Abstract
The envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) function as a trimer composed of three gp120 exterior glycoproteins and three gp41 transmembrane proteins. Soluble gp140 glycoproteins composed of the uncleaved ectodomains of gp120 and gp41 form unstable, heterogeneous oligomers, but soluble gp140 trimers can be stabilized by fusion with a C-terminal, trimeric GCN4 motif (X. Yang et al., J. Virol. 74:5716-5725, 2000). To understand the influence of the C-terminal trimerization domain on the properties of soluble HIV-1 envelope glycoprotein trimers, uncleaved, soluble gp140 glycoproteins were stabilized by fusion with another trimeric motif derived from T4 bacteriophage fibritin. The fibritin construct was more stable to heat and reducing conditions than the GCN4 construct. Both GCN4- and fibritin-stabilized soluble gp140 glycoproteins exhibited patterns of neutralizing and nonneutralizing antibody binding expected for the functional envelope glycoprotein spike. Of note, two potently neutralizing antibodies, immunoglobulin G1b12 and 2G12, exhibited the greatest recognition of the stabilized, soluble trimers, relative to recognition of the gp120 monomer. The observed similarities between the GCN4 and fibritin constructs indicate that the HIV-1 envelope glycoprotein ectodomains dictate many of the antigenic and structural features of these fusion proteins. The melting temperatures and ligand recognition properties of the GCN4- and fibritin-stabilized soluble gp140 glycoproteins suggest that these molecules assume conformations distinct from that of the fusion-active, six-helix bundle.
Human immunodeficiency virus type 1 (HIV-1) encodes a 160-kDa envelope glycoprotein (gp160) precursor, which is proteolytically cleaved into the exterior (gp120) and transmembrane (gp41) glycoproteins (1, 21, 34). The gp120 glycoprotein remains associated with the mature envelope glycoprotein complex through a noncovalent interaction with the gp41 ectodomain (44). The HIV-1 envelope glycoprotein complex consists of three gp120 and three gp41 subunits and is anchored in the viral or infected cell membrane by the gp41 transmembrane region (22, 29, 33, 44). As the sole HIV-1 components exposed on the virion surface, the envelope glycoproteins represent the only realistic viral target for vaccine-induced neutralizing antibody responses. Monomeric HIV-1 gp120 and derivatives were initially considered to be principal vaccine candidates. However, HIV-1 gp120 has repeatedly proven to be an ineffective immunogen in eliciting neutralizing antibodies against clinical HIV-1 isolates (4, 5, 7, 12, 30, 43, 47). Few of the antibodies raised by gp120 monomers effectively bind assembled HIV-1 envelope glycoprotein trimers (36, 37). Therefore, in an attempt to better elicit such antibodies, candidate HIV-1 envelope glycoproteins that mimic the functional trimer have been sought. Initial efforts to express HIV-1 glycoprotein oligomers disrupted the proteolytic cleavage site between gp120 and gp41 and deleted the transmembrane region and intracytoplasmic tail of gp41 (6, 19, 20, 42). The resulting soluble gp140 products do form oligomers. However, such oligomers are invariably quite heterogeneous and are composed of dimers and other higher-order forms. Studies have shown that these soluble gp140 “oligomers” do not exhibit improved immunogenicity compared with that of the gp120 monomer. Efforts to prepare more homogeneous oligomers from these mixtures by biophysical and biochemical means have produced only limited improvements in the immunogenicity of these proteins (3). Moreover, the inefficiency of such approaches largely precludes their practical use. Fusing a GCN4 trimeric motif to the C-terminal end of the gp41 ectodomain, along with disruption of the proteolytic cleavage site between gp120 and gp41, can promote the production of stable, soluble gp140 trimers that appear to be homogeneous (48, 49). Our previous results have shown that these trimers exhibit an antigenic profile similar to that expected of the HIV-1 envelope glycoprotein spike. The GCN4-stabilized HIV-1 envelope glycoprotein trimers elicited neutralizing antibodies more effectively than gp120 monomers (50).
During virus attachment to the target cell, gp120 interacts sequentially with the host cell receptors, CD4, and the chemokine receptors (2, 11, 13, 14, 16, 17, 28, 31, 41). Receptor binding is thought to trigger conformational changes in the envelope glycoprotein complex that eventually promote the fusion of the viral and target cell membranes by the gp41 glycoprotein. The N terminus of gp41 contains a hydrophobic “fusion peptide,” which is thought to insert into the target cell membrane, and an N36 region, which can form a trimeric coiled coil (9, 10, 25, 31, 39, 45). Structures of gp41 ectodomain segments indicate that a gp41 region (designated C34) near the viral membrane-spanning domain can form a helix that packs into the grooves of the N36 coiled coil (10, 39, 45). The formation of this six-helix bundle (“the fusion-active conformation”) is believed to provide the energy necessary to approximate the viral and target cell membranes. The ability of C34 peptides to block HIV-1 envelope glycoprotein-mediated fusion suggests that, in the prefusogenic envelope glycoprotein complex, gp41 exists in a conformation other than that of the six-helix bundle (23, 27, 46). Structural details of this prefusogenic conformation are lacking. The utility of soluble, stabilized gp140 trimers in investigating structural, biochemical, and immunological features of the functional HIV-1 envelope glycoprotein complexes is dependent upon the degree to which they accurately resemble the prefusogenic entity or entities.
Previously, because our studies were limited to soluble gp140 trimers stabilized by the trimeric GCN4 motif, the effect of the C-terminal GCN4 sequences on the conformation of the envelope glycoprotein portions of the construct could not be readily assessed. Since membrane fusion-related conformational transitions in the gp41 ectodomain may involve the formation of new helical structures (26, 45), we were concerned about the possibility that the GCN4 coiled coil might drive the formation of helices in the adjacent gp41 segments, thereby promoting fusion-active conformations in these soluble trimers. The fibritin carboxy-terminal domain assumes a globular configuration and is able to promote the trimerization of heterologous proteins (24, 40). Moreover, the hydrophilic surface of the fibritin C-terminal trimer should improve the solubility characteristics of the gp140(−/GCN4) trimers, which exhibit some aggregation at high concentrations (40). Here we fused the carboxy-terminal domain of T4 bacteriophage fibritin to the C terminus of the cleavage-defective gp140 envelope ectodomains, by using a design analogous to that used to engineer the gp140(−/GCN4) trimers. We compared the antigenic and biochemical properties of the GCN4- and fibritin-stabilized trimers, hoping to gain insights into the properties of the soluble gp140 trimers that are intrinsic to the HIV-1 glycoprotein components of the engineered constructs.
Stabilization of soluble HIV-1 gp120 glycoproteins by fusion with the fibritin trimeric motif.
All of the soluble gp140 glycoproteins used in this study were derived from the YU2 primary R5 HIV-1 isolate, consist of the complete gp120 and gp41 ectodomains, and contain alterations in the gp120/gp41 proteolytic cleavage site (arginines 508 and 511 to serines) (Fig. 1A). The gp140(−) glycoprotein has been shown to form only a small proportion of relatively unstable oligomers, most of which are dimers (20). In contrast, the gp140Δ683(−/GCN4) glycoprotein is almost exclusively trimeric (49). To compare the properties of GCN4-stabilized gp140 trimers with those of gp140 stabilized by another means, we employed a trimerization domain from the C terminus of bacteriophage T4 fibritin. In the gp140Δ683(−/FT) construct, the fibritin motif (GYIPEAPRDGQAYVRKDGEWVLLSTFL) replaces the trimeric GCN4 motif in the gp140Δ683(−/GCN4) protein. HIV-1 envelope glycoprotein genes were cloned into the pSVIIIenv expression vector (38), and the open reading frames encoding HIV-1 envelope proteins were sequenced to verify the presence of the designed changes and the absence of unintended mutations.
FIG. 1.
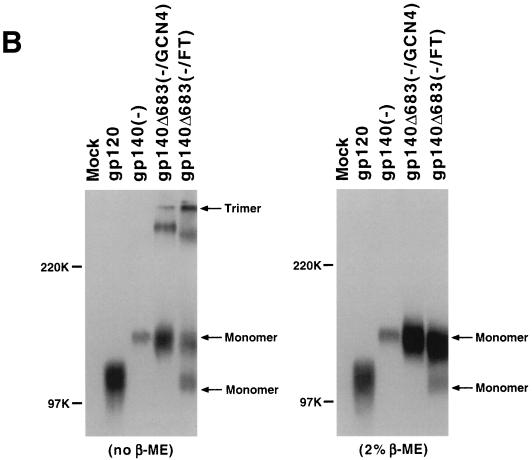
The fibritin trimeric (FT) motif stabilizes uncleaved HIV-1 gp140 glycoprotein trimers. (A) The wild-type HIV-1 gp160 glycoprotein is represented at the top, with the helical N36 and C34 regions and transmembrane (TM) region noted. The gp120, gp140(−), and gp140Δ683(−/GCN4) constructs have been previously described (48-50). The gp140Δ683(−/FT) glycoprotein contains the first 683 amino acids of the HIV-1 YU2 envelope glycoprotein with the arginines at positions 508 and 511 altered to serines (represented by SS). Two glycine residues and the 27-amino-acid trimeric motif from T4 bacteriophage fibritin (FT) immediately follow lysine 683 of the envelope glycoprotein in the gp140Δ683(−/FT) construct. (B) 293T cells in 100-mm tissue culture plates were cotransfected with 9 μg of plasmid DNA expressing the soluble envelope glycoproteins and 1 μg of an HIV-1 Tat expression plasmid by using the Lipofectamine Plus kit (Gibco/Life Technologies, Inc.). The proteins were radiolabeled with 200 μCi of [35S]methionine-cysteine for ca. 24 h in 5 ml of labeling medium and then precipitated from 500 μl of the radiolabeled culture medium with 3 μl of pooled sera from HIV-1-infected individuals and 50 μl of protein A-Sepharose (Pharmacia) at room temperature for 3 h. After three washes with 0.5 M NaCl-PBS, the immunoprecipitated proteins were resolved on an SDS-7.5% polyacrylamide gel after being boiled in sample buffer without β-ME (left panel) or with 2% β-ME (right panel). The positions of the molecular weight markers are shown on the left, and the positions of the trimeric and monomeric HIV-1 glycoproteins were deduced by comparison with previously characterized oligomers (48-50). (C) The 35S-labeled glycoproteins prepared as in panel B were concentrated two- to fivefold by using a Centriprep-30 filter (Amicon). Approximately 750 μl of the concentrated sample was loaded onto a 10 to 25% continuous sucrose gradient, which wascentrifuged in a Beckman SW41 rotor at 40,000 rpm for 20 h at 4°C. Ten 1.1-ml fractions were collected manually from the bottom of the gradient, and the glycoproteins were immunoprecipitated by pooled sera from HIV-1-infected individuals. The precipitates were resolved on an SDS-7.5% polyacrylamide gel after being boiled in sample buffer containing 2% β-ME.
Although large quantities of the purified soluble gp140 glycoproteins were not yet available, sufficient amounts of protein for analysis were produced by transient expression in 293T cells. The soluble gp140 glycoproteins, as well as a control gp120 glycoprotein, were expressed in 293T cells as described in a previous report (48). After cotransfection with the pSVIIIenv expression vector and an HIV-1 Tat expression plasmid, 293T cells were labeled with [35S]cysteine-methionine for ca. 24 h, and the radiolabeled proteins were precipitated from cell supernatants by pooled sera from HIV-1-infected individuals. After the precipitates were boiled in Laemmli sample buffer, the proteins were analyzed under nonreducing conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1B, left panel). Compared to the gp120 and gp140(−) glycoproteins, which migrated as monomers under these conditions, monomeric, dimeric, and trimeric forms of the gp140Δ683(−/GCN4) glycoprotein were evident, as expected (49). Gel-stable oligomers of the gp140Δ683(−/GCN4) glycoprotein have been shown by cross-linking, sucrose density gradient analysis, and molecular exclusion chromatography to be trimers (49). With the use of the migration of the previously characterized gp120, gp140(−) and gp140Δ683(−/GCN4) proteins as benchmarks, the gp140Δ683(−/FT) protein appeared as trimers, dimers, and monomers under these conditions. The gp140Δ683(−/FT) trimer was more abundant than that of the gp140Δ683(−/GCN4) protein. An additional band of ca. 120 kDa, which comigrated with monomeric gp120 and therefore likely represents a product of proteolytic cleavage at a point close to the original gp120/gp41 cleavage site, was evident in the gp140Δ683(−/FT) lane. The appearance of a gp120 form in a construct containing a modification of the primary proteolytic cleavage site between gp120 and gp41 is not uncommon, as less efficiently utilized secondary cleavage sites immediately N terminal to the primary site have been documented (18, 32). As expected (48), no higher-order forms of this gp120 product were apparent.
The precipitates were also boiled under reducing conditions (2% β-mercaptoethanol) and analyzed (Fig. 1B, right panel). The gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins migrated as monomeric proteins under these conditions. A major band corresponding to the unprocessed protein and a minor, faster-migrating 120-kDa band were seen for the gp140Δ683(−/FT) construct.
The gp120 and soluble gp140 glycoproteins were analyzed by sucrose density gradient centrifugation. The gp120 glycoprotein sedimented primarily in fractions 6 and 7, whereas the gp140Δ683(−/GCN4) glycoprotein appeared predominantly in fractions 3 and 4 (Fig. 1C). The uncleaved form of the gp140Δ683(−/FT) glycoprotein also sedimented primarily in fractions 3 and 4. The small amount of proteolytically processed gp140Δ683(−/FT) protein migrated in fractions 6 and 7, equivalent to the gp120 monomer. It is noteworthy that little or no uncleaved gp140Δ683(−/FT) protein sedimented in fractions 6 and 7, where previous studies indicated that uncleaved soluble gp140 monomers are expected to migrate (49). This suggests that, once one subunit of the trimer is cleaved, the other subunits are also efficiently cleaved. As has been previously observed for HIV-1 and simian immunodeficiency virus (SIV) soluble gp140 envelope glycoproteins, proteolytically processed molecules do not remain associated in stable trimers (49). This point is underscored by our studies of the gp140Δ683(FT) construct, which is identical to the gp140Δ683(−/FT) glycoprotein except that the gp120/gp41 proteolytic cleavage site is wild type in sequence. The gp140Δ683(FT) glycoprotein was efficiently processed and secreted and behaved similarly to a gp120 monomer on sucrose density gradients and nonreducing SDS-polyacrylamide gels (data not shown). Apparently, the lability of the gp120-gp41 associations in the soluble trimers leads to the dissolution of these complexes once proteolytic cleavage occurs; this lability may also underlie the tendency to “shed” gp120 subunits from the mature HIV-1 envelope glycoprotein spikes.
Chemical and thermal stability of soluble envelope glycoprotein trimers.
The relative amounts of the trimeric and monomeric forms of the soluble gp140 glycoproteins observed in Fig. 1B suggested that the gp140Δ683(−/FT) trimer might be more stable than the gp140Δ683(−/GCN4) trimer. To examine this point further, the gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins were eluted from immunoprecipitates into 1× Laemmli buffer at 37°C for 30 min in the presence of differing concentrations of β-mercaptoethanol. The eluted proteins were analyzed on an SDS-polyacrylamide gel (Fig. 2). At 37°C, almost all of the uncleaved gp140Δ683(−/FT) glycoprotein migrated as expected for a trimer, even in β-mercaptoethanol concentrations up to 2%. In contrast, the gp140Δ683(−/GCN4) glycoprotein was only partially trimeric in the absence of β-mercaptoethanol and was fully reduced to the monomeric form in the presence of 1% β-mercaptoethanol. These results suggest that, under the conditions examined, the gp140Δ683(−/FT) trimers are more stable than the gp140Δ683(−/GCN4) trimers.
FIG. 2.
Resistance of the stabilized HIV-1 envelope glycoprotein trimers to reducing conditions. Radiolabeled envelope glycoproteins in the supernatants of transfected 293T cells were immunoprecipitated as in Fig. 1B. After the 0.5 M NaCl-PBS washes, the protein-bead complexes were thoroughly desiccated and then incubated with 50 μl of 1× Laemmli buffer with the designated concentrations of β-ME for 30 min at 37°C to elute the bound envelope proteins. The eluted proteins were then resolved on a SDS-7.5% polyacrylamide gel (Ready-Gel; Bio-Rad).
We took advantage of the stable association of the soluble gp140 trimers at 37°C in the absence of β-mercaptoethanol to evaluate the effects of temperature on the integrity of gel-stable trimers. The gp140Δ683(−/GCN4), gp140Δ683(−/FT), and Δ528 proteins were compared. The Δ528 glycoprotein contains amino acid residues 528 to 679 of the HXBc2 HIV-1 glycoprotein and thus includes most of the gp41 ectodomain (49). The Δ528 glycoprotein forms six-helix bundles that mimic the fusion-active conformation of gp41 and that are extremely resistant to heat and denaturing agents. The Δ528 protein was included to gauge the behavior of authentic six-helix bundles of gp41 in our experimental system. Radiolabeled proteins were immunoprecipitated and eluted into 1× Laemmli buffer without β-mercaptoethanol at 37°C. The protein samples were incubated at increasing temperatures for 30 min before being resolved on SDS-polyacrylamide gels. The trimeric and monomeric forms of each protein were quantitated, and the percentage of gel-stable trimers in each sample at a given temperature of incubation was calculated (Fig. 3). Approximately 73% of the Δ528 protein remained trimeric at 37°C under these experimental conditions, and half of these trimers could withstand temperatures of 97°C. Although our experimental conditions differ from those employed by others studying HIV-1 or SIV gp41 six-helix bundles, the level of thermal resistance that we observed is consistent with the melting temperatures of ca. 86°C reported for these structures (29). The melting curve for the gp140Δ683(−/FT) glycoprotein was biphasic. At 37°C, under our experimental conditions, 83% of the gp140Δ683(−/FT) glycoprotein was trimeric, and this value was reduced by half at temperatures near 50°C. An inflection in the curve was consistently observed in the temperature range of 52 to 58°C, and trimers present in the higher temperature ranges exhibited a melting temperature of 87°C. The gp140Δ683(−/GCN4) protein exhibited only 40% trimers at 37°C, and these further disassociated into monomers at increasing temperatures. The melting curve for the gp140Δ683(−/GCN4) protein also exhibited an inflection in the 50 to 60°C temperature range but did not appear to be biphasic. These results suggest that, at temperatures up to 50°C, a significant portion of the gp140Δ683(−/FT) and gp140Δ683(−/GCN4) trimers are less stable than six-helix bundles and probably assume conformations distinct from that of the fusion-active form.
FIG. 3.
Thermal resistance of trimeric HIV-1 envelope glycoproteins. A 5-ml aliquot of the 35S-labeled HIV-1 envelope glycoproteins was precipitated with 20 μl of pooled sera from HIV-1-infected individuals and 200 μl of protein A-Sepharose for 16 h at 4°C. After three washes with 2 ml of 0.5 M NaCl-PBS, the beads were desiccated thoroughly by using an aspirator. The beads were then incubated with 500 μl of 1× Laemmli buffer without β-ME for 30 min at 37°C with shaking to elute the immunoprecipitated HIV-1 envelope glycoproteins. The supernatants containing the eluted proteins were harvested after centrifugation at room temperature for 5 min at 14,000 rpm by using a microcentrifuge, divided into 15 30-μl aliquots in PCR tubes (Fisher Brand), and kept in a 37°C water bath. The samples were then incubated for 30 min in PCR thermal cyclers (GeneAmp 2400; Perkin-Elmer) prewarmed to the designated temperatures for 30 min in groups of five, and the tubes were immediately returned to the 37°C water bath until SDS-PAGE analysis. After all 15 tubes were treated as described above, 15 μl of each sample was analyzed on an SDS-polyacrylamide gel (Ready-Gel; Gibco/Life Technologies). The Δ528 glycoprotein was analyzed on an SDS-15% polyacrylamide gel, whereas the gp140Δ683(−/FT) and gp140Δ683(−/GCN4) glycoproteins were resolved on SDS-5% polyacrylamide gels. After processing of the gels and autoradiography, the bands corresponding to trimeric and monomeric gp140 of each sample were quantified by using a Storm PhosphorImager (Molecular Dynamics). The percentage of the total monomer and trimer that was represented by the trimer in each sample was calculated and is presented. The results shown are representative of those obtained in four experiments with the gp140Δ683(−/FT) protein and three experiments with the gp140Δ683(−/GCN4) and Δ528 proteins. The multiple experiments yielded almost identical melting curves for each of the three proteins.
CD4 and CCR5 binding by the gp140(−/FT) trimers.
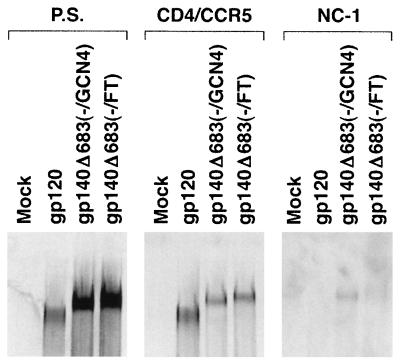
HIV-1 attaches to target cells through the interaction between the gp120 glycoprotein and the cellular CD4 receptor (13, 28, 31). CD4 binding induces structural changes in gp120 that allow binding to the chemokine receptor, normally CCR5 or CXCR4 (2, 11, 14, 16, 17, 41). Previously, we showed that the gp140Δ683(−/GCN4) protein could bind to soluble CD4 and cell surface CCR5, albeit at a reduced level relative to monomeric gp120 (49). Similar experiments were performed to evaluate the ability of the gp140Δ683(−/FT) trimers to bind CD4 and CCR5. Four-milliliter volumes of radiolabeled cell supernatants containing the gp140Δ683(−/FT) and gp140Δ683(−/GCN4) proteins were concentrated ∼10-fold with Centriprep-30 concentrations (Amicon). The concentrated envelope glycoproteins were then incubated with 1 μg of soluble CD4 and 3 × 106 Cf2ThsynCCR5 cells, which express human CCR5 (31), at room temperature for 1 h. After three washes with phosphate-buffered saline (PBS), the cells were lysed, and the bound, 35S-labeled envelope proteins were detected by immunoprecipitation with pooled sera from HIV-1-infected individuals. The gp140Δ683(−/FT) and gp140Δ683(−/FT) trimers bound to the Cf2ThsynCCR5 cells at similar levels, which were slightly reduced compared with that of monomeric gp120 (Fig. 4, middle panel). Similar levels of all three envelope proteins were used in the binding assay, based upon the precipitation of radiolabeled proteins from the unconcentrated cell supernatants by a mixture of sera from HIV-1-infected individuals (Fig. 4, left panel).
FIG. 4.
Recognition of the gp140 trimers by CD4 and CCR5 and by the NC-1 monoclonal antibody. The 35S-labeled envelope glycoproteins in cell supernatants were concentrated 10-fold by using a Centriprep-30 filter and kept on ice. Cf2ThsynCCR5 cells, which stably express high levels of human CCR5 (31), were harvested by treatment with 10 mM EDTA-PBS for 5 min at room temperature. After one wash with PBS, the cells were resuspended in PBS, divided into aliquots in microtubes containing ca. 3 × 106 cells in 400 μl, and incubated with 1 μg of soluble CD4 and 400 μl of the concentrated envelope glycoproteins for 1 h at room temperature. The cells were then washed with PBS three times and lysed with 1 ml of lysis buffer (0.5 M NaCl, 0.1% Triton X-100, 50 mM Tris-HCl [pH 7.5]) with 1× protease inhibitors (Pharmacia) for 30 min at 4°C. The lysates were harvested after centrifugation at 14,000 rpm for 30 min at 4°C. The bound envelope proteins were precipitated for 16 h at 4°C from 400 μl of the above lysates with 3 μl of pooled sera from HIV-1-infected individuals and 50 μl of protein A-Sepharose. After three washes with lysis buffer, the proteins were resolved on an SDS-7.5% polyacrylamide gel after boiling in 1× Laemmli buffer with 2% β-ME (middle panel). In parallel with these studies, the unconcentrated cell supernatants containing the radiolabeled envelope glycoproteins were precipitated by a mixture of sera from HIV-1-infected individuals (left panel) or by the NC-1 monoclonal antibody (right panel).
Antigenicity of the gp120 subunits of the gp140Δ683(−/FT) trimers.
Previous studies (49) showed that the epitopes for virus-neutralizing antibodies are well exposed on the gp140Δ683(−/GCN4) trimers, whereas the epitopes for antibodies with little or no neutralizing ability are less accessible. In contrast, both neutralizing and nonneutralizing antibody epitopes on gp120 and gp140(−) monomers are well exposed. The antigenic differences between soluble trimers and monomers are most evident in an assay in which both radiolabeled proteins are incubated with an antibody and the ratio of precipitated proteins is compared. These ratios are normalized to those seen with a polyclonal mixture of sera from HIV-1-infected individuals. The results of such a competition assay performed with the gp120 monomer and the gp140Δ683(−/FT) glycoprotein is shown in Fig. 5 A. A striking result was the preferential recognition of the gp140Δ683(−/FT) glycoprotein by two monoclonal antibodies, immunoglobulin G1b12 and 2G12, that are among the small group of antibodies exhibiting potent neutralizing activity against primary HIV-1 isolates (8, 41a). The relative recognition of the gp140Δ683(−/FT) glycoprotein by the F105, F91, 17b, 48d, and 39F antibodies was intermediate in the competition assay. These antibodies exhibit more typical levels of neutralizing potency and are only of limited efficacy in inhibiting many primary HIV-1 isolates (49). Relative to their ability to precipitate the monomeric gp120 glycoprotein, the nonneutralizing antibodies C11, A32, and 30D minimally recognized the gp140Δ683(−/FT) glycoprotein. In parallel experiments, the relative recognition of the gp140Δ683(−/GCN4) glycoprotein by this panel of monoclonal antibodies was similar to that seen for the gp140Δ683(−/FT) glycoprotein (Fig. 5B). Thus, the ability of an antibody to neutralize HIV-1 is closely correlated with its affinity for the soluble trimeric glycoproteins relative to the affinity for the gp120 monomer.
FIG. 5.
Antibody recognition of stabilized gp140 trimers relative to that of gp120 monomers. The 35S-labeled HIV-1 envelope glycoproteins in labeling media were first quantified by precipitation with 3 μl of pooled sera from HIV-1-infected individuals, SDS-PAGE and analysis on a PhosphorImager (Molecular Dynamics). Equivalent amounts of gp120 and the stabilized gp140 trimers were mixed and precipitated either with 3 μl of pooled sera from HIV-1-infected individuals (P.S.) or with 1 μg of each of the indicated monoclonal antibodies for 3 h. The precipitated proteins were boiled in 1× Laemmli buffer with 2% β-ME and resolved on an SDS-7.5% polyacrylamide gel. The ratio of gp140 to gp120 in each sample was quantified by using a Storm PhosphorImager. The value shown represents the gp140/gp120 ratio for each monoclonal antibody divided by the ratio obtained with the pooled sera. Three independent experiments yielded similar results; the results of one of the experiments are shown. The potency withwhich the monoclonal antibodies neutralize HIV-1 is indicated. (A) gp140Δ683 (−/FT) versus gp120. (B) gp140Δ683 (−/GCN4) versus gp120.
Recognition of soluble trimers by the five-helix protein and by the NC-1 monoclonal antibody.
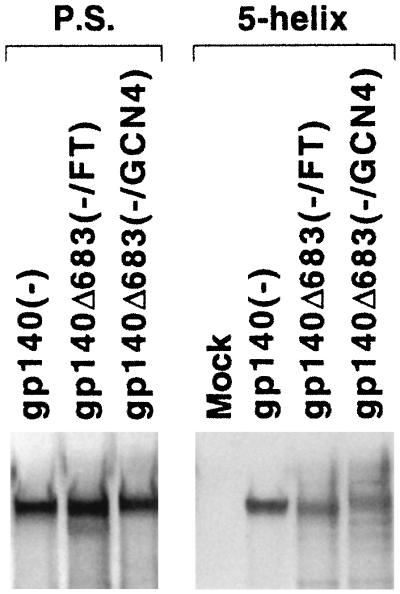
The studies of thermal stability described above suggested that at least a portion of the gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins assumes a conformation distinct from that of a six-helix bundle. To investigate this further, we tested the ability of the five-helix protein to recognize these stabilized trimers and the gp140(−) glycoprotein. The five-helix protein consists of five of the helices (three N36 helices and two C34 helices) in the HIV-1 gp41 six-helix bundle; these five helices are joined by linker segments in the five-helix protein (35). The five-helix protein interacts with the C34 region of the HIV-1 gp41 glycoprotein and thereby inhibits the function of the HIV-1 envelope glycoproteins but is not expected to interact with the fusion-active six-helix bundle. The His6-tagged five-helix polypeptide was expressed in bacteria by using an expression vector kindly provided by P. Kim at the Whitehead Institute, Massachusetts Institute of Technology. About 0.5 μg of the five-helix protein in the bacterial lysates was bound to Ni-nitrilotriacetic acid (NTA) gel (Qiagen) through its His6 tag at 4°C for 16 h. After being washed with lysis buffer, the protein-gel complexes were incubated with radiolabeled gp140(−), gp140Δ683(−/GCN4), and gp140Δ683(−/FT) glycoproteins in 293T cell supernatants at 37°C for 3 h. After five washes with lysis buffer containing 15 mM imidazole, the bound glycoproteins were boiled in 1× Laemmli buffer with 2% β-mercaptoethanol (β-ME) and resolved on an SDS-7.5% polyacrylamide gel. Figure 6 shows that all three soluble gp140 glycoproteins were recognized by the five-helix protein. Under identical conditions, a negative control protein, a His6-tagged HIV-1 Tat protein, did not precipitate any of the soluble gp140 glycoproteins (data not shown).
FIG. 6.
Recognition of soluble gp140 glycoproteins by the five-helix protein. The plasmid expressing the His6-tagged five-helix protein was used to transform Escherichia coli BL21. One of the resulting colonies was inoculated into 50 ml of Luria-Bertani medium with 50 μg of ampicillin/ml and cultured for 16 h at 37°C with shaking. The culture was then diluted in 500 ml of Luria-Bertani medium plus ampicillin and incubated for 3 h before 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce the expression of the five-helix protein. After 4 h, the bacteria were harvested and washed once with lysis buffer (50 mM Tris-HCl, pH 8.0; 120 mM NaCl; 0.5% NP-40; 5 mM dithiothreitol). The bacteria were lysed by 2 mg of lysozyme (Sigma)/ml in 10 ml of lysis buffer plus 1× protease inhibitor cocktail (Pharmacia) and by five 1-min sonications at scale 9 by using a sonicator (Branson). The protein lysate was finally harvested after two centrifugations of 14,000 rpm for 15 min at 4°C and stored at −20°C. To perform the binding assay, 500 μl of the above lysate, containing ∼0.5 μg of the five-helix protein (data not shown), was first incubated with 100 μl of Ni-NTA gel (Qiagen) for 16 h at 4°C with shaking. After three washes with ice-cold lysis buffer, the protein-Ni-NTA complex was incubated with 500 μl of the indicated 35S-labeled HIV-1 envelope glycoproteins for 3 h at 37°C with shaking. The resulting gel complexes were then washed five times with the lysis buffer plus 15 mM imidazole. The samples were boiled in 1× Laemmli buffer with 2% β-ME and resolved on an SDS-7.5% polyacrylamide gel. In parallel, the labeled HIV-1 envelope glycoproteins were precipitated by pooled sera (P.S.) from HIV-1-infected individuals.
The NC-1 monoclonal antibody has been shown to be relatively specific for the six-helix bundle structure of HIV-1 gp41 (27). In our previous studies, ca. 15% of the gp140Δ683(−/GCN4) glycoprotein could bind to 1 μg of the NC-1 monoclonal antibody in a standard immunoprecipitation assay performed at room temperature for 3 h. In such an assay, the gp140Δ683(−/FT) glycoproteins bound to the NC-1 antibody at a significantly reduced level compared with the gp140Δ683(−/GCN4) trimer (Fig. 4, right panel). This result is consistent with the above observation with the five-helix protein, suggesting that majority of the gp140Δ683(−/FT) glycoprotein is not in a conformation containing the six-helix bundle structure, i.e., the fusogenic conformation. Moreover, the lower level of NC-1 recognition of the gp140Δ683(−/FT) glycoprotein indicates that the gp140Δ683(−/FT) trimer is more homogeneous than the gp140Δ683(−/GCN4) glycoprotein.
The availability of soluble forms of the HIV-1 envelope glycoproteins that effectively mimic the conformation of these proteins on the virus or infected cell surface is critical for attempts to obtain detailed information on the structure and function of these key molecules. The multiple oligomeric forms and instability of soluble HIV-1 gp140 preparations have created challenges for their use as reagents and as immunogens. Removal of the gp120-gp41 proteolytic cleavage site is insufficient to address these problems (6, 19, 20, 42); however, considerable increases in homogeneity result from further addition of C-terminal GCN4 trimeric motifs (48-50). The trimeric globular domain of bacteriophage fibritin conferred even greater stability to heat, reducing agents, and detergents, a finding consistent with the increased stability of fibritin compared with GCN4 in other contexts (24, 40). The gp140Δ683(−/FT) glycoprotein exhibited greater homogeneity than the gp140Δ683(−/GCN4) trimer, as demonstrated by the reduced recognition by the NC-1 monoclonal antibody. Whether the hydrophilic surface of the fibritin trimeric motif will result in higher solubility and a lower tendency to aggregate during high-level production and concentration has yet to be tested.
The utility of soluble HIV-1 envelope glycoprotein trimers is dependent upon the extent to which the envelope glycoprotein ectodomains assume native conformations and intersubunit associations. The antigenic similarities between the gp140Δ(−/GCN4) and gp140Δ683(−/FT) glycoproteins suggest that the envelope glycoprotein ectodomains, rather than the appended trimeric motifs, dictate not only the folding of the proteins but also the orientation of the subunits within the soluble complex. Particularly reassuring was our observation that the potently neutralizing monoclonal antibodies immunoglobulin G1b12 and 2G12 exhibited a preference for binding the soluble gp140 trimers compared to the gp120 monomer. Less potently neutralizing antibodies demonstrated much less of a preference for trimer binding, and nonneutralizing antibodies bound monomeric gp120 significantly better than either of the soluble gp140 glycoprotein trimers. This finding is consistent with the expectation that the affinity of antibody binding to a structural mimic of the functional envelope glycoprotein complex should correlate with neutralization efficiency. These observed patterns of antibody recognition and the striking similarity between antigenic profiles of the gp140Δ(−/GCN4) and gp140Δ683(−/FT) trimers, in addition to the formation of intersubunit disulfide bonds in GCN4-stabilized soluble gp140 variants containing appropriately positioned cysteine substitutions (22, 49), suggest that at least some of the interactions among the subunits of the soluble trimers resemble those on the native HIV-1 envelope glycoproteins.
There are thought to exist different conformational states of the native HIV-1 envelope glycoproteins depending upon whether receptor has been bound and the extent of progression along the pathway leading to membrane fusion (15). Stabilization of trimeric forms of the envelope glycoproteins could potentially increase the opportunity for the formation of the energetically favored six-helix bundle that is thought to represent a fusogenic conformation (10, 29, 39, 45). Our data suggest that at least some of the gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins exist in a conformation distinct from that of the six-helix bundle. The trimeric form of the Δ528 glycoprotein has been shown to be recognized by antibodies, like NC-1, that are specific for the six-helix bundle (29, 49); consistent with this, theΔ528 glycoprotein demonstrated stability at temperatures up to 85 to 95°C in our assays. At lower temperatures, the interactions among the subunits of the six-helix bundle must be energetically more favorable than the potential interactions between the monomers and the medium components, which include SDS. In contrast, significant fractions of the gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins were monomers after heating to 50 to 55°C under these conditions. The ability of the five-helix protein to precipitate the gp140Δ683(−/GCN4) and gp140Δ683(−/FT) glycoproteins suggests that the N36-binding surface of the C34 helix is accessible on these proteins, further indicating that conformations other than a six-helix bundle are assumed by these molecules. The inflections and biphasic nature of the melting curves observed for the stabilized trimers hint that these glycoproteins may undergo transitions to alternative conformations or that heterogeneous forms of these proteins exist. We expect that some of the stabilized trimers resemble the uncleaved form of the gp160 envelope glycoprotein as it exists in the Golgi apparatus prior to proteolytic activation. Further studies need to be conducted to clarify the nature of the conformational state of the stabilized soluble envelope glycoprotein trimers and to assess their suitability for detailed structural investigation. An understanding of the structure of these trimeric molecules should assist optimization of their potential as immunogens.
Acknowledgments
We thank Y. McLaughlin, E. Morrison, and S. Farnum for manuscript preparation. We thank Richard Kammerer and Jurgen Engel for the fibritin plasmid and Lawrence Shapiro for helpful discussions.
This work was supported by NIH grants AI24755, AI31783, and AI39420 to J.S. and by NIH CFAR grant AI28691. We also acknowledge the support of the G. Harold and Leila Mathers Foundation, the Friends 10, Douglas and Judith Krupp, and the late William F. McCarty-Cooper. P.D.K. was a recipient of a Burroughs Wellcome Career Development Award.
REFERENCES
- 1.Alan, J. S., J. E. Coligan, F. Barin, J. Sodroski, C. A. Rosen, W. A. Haseltine, T.-H. Lee, and M. Essex. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091-1094. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, S. W., S. Lu, L. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, L. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, S. W., S. Rajasekar, H. Legg, B. Doe, D. H. Fuller, J. R. Haynes, C. M. Walker, and K. S. Steimer. 1997. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine 15:869-873. [DOI] [PubMed] [Google Scholar]
- 5.Belshe, R. B., G. J. Gorse, M. J. Mulligan, T. G. Evans, M. C. Keefer, J.-L. Excler, A. M. Duliege, J. Tartaglia, W. I. Cox, J. McNamara, K.-L. Hwang, A. Bradney, D. C. Montefiori, and K. J. Weinhold. 1998. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS 12:2407-2415. [DOI] [PubMed] [Google Scholar]
- 6.Broder, C. C., P. L. Earl, D. Long, S. T. Abedon, B. Moss, and R. W. Doms. 1994. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. USA 91:11699-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R., and J. P. Moore. 1998. Why do we not have an HIV vaccine and how can we make one? Nat. Med. 4(Suppl. 5):495-498. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Koduri, G. B. Thornton, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 (HIV-1) gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 12.Connor, R. I., B. T. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, R. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalam, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 14.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 15.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 17.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 18.Dubay, J. W., S. R. Dubay, H. J. Shin, and E. Hunter. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J. Virol. 69:4675-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type-1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 24.Frank, S., R. A. Kammerer, D. Mechling, T. Schulthess, R. Landwehr, J. Bann, Y. Gao, A. Lustig, H. P. Bachinger, and J. Engel. 2001. Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308:1081-1089. [DOI] [PubMed] [Google Scholar]
- 25.Freed, E. O., D. J. Myers, and R. Risser. 1990. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc. Natl. Acad. Sci. USA 87:4650-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 28.Klatzmann, D., E. Chamoagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 29.Lu, M., S. Blackow, and P. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 30.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, and The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 31.McDougal, J. S., J. K. Nicholson, G. D. Cross, S. P. Cort, M. S. Kennedy, and A. C. Mawle. 1986. Binding of the human retrovirus HTLV-III/LAV/ARV/HIV to the CD4 (T4) molecule: conformation dependence, epitope mapping, antibody inhibition, and potential for idiotypic mimicry. J. Immunol. 137:2937-2944. [PubMed] [Google Scholar]
- 31a.Mirzabekov, T., N. Bannert, M. Farzan, W. Hofmann, P. Kolchinsky, L. Wu, R. Wyatt, and J. Sodroski. 1999. Enhanced expression, native purification and characterization of CCR5, a principal HIV-1 coreceptor. J. Biol. Chem. 274:28745-28750. [DOI] [PubMed] [Google Scholar]
- 32.Morikawa, Y., E. Barsov, and I. Jones. 1993. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J. Virol. 67:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 34.Robey, W. G., B. Safai, S. Oroszlan, L. Q. Arthur, M. A. Gonda, R. C. Gallo, and P. J. Fischinger. 1985. Characterization of envelope and core gene products of HTLV-III with sera from AIDS patients. Science 228:593-595. [DOI] [PubMed] [Google Scholar]
- 35.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 36.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schonning, K., A. Bolmstedt, J. Novotny, O. S. Lund, S. Olofsson, and J. Hansen. 1998. Induction of antibodies against epitopes inaccessible on the HIV type 1 envelope oligomer by immunization with recombinant monomeric glycoprotein 120. AIDS Res. Hum. Retrovir. 14:1451-1456. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan, N., S. Ying, Q. Sattenteu, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, K., J.-H. Lee, J.-H. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao, Y., S. V. Strelkov, V. V. Mesyanzhinov, and M. G. Rossmann. 1997. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure 15:789-798. [DOI] [PubMed] [Google Scholar]
- 41.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 41a.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanCott, T. C., J. R. Mascola, L. D. Loomis-Price, F. Sinangil, N. Zitomersky, J. McNeil, M. L. Robb, D. L. Birx, and S. Barnett. 1999. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J. Virol. 73:4640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veronese, F. D., A. L. DeVico, T. D. Copeland, S. Oroszlan, R. S. Gallo, and M. G. Sarngadharan. 1985. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science 229:1402-1405. [DOI] [PubMed] [Google Scholar]
- 45.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 46.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrin, T., and J. H. Nunberg. 1994. HIV-1 MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS 8:1623-1666. [DOI] [PubMed] [Google Scholar]
- 48.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralization antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoproteins. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]