The amounts of CAT transcripts in the cells transfected with pRN-SA, pRN-SD, or pRN-SE were 17, 63, and 85% of that in cells expressing wt N (pRN), respectively. The amounts of genomic analogues in the cells transfected with pRN-SA, pRN-SD, or pRN-SE were 12, 55, and 95% of the amount in cells expressing wt N (pRN), respectively, demonstrating that the synthesis of both viral transcripts and genomic analogues in the minigenome system is affected by N phosphorylation.
FIG. 3.
Rabies virus N phosphorylation affects both viral RNA transcription and replication in the minigenome system. BSR cells infected and transfected as described for Fig. 1 were harvested for RNA or protein analysis. mRNA was purified from total RNA, and genomic analogues were purified from immunoprecipitated RNP with polyclonal anti-N antibodies. Northern blots prepared from these two types of RNA preparations were hybridized with a CAT cDNA probe to assay mRNA transcripts (CAT mRNA) and genomic RNA (both genomic and antigenomic) analogues. [35S]methionine- or [32P]phosphoric acid-labeled N protein expressed in the cells was immunoprecipitated with polyclonal anti-N antibodies and analyzed by SDS-PAGE.
To exclude the possibility that the reduced transcription and replication in the cells transfected with mutant N-expressing plasmids were caused by different levels of N synthesized, transfected cells were labeled with [35S]methionine and lysed with RIPA buffer. The labeled N was immunoprecipitated with anti-N antibodies and analyzed by SDS-PAGE. As shown in Fig. 3, similar amounts of N and mutant N were immunoprecipitated in the cells transfected with different N constructs. To confirm that the N mutants are indeed not phosphorylated, transfected cells were labeled with [32P]phosphoric acid. After lysis with RIPA buffer, the labeled N was immunoprecipitated with anti-N antibodies and analyzed by SDS-PAGE. Only the wt N was phosphorylated, whereas all the mutant N proteins were not (Fig. 3).
Construction and selection of mutant rabies viruses.
In the minigenome system, viral proteins necessary for viral transcription and replication were synthesized by T7 polymerase, and thus their synthesis was not under the control of rabies virus regulatory machinery. Therefore, it was necessary to determine the effects of rabies virus N phosphorylation on viral transcription and replication in the full-length infectious virus. To this end, we introduced mutations of S 389 on the N to A, D, and E into the full-length infectious clone (18). After transfection of these clones into BSR cells, virus L16 wt as well as mutant viruses L16D and L16E were obtained. However, L16A was not rescued in BSR cells. Therefore, we used BSR T7/5 cells (4) for selection of L16A, and L16A was successfully rescued. Reverse transcription-PCR and direct sequencing confirmed that these mutant viruses contained the desired mutations. The genomic RNA of the wt virus (L16) retained the codon for S (UCU) at position 389, whereas the genomic RNAs from L16A, L16D, and L16E viruses have the S (UCU) replaced with A (GCU), D (GAU), and E (GAA), respectively. To confirm that the mutant viruses express unphosphorylated N, BSR cells infected with each of the viruses were labeled with either [35S]methionine or [32P]phosphoric acid and subjected to immunoprecipitation and PAGE analysis. As for the minigenome system, [35S]methionine-labeled N was detected in BSR cells infected with each of the viruses, whereas [32P]phosphoric acid-labeled N was detected only in BSR cells infected with L16 (data not shown), indicating that mutation of serine at position 389 abolishes N phosphorylation in the full-length infectious virus.
The mutant rabies viruses replicate more slowly than the wt virus.
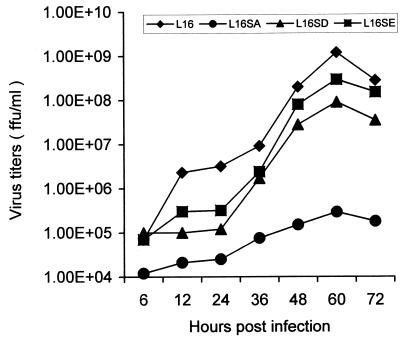
Initially, wt rabies virus (L16) and mutant viruses L16D and L16E grew to high titers (>107 FFU/ml) in BSR cells at 37°C, but L16A grew poorly (titers of <104 FFU/ml). To overcome this difficulty, L16A was propagated in BSR cells at 31°C as described for mutant VSV (26), and L16A grew to higher titers (>105 FFU/ml). The growth curves of the wt and mutant rabies viruses were investigated by infecting BSR cells at 37°C with each of the viruses at a MOI of 1 FFU per cell. As shown in Fig. 4, the wt virus (L16) consistently grew better than the mutant viruses and reached a titer of more than 109 FFU/ml at the peak of virus production (60 h). At this time, the rate of growth for the mutant viruses lagged behind, particularly that of L16A. Its yield was only 105 PFU/cell, at least 4 log units (10,000 times) lower than that of the wt virus. These data indicate that N phosphorylation promotes virus production, possibly through regulation of rabies virus transcription and replication.
FIG. 4.
Virus growth curves. BSR cells were infected with wt (L16) and mutant virus (L16A, L16D, or L16E) at a MOI of 1 FFU/cell, and virus aliquots were removed at indicted time points and subjected to virus titration as described in Materials and Methods.
N phosphorylation modulates both viral transcription and replication in the full-length infectious rabies virus.
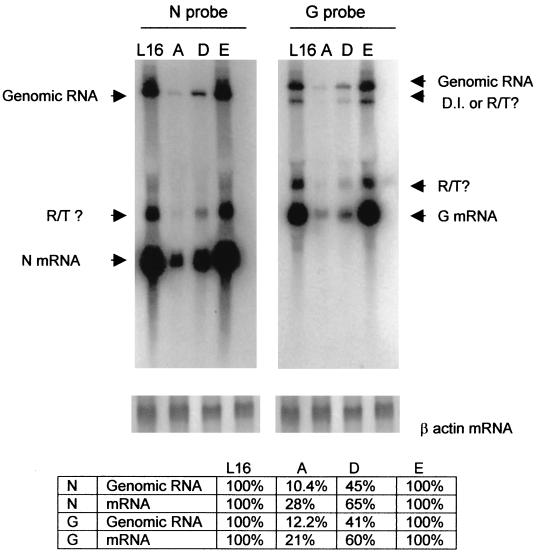
The growth curve data presented in Fig. 4 indicate that growth of the mutant viruses with unphosphorylated N, particularly mutant virus L16A, was severely reduced. This could result from the inhibition of viral transcription or replication or both. To investigate if both viral transcription and replication are affected in the mutant infectious rabies viruses, BSR cells were infected at a MOI of 1 with wt virus L16 as well as mutant viruses L16A, L16D, and L16E. Total RNA isolated 40 h postinfection was analyzed by Northern blot hybridization using probes made from both the N (10) and the G cDNAs (11). These probes can distinguish the N and G transcripts (1.4 and 1.8 kb) from the genomic RNA (12 kb). As illustrated in Fig. 5, each probe detected both genomic RNA and N or G transcripts. In addition, readthrough (RT) transcripts or defective-interfering (DI) RNA or both were also detected. The bands immediately above the N or the G transcripts, when hybridized with either G or N probes, may represent RT transcripts. The band immediately below the genomic RNA, when hybridized with the G probe, may represent an RT transcript (G or L) or DI RNA. For the L16A virus, the amounts of viral genomic RNA and transcripts were 10 to 12% and 21 to 28% of those for the wt virus, respectively. For the L16D virus, the amounts of viral genomic RNA and transcripts were 41 to 45% and 60 to 65% of those for the wt virus, respectively. For the L16E virus, the amounts of viral genomic RNA and transcripts were 100% of those for the wt virus. Generally, the levels of inhibition in viral replication (10 to 12% and 41 to 54%) were more severe than those of inhibition in transcription (21 to 28% and 60 to 65%) in cells infected with L16A and L16D viruses. These observations, together with the data obtained from the minigenome system, indicate that unphosphorylated N results in reduction of viral replication. In the minigenome system viral replication is not dependent on viral transcription because N transcription was under control of T7 polymerase and the level of unphosphorylated N was similar to that of the wt N. Yet the amounts of genomic RNA are reduced when the N is not phosphorylated (Fig. 2), demonstrating that N phosphorylation is required for optimal viral replication.
FIG. 5.
Detection of viral transcripts and genomic RNA with either rabies virus N or G probe. Total RNA was prepared from BSR cells infected with L16, L16A (A), L16D (D), or L16E (E) and was hybridized with the N probe (left) or the G probe (right). The respective mRNAs, the genomic RNA, and the possible RT transcripts and DI RNA are also indicated. The total RNA was also hybridized with a β actin probe (bottom). The amounts of N and G transcripts and genomic RNA products in relation to those for the wt virus were quantitated by densitometry.
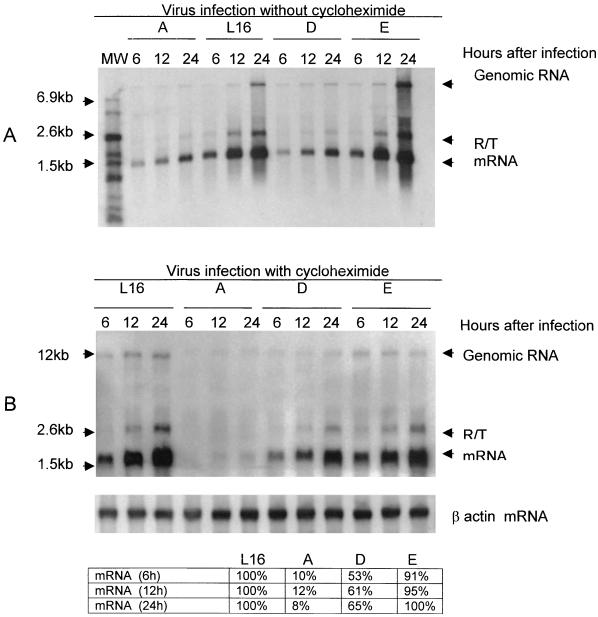
To further determine if N phosphorylation also affects viral transcription, we sought to uncouple the transcription process from the replication process by inhibiting de novo protein synthesis in the infected cells with CHX. BSR cells were infected with each of the viruses at a MOI of 3 FFU per cell, and at 1 h after infection CHX was added to the culture medium at a final concentration of 150 μg/ml to completely inhibit protein synthesis (3). Infected cells without treatment with CHX were included as controls. At 6, 12, and 24 h after infection, cells were harvested and RNA was extracted for Northern blot hybridization with the rabies virus N probe. Without CHX, all the viruses replicated because the amounts of genomic RNA increased for each of the viruses as a function of time (Fig. 6A). The efficiency in viral transcription and replication for each of the viruses was similar to that shown in Fig. 5 at each time point. With the addition of CHX to the culture medium, virus replication was inhibited because the amount of genomic RNA did not increase for any of the viruses at each of the time points (Fig. 6B). Furthermore, rabies virus N protein synthesis was inhibited in BSR cells treated with CHX (data not shown). However, the amounts of transcripts for each of the viruses increased during the same period of time. Quantitation of the transcripts at each time point for each of the viruses in relation to the amount for wt L16 indicated that the transcription efficiencies for L16A, L16D, and L16E were 8 to 12%, 53 to 65%, and 91 to 100%, respectively, of that for wt virus L16. The results demonstrate definitively that unphosphorylated N, except the N with mutation from S to E, also inhibits viral transcription.
FIG. 6.
N phosphorylation also modulates viral transcription. BSR cells were infected with L16, L16A (A), L16D (D), or L16E (E); treated 1 h later either without (A) or with (B) CHX; and harvested at the indicated time points for total-RNA isolation. The RNA was hybridized with an N probe. The respective mRNAs, the genomic RNA, and the possible RT transcripts are also indicated. The total RNA was also hybridized with a β actin probe (bottom). The amounts of N transcripts in relation to that for the wt virus were quantitated by densitometry.
Unphosphorylated N did not alter the ratio between the genomic RNA and the antigenomic RNA.
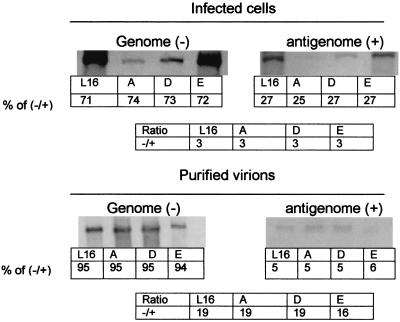
We previously showed that unphosphorylated N bound more strongly to the leader RNA than the phosphorylated N (28). Therefore, it is possible that N phosphorylation plays a role in viral transcription and replication by encapsidating more antigenomic RNA than genomic RNA. This is because the unphosphorylated N, due to its stronger binding to the antigenomic RNA (28), the first step in viral replication, may reduce the synthesis of the progeny negative-strand genomic RNA, thus changing the ratio of genomic to antigenomic RNA. To test this hypothesis, total RNA extracted from BSR cells infected with the wt or mutant viruses were subjected to Northern blot hybridization with sense (transcribed by T7 polymerase) or antisense (transcribed by SP6 polymerase) riboprobes made from the pRN template. RNA was also prepared from virions purified from BSR cells infected with each of the viruses. The ratios of genomic RNA to antigenomic RNA in the infected cells for the wt and mutant viruses were roughly similar (3:1) (Fig. 7). The ratios for these viruses found in the purified virions were also similar (16:1 to 19:1). These data indicate that N phosphorylation probably does not affect the encapsidation and packaging of the genomic RNA and the antigenomic RNA.
FIG. 7.
Quantitation of the ratio between genomic RNA and antigenomic RNA in the infected cells and purified virions. Total RNAs from infected cells or purified virions were hybridized with either the sense probe or the antisense riboprobes prepared from pRN by in vitro transcription. The levels of genomic and antigenomic RNA were determined by densitometry.
DISCUSSION
The major finding of the present study, i.e., that both rabies virus transcription and replication are reduced when the phosphorylated serine of the N is mutated, has been demonstrated both in the minigenomic system and with infectious viruses. When N was unphosphorylated, the rates of transcription and replication were reduced as much as 10-fold compared to those with phosphorylated N. The viral yield for these mutant viruses was reduced as much as 10,000-fold, particularly when the phosphorylated S was mutated to A. These data suggest that phosphorylation of rabies virus N, although not absolutely necessary, is important in modulation of rabies virus transcription and replication. Furthermore, interpretation of the data also suggests that the effects of phosphorylation of rabies virus N on viral transcription and replication are due to a combination of the structure of the serine and the net negative charge of the phosphate moiety. Mutation of the phosphorylated serine to neutral amino acids A and G reduced viral transcription and replication by as much as 10-fold. Alternatively, mutation of the phosphoserine to D or E, both of which contain acidic side chains that are negatively charged at physiological pH, restored the viral transcription and replication activities to more than 60 and 80% of those with phosphorylated N. However, when the S was mutated to N or Q, both of which have structures similar to those of D and E but lack the negative charge, viral transcription activities were reduced by at least one-third but were still higher than those resulting from S to A or S to G mutations. Furthermore, the fact that mutant virus L16A grow better at 31°C than at 37°C indicates the temperature sensitivity of the mutant virus. These results suggest that both the structure of the amino acid and the net negative charge of the phosphate moiety are important for viral transcription and replication.
Rabies virus N, like its counterpart in VSV, plays vital roles in regulating viral RNA transcription and replication by encapsidating de novo-synthesized viral genomic RNA (23, 25, 27). The fact that rabies virus N, but not VSV N, is phosphorylated has raised questions as to how the phosphorylation is involved in the regulation of rabies virus RNA transcription and replication (27). One possible scenario is that N phosphorylation results in a switch from the viral transcription mode to the replication mode. If that is the case, then unphosphorylated N would not affect or might even stimulate viral transcription but could inhibit viral replication. Thus, it is important for us to address whether N phosphorylation affects viral transcription, replication, or both. Quantitation of viral transcripts and genomic RNA in the minigenome and the infectious virus indicated that both viral transcript and genomic RNA (or analogue) levels were reduced when N was not phosphorylated (Fig. 2 and 5). However, these data do not necessarily mean that phosphorylated N favors both viral transcription and replication because of the inherent complexity of viral transcription and replication in the infected cells (23). Reduction in viral genome replication results in fewer templates for transcription, and this likely decreases the accumulation of viral mRNA. On the other hand, reduction in transcription reduces the N pool and eventually leads to reduction in replication. Nevertheless, it could be concluded from the data, particularly those for the minigenome system, that phosphorylated N favors viral replication. We have observed that viral replication is reduced when the N is not phosphorylated in the minigenome system (Fig. 2) despite the fact that, in the minigenome system, viral replication is not dependent on viral transcription because N transcription is under the control of T7 polymerase. Indeed, immunoprecipitation indicated that the level of unphosphorylated N was similar to that of the wt N in the minigenomic system.
To further demonstrate if and how N phosphorylation also affects viral transcription, we uncoupled viral transcription from viral replication by inhibiting protein synthesis by treating infected cells with CHX. Viral replication is dependent on the de novo synthesis of viral N protein while transcription is not (25). Under these conditions, viral replication was reduced but transcription was not (Fig. 6), which allowed us to assess the effects of N phosphorylation on viral transcription independent of viral replication. Our data demonstrate that N phosphorylation also modulates viral transcription because viral transcription was inhibited by almost 90% when N was not phosphorylated, particularly when the phosphorylated S was mutated to A. Thus our data demonstrate that N phosphorylation favors both viral transcription and replication.
As in all the single-stranded negative-sense RNA viruses, in rabies virus the RNP complex is the infectious unit (27). The complicated interaction between the components within the RNP complex brings about rabies virus transcription and replication (23, 25). Previously, we demonstrated that dephosphorylated rabies virus N encapsidated more leader RNA than phosphorylated N (28). Mutation of the serine at position 389 of the rabies virus N to alanine also resulted in increased binding to leader RNA in comparison to that for wt N. In the present study, we demonstrated that N phosphorylation affects both viral transcription and replication. It is thus possible that strong binding of unphosphorylated N to RNA may prevent L from gaining access to the genomic RNA to initiate viral transcription and replication. Although N remains bound to genomic RNA during the transcription and replication processes through the phosphate backbone (8), the template-associated N has to unfold transiently so that L can gain contact with the template RNA (1). We propose that N phosphorylation weakens the interaction between N and genomic RNA and therefore enables the L to gain access to and bind the RNA template to initiate transcription and replication. This hypothesis is supported by our data for both the minigenomic system and the infectious virus. When the phosphorylated S was mutated to neutral amino acids A and G, viral transcription and replication were reduced the most. When the phosphorylated serine was mutated to the negatively charged D or E, transcription and replication activities were restored to more than 60 to 90% of the levels with wt N. The amounts of transcription and replication products in cells infected with mutant virus L16E (S changed to E) were essentially equivalent to those in cells infected with the wt virus L16. This occurred even though the virus production in cells infected with L16E was slightly less than that in cells infected with L16. Because the genomic RNA is the template for both transcription and replication (23), it is conceivable that phosphorylated N facilitates the initiation of both transcription and replication.
Because N phosphorylation affects its efficiency for encapsidating leader RNA (28), it is possible that N phosphorylation leads to encapsidation of more antigenomic than genomic RNA (25). Unphosphorylated N, by strongly binding to the antigenomic RNA, the first step in viral replication, may reduce the synthesis of the genomic RNA. We determined the ratio between genomic RNA and antigenomic RNA in the infected cells as well as in the purified virions by Northern hybridization, using sense or antisense riboprobes. The ratio between genomic RNA and antigenomic RNA remained constant in cells (approximately 3:1) infected with the wt virus or with one of the mutant viruses. The ratios between genomic and antigenomic RNA in the purified virions from all of the viruses were also similar (approximately 20:1). The ratio of the genomic RNA to antigenomic RNA measured in this study was different from that (50:1) reported previously (9). The discrepancy may be due to the methods of quantitation used in these two studies. The RNA was quantitated by densitometry in the present study, whereas the RNA was quantitated by phosphorimaging in the previously reported study (9). Nevertheless, the data reported in this study indicate that N phosphorylation does not affect the encapsidation of either the genomic or the antigenomic RNA.
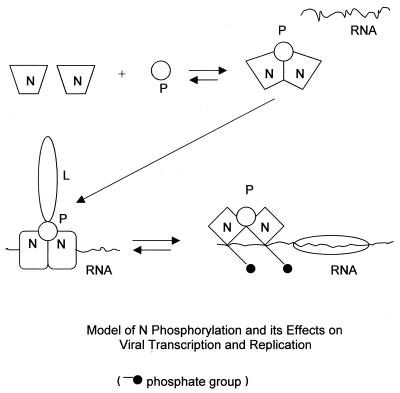
Recently, Kawai et al. (15) reported that phosphorylated N is detected only in the nucleocapsid, whereas N in the free-N pool (mostly in the N-P complex) is not phosphorylated. Based on that study and our data presented in this paper, a model to explain how rabies virus N is phosphorylated and how phosphorylation of rabies virus N modulates viral transcription and replication emerges (Fig. 8). N is not phosphorylated as free N or as part of the N-P heterocomplex, possibly because of its conformation. It is possible that the phosphorylation site is buried at this stage. It is advantageous for the N not to be phosphorylated before encapsidating genomic RNA because unphosphorylated N has higher affinity for genomic RNA than does phosphorylated N (28). The interaction (encapsidation) of genomic RNA with the N-P complex may induce conformational changes of N, enabling N to interact with kinase or expose serine 389 for phosphorylation or both. Phosphorylation of N in turn may affect the interaction between N and the genomic RNA. Following phosphorylation, the charge repulsion between the negatively charged phosphoserine and the negatively charged RNA may weaken the interaction between N and RNA. This could enable L to gain access and bind to genomic RNA, thereby initiating viral RNA transcription and replication. Evidence that supports this model comes from studies showing that unphosphorylated N binds to genomic RNA more strongly than does phosphorylated N (28) and that the phosphorylation site (residue 389) is close to the putative RNA-binding domain (residues 289 to 352) (16). Unphosphorylated N, because of tighter binding with RNA, could prevent the L from gaining access to the genome template. Consequently, the efficiency of viral RNA transcription and replication is reduced.
FIG. 8.
Proposed model of N phosphorylation and its function in viral transcription and replication. N, once synthesized, interacts with P and/or L. At this stage, the N is not phosphorylated. It is possible that through the interaction of N with (encapsidation of) genomic RNA, the N goes through conformational changes, which expose the site for phosphorylation. Once the N is phosphorylated, the charge repulsion between the genomic RNA and N helps the L gain access to the genomic template for the initiation of viral RNA transcription and replication.
Acknowledgments
This work was supported partially by Public Health Service grant AI-51560 (Z.F.F.) from the National Institute of Allergy and Infectious Diseases and a grant from Ft. Dodge Animal Health Laboratories.
We thank Klaus Conzelmann for supplying plasmids expressing the minigenome and the full-length infectious virus, Bernie Moss for the recombinant vaccinia virus vTF7-3, and Kathy Spindler and Bruce Seal for critically reading the manuscript.
REFERENCES
- 1.Banerjee, A. K., and D. Chattopadhyay. 1990. Structure and function of the RNA polymerase of vesicular stomatitis virus. Adv. Virus Res. 38:99-124. [DOI] [PubMed] [Google Scholar]
- 2.Barr, J., C. R. Chambers, C. R. Pringle, and A. J. Easton. 1991. Sequence of the major nucleocapsid protein gene of pneumonia virus of mice: sequence comparisons suggest structural homology between nucleocapsid proteins of pneumoviruses, paramyxoviruses, rhabdoviruses and filoviruses. J. Gen. Virol. 72:677-685. [DOI] [PubMed] [Google Scholar]
- 3.Boudinot, P., S. Salhi, M. Blanco, and A. Benmansour. 2001. Viral haemorrhagic septicaemia virus induces vig-2, a new interferon-responsive gene in rainbow trout. Fish Shellfish Immunol. 11:383-397. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conzelmann, K.-K., J. Cox, L. G. Schneider, and H.-J. Theil. 1990. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. J. Virol. 175:484-499. [DOI] [PubMed] [Google Scholar]
- 6.Conzelmann, K.-K., and M. Schnell. 1994. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J. Virol. 68:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietzschold, B., M. Lafon, H. Wang, L. Otvos, E. Celis, W. H. Wunner, and H. Koprowski. 1987. Localization and immunological characterization of antigenic domains of rabies virus internal N and NS proteins. Virus Res. 8:103-125. [DOI] [PubMed] [Google Scholar]
- 8.Emerson, S. U. 1982. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31:635-642. [DOI] [PubMed] [Google Scholar]
- 9.Finke, S., and K. K. Conzelmann. 1997. Ambisense gene expression from recombinant rabies virus: random packaging of positive- and negative-strand ribonucleoprotein complexes into rabies virions. J. Virol. 71:7281-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu, Z. F., B. Dietzschold, C. L. Schumacher, W. H. Wunner, H. C. J. Ertl, and H. Koprowski. 1991. Rabies virus nucleoprotein expressed in and purified from insect cells is efficacious as a vaccine. Proc. Natl. Acad. Sci. USA 88:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, Z. F., R. Rupprecht, B. Dietzschold, P. Saikumar, H. S. Niu, I. Babka, W. H. Wunner, and H. Koprowski. 1993. Oral vaccination of raccoons (Procyon lotor) with baculovirus-expressed rabies virus glycoprotein. Vaccine 11:925-928. [DOI] [PubMed] [Google Scholar]
- 12.Fu, Z. F., E. Wickstrom, M. Jiang, S. Corisdeo, J. Yang, B. Dietzschold, and H. Koprowski. 1996. Inhibition of rabies virus infection by an oligodeoxynucleotide complementary to rabies virus genomic RNA. Antisense Nucleic Acid Drug Dev. 6:87-93. [DOI] [PubMed] [Google Scholar]
- 13.Fu, Z. F., Y. M. Zheng, W. H. Wunner, H. Koprowski, and B. Dietzschold. 1994. Both the N- and C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology 200:590-597. [DOI] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai, A., H. Toriumi, T. S. Tochikura, T. Takahashi, Y. Honda, and K. Morimoto. 1999. Nucleocapsid formation and/or subsequent conformational change of rabies virus nucleoprotein (N) is a prerequisite step for acquiring the phosphatase-sensitive epitope of monoclonal antibody 5-2-26. Virology 263:395-407. [DOI] [PubMed] [Google Scholar]
- 16.Kouznetzoff, A., M. Buckle, and N. Tordo. 1998. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J. Gen. Virol. 79:1005-1013. [DOI] [PubMed] [Google Scholar]
- 17.Schnell, M. J., H. D. Foley, C. A. Siler, J. P. McGettigan, B. Dietzschold, and R. J. Pomerantz. 2000. Recombinant rabies virus as potential live-viral vaccines for HIV-1. Proc. Natl. Acad. Sci. USA 97:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnell, M. J., T. Mebatsion, and K.-K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith, J. S., D. B. Fishbein, C. S. Rupprecht, and K. Clark. 1991. Unexplained rabies in three immigrants in the United States. A virological investigation. N. Engl. J. Med. 324:205-211. [DOI] [PubMed] [Google Scholar]
- 20.Sokol, F., and H. F. Clark. 1973. Phosphoproteins, structural components of rhabdoviruses. Virology 52:246-263. [DOI] [PubMed] [Google Scholar]
- 21.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tordo, N., O. Poch, A. Ermine, and G. Keith. 1986. Primary structure of leader RNA and nucleoprotein genes of rabies genome: segmented homology with VSV. Nucleic Acids Res. 14:2671-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner, R. R., and J. K. Rose. 1996. Rhabdoviridae: the viruses and their replication, p. 1121-1136. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 24.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 25.Wertz, G. W., N. L. Davies, and J. Patton. 1987. The role of proteins in vesicular stomatitis virus RNA replication, p. 271-296. In R. R. Wagner (ed.), The rhabdoviruses. Plenum Press, New York, N.Y.
- 26.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wunner, W. H. 1991. The chemical composition and molecular structure of rabies viruses, p. 31-67. In G. M. Baer (ed.), Natural history of rabies, 2nd ed. CRC Press, Inc., Boca Raton, Fla.
- 28.Yang, J., H. Koprowski, B. Dietzschold, and Z. F. Fu. 1999. Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J. Virol. 73:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]