Abstract
The nucleocapsid protein (NC) of human immunodeficiency virus type 1 has two zinc fingers, each containing the invariant CCHC zinc-binding motif; however, the surrounding amino acid context is not identical in the two fingers. Recently, we demonstrated that zinc coordination is required when NC unfolds complex secondary structures in RNA and DNA minus- and plus-strand transfer intermediates; this property of NC reflects its nucleic acid chaperone activity. Here we have analyzed the chaperone activities of mutants having substitutions of alternative zinc-coordinating residues, i.e., CCHH or CCCC, for the wild-type CCHC motif. We also investigated the activities of mutants that retain the CCHC motifs but have mutations that exchange or duplicate the zinc fingers (mutants 1-1, 2-1, and 2-2); these changes affect amino acid context. Our results indicate that in general, for optimal activity in an assay that measures stimulation of minus-strand transfer and inhibition of nonspecific self-priming, the CCHC motif in the zinc fingers cannot be replaced by CCHH or CCCC and the amino acid context of the fingers must be conserved. Context changes also reduce the ability of NC to facilitate primer removal in plus-strand transfer. In addition, we found that the first finger is a more crucial determinant of nucleic acid chaperone activity than the second finger. Interestingly, comparison of the in vitro results with earlier in vivo replication data raises the possibility that NC may adopt multiple conformations that are responsible for different NC functions during virus replication.
The nucleocapsid protein (NC) of human immunodeficiency virus type 1 (HIV-1) is a small, basic, nucleic acid-binding protein which associates with genomic RNA in the mature virion core (14, 15, 54); the mature protein is generated by proteolytic cleavage of the Gag precursor (36, 47, 63). Structural studies have revealed that free HIV-1 NC in solution has two rigid zinc-binding domains or zinc fingers, each containing the invariant CCHC metal ion-binding motif (30, 37, 59, 61). The two fingers are covalently linked to each other by a short flexible basic amino acid region and are flanked by flexible N- or C-terminal “tails” (49-51, 59, 60, 62). The Summers group has recently solved the three-dimensional structures of HIV-1 NC bound to the SL2 (3, 4) and SL3 (18) RNA stem-loops that form part of the larger HIV-1 packaging signal, by nuclear magnetic resonance (NMR) analysis.
The two NC zinc fingers are located in close proximity (45, 46, 49, 50) but exhibit only weak interactions with one another (13, 43, 46, 49, 66). Interestingly, their structures are similar (58), despite differences in the amino acid sequences surrounding the CCHC motifs (37, 54). Moreover, the biochemical properties (8, 45) and biological activities of the two fingers are not equivalent, and the presence of both fingers is critical for production of replication-competent virus (9, 21, 26, 28, 29, 48, 72); in addition, the positions of the zinc fingers cannot be exchanged (21, 26).
NC function in virus replication is dependent on its dynamic interaction with nucleic acids. Acting as a nucleic acid chaperone, NC can catalyze nucleic acid conformational rearrangements that lead to the formation of the most thermodynamically stable structure (65; reviewed in references 15, 38, and 54). For example, NC chaperone activity plays a major role in the two strand transfer steps that occur during viral DNA synthesis. During minus-strand transfer, minus-strand strong-stop DNA [(−) SSDNA] is translocated to the 3′ terminus of the viral RNA genome in a reaction facilitated by annealing of the complementary repeat (R) regions at the 3′ ends of the RNA and DNA reactants (14). In the case of HIV-1, we and others have found that NC stimulates minus-strand transfer (2, 10, 16, 17, 20, 31, 32, 40, 52, 55, 71) by increasing the rate and extent of annealing (17, 32, 33, 42, 71) and by blocking nonspecific self-priming induced by the complementary TAR sequence at the 3′ end of (−) SSDNA (10, 22, 31, 32, 41, 44). In addition, it has been reported that the efficiency of murine leukemia virus (MuLV) (2) and Rous sarcoma virus (67) minus-strand transfer is increased in the presence of MuLV (2) and HIV-1 NC (67), respectively.
HIV-1 plus-strand transfer is also stimulated by NC (5, 69). Thus, HIV-1 NC promotes removal of the tRNA3Lys primer from the 5′ end of minus-strand DNA (32, 69) as well as annealing of the 18-nucleotide (nt) complementary DNA primer-binding site (PBS) sequences at the 3′ ends of minus-strand DNA and plus-strand strong-stop DNA [(+) SSDNA] (32, 69). A recent NMR study demonstrated that the nucleic acid chaperone activity of NC destabilizes the relatively stable stem-loop formed by the 18-nt minus-strand PBS DNA (39). These results indicated that NC facilitates plus-strand annealing by exposing nucleotides in the stem of the minus-strand PBS DNA structure so that they can base pair with the complementary 18-nt plus-strand PBS sequence.
For many years, it was not clear which NC functions, in addition to viral RNA packaging (7), required zinc coordination. Recently, the importance of the zinc fingers for NC activity in HIV-1 reverse transcription in vivo was shown in studies of viral mutants. Virions with NCs having alternative zinc-coordinating motifs that still package genomic RNA but synthesize reduced amounts of viral DNA and are noninfectious have been described (28, 64; R. J. Gorelick and J. Buckman, unpublished observations). A similar class of MuLV mutants (25, 27, 73) produces viral DNA with major defects in terminal sequences (27).
In earlier studies with in vitro systems, an effect of zinc finger mutations on extension of the tRNA3Lys primer (56), minus-strand DNA elongation (23, 70), and integration (11) was observed. More recently, we investigated the effects of an HIV-1 NC mutation that eliminates zinc coordination by changing all of the cysteine residues to serine (SSHS NC). This work demonstrated that while stimulation of plus-strand annealing by NC is not dependent on the zinc finger structures, the fingers facilitate tRNA primer removal in plus-strand transfer and are crucial for stimulation of annealing and prevention of self-priming in minus-strand transfer (32). These results led us to conclude that the zinc fingers in NC contribute to transient destabilization of highly structured nucleic acid strand transfer intermediates, such as TAR-containing (−) SSDNA.
On the basis of data from single-molecule DNA stretching experiments using HIV-1 wild-type and SSHS NC proteins, Williams et al. (68) reported that wild-type NC significantly destabilizes double-stranded DNA, presumably due to the preferential binding of NC to single-stranded nucleic acids, and reduces the cooperativity of the helix-coil transition in double-stranded DNA structures; in contrast, SSHS NC stabilizes double-stranded DNA and has no effect on cooperativity. In addition, Hargittai et al. (35) found that the zinc finger motifs are also important for NC-induced changes in the tertiary structure of the tRNA3Lys primer. These observations underscore the critical contribution of the zinc fingers to NC nucleic acid chaperone activity under conditions in which there is a conformational rearrangement of complex nucleic acid structures.
In the present work, we have extended our studies on the role of the zinc fingers in strand transfer reactions using NC mutants with subtle changes that may not have as drastic an effect on zinc finger structure as the SSHS mutation. Our results indicate that, in general, for optimal NC nucleic acid chaperone activity, (i) the CCHH residues cannot be replaced by alternative zinc-coordinating residues, i.e., CCHH and CCCC, found in certain cellular proteins (6) and (ii) the amino acid context surrounding the CCHC motifs cannot be changed by duplicating or exchanging the zinc fingers. We also found that the CCHC zinc-binding motif and amino acid context of the first zinc finger are more critical determinants of chaperone activity than the corresponding features in the second finger. Interestingly, for most of the zinc finger mutants, it is possible to correlate in vivo parameters of virus replication with the observed in vitro nucleic acid chaperone activity.
MATERIALS AND METHODS
Materials.
RNA, RNA-DNA, and DNA oligonucleotides were purchased from the sources listed by Guo et al. (32). [γ-32P]ATP (3,000 Ci/mmol) was obtained from Amersham Pharmacia Biotech (Piscataway, N.J.).
Wild-type and mutant NC proteins.
Wild-type HIV-1 NC was prepared as a recombinant protein, which was expressed and purified as described previously (70). Mutant NC proteins were prepared essentially as described in references 11 and 32. A list of the mutants used in this study is given in Table 1.
TABLE 1.
In vitro and in vivo activities of HIV-1 NC zinc finger mutantsa
| NC class | Compositionb
|
In vivo activityc
|
In vitro chaperone activityd | |||
|---|---|---|---|---|---|---|
| F1 | F2 | RNA packaging | DNA synthesis | Infectivity | ||
| Wild type | CCHC | CCHC | 4+ | 4+ | 4+ | 4+ |
| Class 1 | ||||||
| CCHC/CCHH | CCHC | CCHH | 4+ | 4+ | 1+ | 4+ |
| Class 2 | ||||||
| 2-1 | F2 | F1 | 1+ | n.d. | — | <1+ |
| 2-2 | F2 | F2 | 1+ | n.d. | — | <1+ |
| SSHSe | SSHS | SSHS | 1+ | — | — | 1+ |
| Class 3 | ||||||
| CCHH/CCHH | CCHH | CCHH | 1+ | — | — | 3+ |
| CCHH/CCHC | CCHH | CCHC | 2+ | — | — | 2+ |
| Class 4 | ||||||
| 1-1 | F1 | F1 | 3+ | n.d. | <1+f | 3+ |
| Class 5 | ||||||
| CCCC/CCHC | CCCC | CCHC | 4+ | 1+g | — | <1+ |
| CCHC/CCCC | CCHC | CCCC | 4+ | 3+ | <1+h | <1+ |
The numerical values refer to percentage of wild-type activity: 4+, 75 to ≥100%; 3+, 50 to 75%; 2+, 25 to 50%; 1+, 1 to 25%.
F1 and F2 refer to zinc fingers 1 and 2, respectively. DNA synthesis, synthesis of 2-LTR circular DNA. Infectivity was based on infectivity in a long-term replication assay. n.d., not determined. —, below limits of detection.
Based on activity in the minus-strand transfer assay system.
Data taken from Guo et al. (32).
Reverted to wild-type phenotype at 21 days postinfection.
Tanchou et al. (64) reported that this mutant synthesized elongated plus-strand DNA, but not 2-LTR circular DNA.
Reverted to wild-type genotype at 35 days postinfection.
Reconstituted minus-strand transfer system.
A detailed description of the assay for minus-strand transfer is given by Guo et al. (31). The sequences of the nucleic acid components in this and all other assays are from HIV-1 NL4-3 (1). Briefly, 0.2 pmol of a 131-nt donor RNA (nt 454 to 584) was annealed at 65°C for 5 min to 0.4 pmol of a 32P-labeled 20-nt DNA primer complementary to nt 565 to 584 in U5. Reaction buffer (50 mM Tris-HCl [pH 8.0], 75 mM KCl, 1 mM dithiothreitol), 0.2 pmol of a 148-nt acceptor RNA (nt 9475 to 9622), and HIV-1 wild-type or mutant NC proteins were then added, and the mixture was incubated for 5 min at 37°C. Reactions (final volume, 20 μl) were initiated by addition of 0.2 pmol of HIV-1 reverse transcriptase (RT) (Worthington Biochemical Corp.), MgCl2 (final concentration, 7 mM), and the four deoxyribonucleoside triphosphates (dNTPs) (final concentration, 100 μM each) and incubated at 37°C for 30 min. Termination of reactions, polyacrylamide gel electrophoresis (PAGE) of DNA products, and PhosphorImager analysis were performed as described before (31).
Annealing reaction.
A total of 0.2 pmol each of unlabeled acceptor RNA and synthetic (−) SSDNA (131 nt), labeled at its 5′ end with 32P (34), were incubated with wild-type or mutant NC proteins (final concentration, 1 μM) in a final volume of 20 μl at 37°C; reaction mixtures were scaled up as needed, and 10-μl aliquots were removed at the specified times (33). The RNA-DNA hybrid was resolved by PAGE in 7.5% polyacrylamide gels (32) and quantified by PhosphorImager analysis (33).
Assay for removal of tRNA3Lys from minus-strand DNA.
The assay for removal of tRNA3Lys from minus-strand DNA models secondary RNase H cleavage during plus-strand transfer (see Fig. 3A) (69). Briefly, a 32-nt minus-strand DNA with an rA at its 5′ end (1 pmol) and a 17-nt RNA (1 pmol) representing the 17 nt remaining at the 3′ end of tRNA3Lys after initial cleavage were annealed at 65°C for 5 min to a 50-nt (+) SSDNA (0.2 pmol) labeled at its 5′ end with 32P (34). Wild-type or mutant NCs, a 48-nt minus-strand acceptor DNA (0.2 pmol), RT (0.2 pmol), MgCl2, and four dNTPs (concentrations as indicated above for minus-strand transfer assay) were then added, and reactions (final volume, 20 μl) were incubated for 60 min at 37°C. The amount of labeled 80-nt plus-strand DNA (the actual readout for RNase H cleavage) was quantified by PhosphorImager analysis of gel data (69).
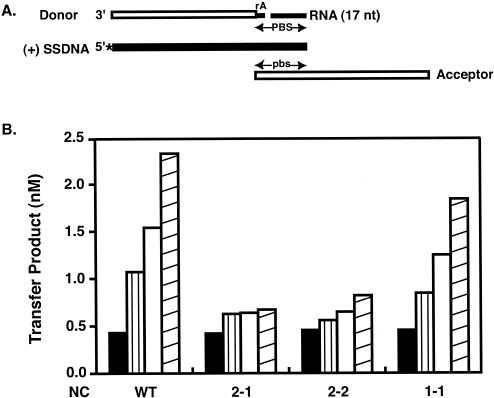
FIG. 3.
Effect of wild-type and mutant NC proteins on complete primer removal. The assay was performed as described in Materials and Methods. (A) Nucleic acid strand transfer intermediates present in the reaction mixture. The minus-strand donor (32 nt) and acceptor (48 nt) DNAs are shown as open rectangles, RNA (rA attached to the donor DNA and 17-nt RNA, representing the 17 nt remaining at the 3′ end of tRNA3Lys following initial RNase H cleavage) is shown as narrow solid rectangles, and (+) SSDNA (50 nt) is shown as a thick solid rectangle. The 32P label at the 5′ end of (+) SSDNA is shown by an asterisk. This diagram is taken from Fig. 7A in reference 69. PBS and pbs, plus- and minus-strand PBS sequences, respectively. (B) Primer removal in reactions containing increasing concentrations of wild-type (WT) NC or the 2-1, 2-2, and 1-1 NC mutants. The concentrations were as follows: solid bars, no NC; vertically striped bars, 0.5 μM (7 nt/NC); open bars, 1.0 μM (3.5 nt/NC); and diagonally striped bars, 2.0 μM (1.75 nt/NC).
RESULTS
HIV-1 mutant NC proteins.
In a recent study (32), we demonstrated that the zinc fingers of HIV-1 NC play a critical role in minus-strand transfer (including stimulation of annealing and inhibition of self-priming) and in promoting the tRNA removal step in plus-strand transfer. To determine whether replacement of the CCHC motifs with alternate zinc-coordinating residues or changes in the positions of the zinc fingers would affect NC function, we tested eight NC mutants (Table 1) in our established strand transfer assay systems (31, 69).
Strand transfer activity of the NC mutants.
The CCHH zinc-coordinating motif is commonly found in cellular transcription factors and is important for recognition of specific sequences in duplex DNA (6). To investigate whether CCHH can be substituted for the CCHC motifs in HIV-1 NC, we assayed minus-strand transfer activity in reactions containing increasing concentrations of wild-type or CCHH mutant NCs. Three mutants with CCHH substitutions in the first (CCHH/CCHC), second (CCHC/CCHH), or both (CCHH/CCHH) zinc fingers (Table 1) were tested (Fig. 1, A-1 and A-2). Reaction products were resolved by PAGE (data not shown) and quantified by PhosphorImager analysis.
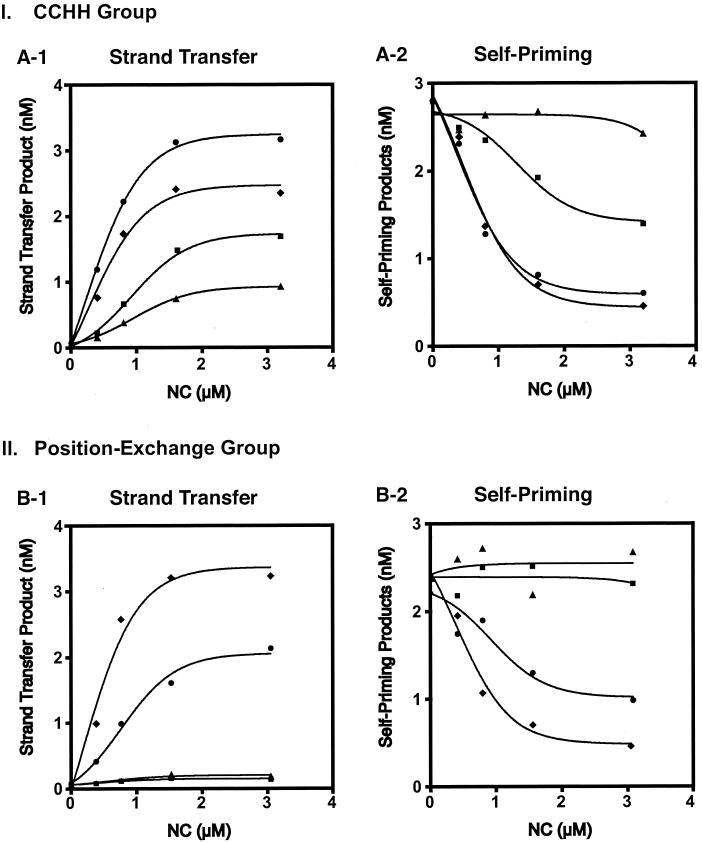
FIG. 1.
Strand transfer activity of CCHH and position exchange mutants. (I) CCHH group (A-1 and A-2). Gel data from four experiments were averaged and quantified by PhosphorImager analysis; the amounts of the strand transfer product (A-1) and self-priming products (A-2) were plotted as a function of NC concentration. Symbols: ⧫, wild type; •, CCHC/CCHH; ▪, CCHH/CCHH; and ▴, CCHH/CCHC. (II) Position exchange group (B-1 and B-2). The amounts of the strand transfer product (B-1) and self-priming products (B-2) from a representative experiment were quantified and plotted as described above. Symbols: ⧫, wild type; •, 1-1; ▪, 2-1; and ▴, 2-2.
As expected (31), wild-type NC greatly stimulated minus-strand transfer and dramatically inhibited self-priming (Fig. 1, A-1 and A-2). Increasing concentrations of the CCHH/CCHH mutant led to moderate stimulation of minus-strand transfer and inhibition of self-priming, whereas the CCHH/CCHC mutant had low levels of activity. With CCHC/CCHH NC, synthesis of strand transfer DNA reached a plateau level slightly higher (1.3-fold) than that of the wild type; however, the extent to which self-priming was inhibited was approximately the same for both of these proteins.
These results demonstrate that the CCHC motif is (i) required for NC chaperone activity in minus-strand transfer and (ii) more strictly required in the N-terminal finger position than it is in the C-terminal position. The fact that the double mutant was more active than CCHH/CCHC NC suggests that the effects of the mutations on activity are not simply additive.
The CCHC motif is invariant in retroviral NC zinc fingers (37). In addition, a database search of 50 HIV-1 strains indicates that within each finger, the amino acid residues surrounding these motifs (i.e., the amino acid context) are highly conserved (J. Guo and J. G. Levin, unpublished observations). However, the amino acid sequences of the two fingers are not identical (37, 54). These differences might be responsible for the functional asymmetry of the two fingers observed with the CCHH mutants (Fig. 1, A-1 and A-2).
To test this hypothesis, the activities of three zinc finger position exchange mutants which retain the CCHC residues in both fingers (Table 1) were also assayed (Fig. 1, B-1 and B-2). Interestingly, the 1-1 mutant (two copies of the first finger) had about 50% of wild-type NC activity with respect to stimulation of minus-strand transfer (Fig. 1, B-1) and inhibition of self-priming (Fig. 1, B-2). However, in both cases where the sequence of the second finger was placed in the N-terminal position (mutants 2-1 [position exchange] and 2-2 [two copies of second finger]), there was a complete loss of activity. These results demonstrate that for maximal activity in minus-strand transfer, the positions of the two zinc fingers in HIV-1 NC cannot be duplicated or exchanged. Moreover, there is a strict requirement for the sequence of finger 1 in the N-terminal position, whereas the identity of the zinc finger in the C-terminal position is less critical. Thus, the two zinc fingers in HIV-1 NC do not exhibit equivalent activity in minus-strand transfer.
In other experiments, CCCC, a zinc coordination motif commonly contained in steroid hormone receptors (6), was substituted in either the first or second finger. Both mutant NC proteins (CCCC/CCHC and CCHC/CCCC) failed to stimulate minus-strand transfer or inhibit self-priming (Table 1 and data not shown).
Annealing activity of mutant NC proteins.
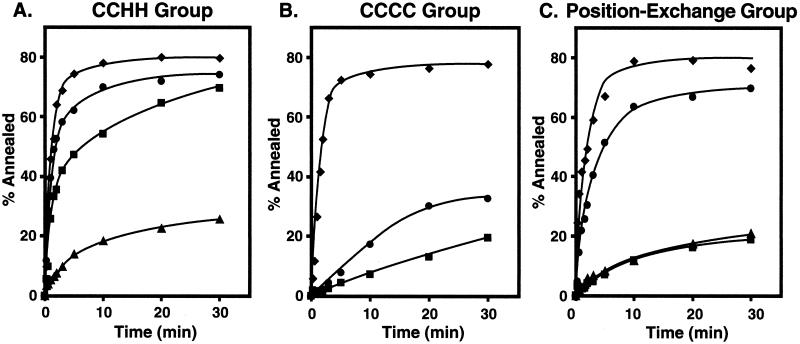
Recently, we established that NC-stimulated annealing in minus-strand transfer is dependent on the presence of the zinc fingers, which participate in destabilization of the complementary TAR structures at the 3′ ends of (−) SSDNA and acceptor RNA (32). To extend these observations, we tested all eight NC mutants in the annealing assay (Fig. 2), as described in Materials and Methods. Figure 2A, B, and C shows the results obtained with the CCHH, CCCC, and position exchange mutants, respectively.
FIG. 2.
Annealing activities of wild-type and mutant NC proteins. Annealing reactions were performed as described in Materials and Methods. The data were plotted as the percentage of (−) SSDNA annealed as a function of time. (A) CCHH group. In this set, the results of four experiments were averaged. Symbols: ⧫, wild type; •, CCHC/CCHH; ▪, CCHH/CCHH; and ▴, CCHH/CCHC. (B) CCCC group. Symbols: ⧫, wild type; •, CCHC/CCCC; and ▪, CCCC/CCHC. (C) Position exchange group. Symbols: ⧫, wild type; •, 1-1; ▪, 2-1; and ▴, 2-2.
The data in Fig. 2A indicate that, compared with wild-type NC, substitution of CCHH in the first finger (CCHH/CCHC) dramatically reduced annealing activity. However, the same change in the second finger (CCHC/CCHH) had only a small effect on the rate and extent of annealing. Interestingly, the mutant having CCHH in both fingers (CCHH/CCHH) had lower annealing activity than wild-type NC, but higher annealing activity than the CCHH/CCHC mutant (CCHH in the first finger only).
Figure 2B shows that replacement of CCHC with CCCC in either the first or second finger greatly reduced stimulation of annealing compared to wild-type NC. However, it is noteworthy that the CCHC/CCCC mutant had low but significant activity, whereas the activity of the CCCC/CCHC mutant reached a plateau value only slightly higher than that observed with a minus-NC control (32, 33).
The data in Fig. 2C show that the 1-1 mutant had relatively high activity in the annealing assay. However, replacement of the first finger with the second finger (mutants 2-1 and 2-2) led to a drastic reduction in annealing activity, with the rate and extent of annealing similar to that seen in the absence of NC (32, 33). These results underscore the conclusion reached from the assay of minus-strand transfer (Fig. 1, B-1 and B-2), indicating that the positions of the two zinc fingers are not interchangeable.
The annealing data were further analyzed by calculating the t1/2 values for the individual reactions. For reactions containing the wild-type and CCHC/CCHH NCs, the t1/2 values were 0.88 and 0.95 min, respectively; the values for the CCHH/CCHH and 1-1 mutants were 1.9 and 2.0 min, respectively. The other NC mutants were inefficient at promoting the annealing reaction, and the t1/2 values were not estimated.
Taken together, the data in Fig. 2 demonstrate that while wild-type NC has higher annealing activity than any of the mutants, annealing is facilitated when the CCHC motif is present in the first finger and the residues surrounding the CCHC motif in the first finger are in the N-terminal position. These results parallel the findings for overall strand transfer and self-priming (Fig. 1). However, it should be noted that the annealing assay is a less sensitive indicator of zinc finger function than assay of strand transfer and self-priming: for example, mutant 1-1 has 50% of wild-type activity in the strand transfer assay (Fig. 1, B-1 and B-2) and 90% of wild-type activity in the annealing assay (Fig. 2C).
Primer removal activity of zinc finger position exchange mutant NC proteins.
During plus-strand transfer, the tRNA3Lys primer must be removed from minus-strand DNA (12). This occurs in two steps (57, 69): (i) RNase H-catalyzed primary cleavage, which removes the 3′ rA of the tRNA; and (ii) removal of an additional 17 nt from the 3′ end of the tRNA, which occurs most efficiently when both RNase H activity and NC, containing the zinc fingers, are present (32, 69).
To determine whether both zinc fingers are equally important for this activity of NC, we tested the position exchange mutants (Table 1) for tRNA removal activity (Fig. 3). In this assay (69), we have modeled the second step in this process (see above; Fig. 3A). Since minus-strand acceptor DNA, NC, and RT are provided in the assay, once primer removal is complete, the 18-nt complementary PBS in (+) SSDNA and acceptor DNA are annealed (Fig. 3A), plus- and minus-strand DNAs are elongated, and the strand transfer product, an 80-bp double-stranded DNA, is formed. Thus, the readout for the assay is synthesis of the 80-bp DNA, which reflects the extent of primer removal (69). However, since only plus-strand DNA is labeled, the product detected by PAGE is actually the 80-nt plus-strand DNA (reference 69 and data not shown).
Figure 3B illustrates the effect of increasing amounts of wild-type and mutant NC proteins on primer removal. With the highest concentration of wild-type NC (2.0 μM, 1.75 nt/NC), the amount of the strand transfer product was 5.4-fold greater than the amount made in the absence of NC (black bar). In the presence of mutant 1-1, a similar stimulation was observed (∼4-fold). In contrast, under the same conditions, the 2-1 and 2-2 mutants showed increases of only 1.6- and 1.8-fold, respectively. These results demonstrate that the two zinc fingers in HIV-1 NC are not exchangeable in this assay and indicate a more critical role of the first finger in the promotion of primer removal.
DISCUSSION
A major goal of the present study was to investigate the zinc finger requirement for NC nucleic acid chaperone activity in HIV-1 minus- and plus-strand transfer and to address the following questions: (i) can the CCHC motifs in the NC zinc fingers be replaced by alternative zinc coordination motifs such as CCHH and CCCC and (ii) can the zinc fingers be exchanged or duplicated? The data indicate that the CCHC motifs and the amino acid context surrounding these motifs must be preserved for optimal NC activity. The results also point to the critical connection between nucleic acid chaperone activity and the presence of a native zinc finger in the N-terminal position.
The CCHH mutation (change from C to H) shortens the length of the peptide-to-metal ion bond. In contrast, the CCCC mutation (change from H to C) increases the length of the peptide-to-metal ion bond. Both of these mutations change the conformation of the peptide in the immediate vicinity of the zinc fingers. This, in turn, alters the surface orientation of the side chains and the peptide backbone (19, 53, 66) and ultimately leads to a loss of biological activity, as discussed below.
For example, the CCHH/CCHC mutant (CCHH substitution in the first finger) has low levels of activity in assays for NC function (Fig. 1, A-1 and A-2; Fig. 2A). Surprisingly, substitution of CCHH in both fingers, which might be expected to lower NC activity to an even greater extent, actually results in less than twofold reduction in minus-strand transfer activity compared with wild-type NC (Fig. 1, A-1 and A-2), relatively high activity in the annealing assay (Fig. 2A), and fourfold greater activity than wild type in an in vitro integration assay (11). These findings indicate that the zinc fingers do not function independently and that the activities of the two fingers are not additive.
In view of the high level of activity exhibited by the CCHC/CCHH mutant in the minus-strand transfer assay (Fig. 1, A-1 and A-2), the lack of activity of the CCHC/CCCC mutant (Table 1 and data not shown) was unexpected, particularly since inadvertent oxidation of the cysteine residues was excluded (E. N. Chertova and L. E. Henderson, unpublished observations). Structural analysis will be needed to resolve the question of why the CCCC and CCHH second-finger replacements lead to such different biological activities.
In earlier in vitro studies of the position exchange group (1-1, 2-1, and 2-2), the mutant NCs were shown to have either equivalent inhibitory effects on NC activity (24, 70) or little or no effect on activity (11). In sharp contrast, the results presented here demonstrate differential activity and parallel in vivo observations (Table 1) (26). Thus, the 1-1 mutant significantly stimulates minus-strand transfer (∼50% of wild-type activity) (Fig. 1, B-1 and B-2), promotes annealing almost as efficiently as wild-type NC (Fig. 2C), and facilitates tRNA removal (∼75% of wild-type activity) (Fig. 3). However, the two other mutants have only minimal activity in all of these assays. It is of interest that when some of the mutants used in this study (including 1-1 and 2-1) were assayed for nucleic acid chaperone activity by the single-molecule DNA stretching technique (68), the results were in excellent agreement with the minus-strand transfer data presented here (M. Williams, R. J. Gorelick, and K. Musier-Forsyth, submitted for publication).
In general, it is desirable to have in vitro assays that reflect the in vivo properties of a given protein. Thus, another objective of this study was to see if the in vitro chaperone activity of the purified mutant proteins could be correlated with markers for virus replication. The results obtained with the position exchange mutants suggested that such a correlation might be possible. We grouped the mutants into several classes with respect to biological function (Table 1). Class 1 consists of mutant CCHC/CCHH, which exhibits in vitro (Fig. 1, A-1 and A-2) and in vivo (28) activities comparable to those of the wild type. Although the mutant has 50% of wild-type infectivity in a single-cycle assay, its titer is 100-fold lower than that of the wild type in a long-term replication assay (28). This suggests that a viral function other than RNA packaging or reverse transcription may be responsible for the replication defect.
Class 2 mutants, 2-1, 2-2 (26), and SSHS NC (32), are severely deficient in RNA packaging, do not synthesize detectable levels of two-long-terminal-repeat (2-LTR) circular DNA, are noninfectious, and have little or no in vitro chaperone activity. Class 3 mutants, CCHH/CCHC and CCHH/CCHH, have a phenotype (28) similar to that of the class 2 mutants except that the class 3 mutants have low (CCHH/CCHC) or moderately high (CCHH/CCHH) chaperone activity (Fig. 1, A-1 and A-2). Class 4 consists of mutant 1-1: it packages high levels of genomic RNA (26) and has relatively high chaperone activity (Fig. 1, B-1 and B-2, Fig. 2C, and Fig. 3). At first this mutant appeared to be replication defective, but after 21 days, it reverted to replication with wild-type kinetics; this suggests that the mutant replicated initially at a low rate (26).
Class 5 includes the two CCCC mutants. The CCCC/CCHC mutant is not infectious (19, 28) and has virtually no nucleic acid chaperone activity in vitro (Fig. 2B, Table 1, and data not shown). However, it packages high levels of genomic RNA (28) and synthesizes a small amount of DNA in vivo (28, 64). It was suggested that the DNA is not only reduced in amount, but may also contain defects that block integration (28, 64; R. J. Gorelick and J. Buckman, unpublished observations). The CCHC/CCCC mutant phenotype is more puzzling. Despite a severe deficiency in its ability to stimulate minus-strand transfer (Table 1 and data not shown), this mutant encapsidates wild-type levels of viral RNA, synthesizes significant amounts of DNA, and is infectious in a single-cycle assay (45% of wild-type value) (28). In a long-term replication assay, replication was initially undetectable, but after ∼4 weeks in culture, a titer measuring 1,000-fold lower than that of the wild type was observed; by day 35, the mutant had reverted to the wild-type genotype (28).
Correlation of in vivo and in vitro activities might be expected to be approximate, since virus replication requires some NC functions that are not directly related to chaperone activity (14). Interestingly, however, Table 1 demonstrates that most of the NC mutants tested here can be grouped into functional classes in which infectivity correlates with in vivo parameters and with in vitro chaperone activity. This is possible because our assays, in particular the assay for minus-strand transfer and self-priming, provide a highly sensitive approach for evaluating NC zinc finger function. Thus, both the in vivo and in vitro assays reveal a requirement for the CCHC zinc-coordinating residues and a wild-type first finger in the N-terminal position.
Another interesting finding that emerges from the data in Table 1 is that reduction or loss of RNA packaging activity does not always correlate with loss or reduction of in vitro nucleic acid chaperone activity (e.g., class 3, CCHH/CCHH, and class 5, CCCC/CCHC). In general, loss of either one of these activities is associated with loss of infectivity, suggesting that both are required for production of infectious virus. This raises an intriguing question: Why do some of the zinc finger mutations abolish either RNA packaging activity or chaperone activity, but not both?
All NC functions clearly derive from the ability of NC to bind to multiple nucleic acids. However, it is known from structural studies that NC binding to different nucleic acids is associated with unique structural configurations, dictated in part by the structure and sequence of the bound molecules (3, 4, 18). This shows that NC has the flexibility to adopt multiple conformations that are appropriate for different functions, including some that may still remain to be identified. Our data suggest that the specific structural distortions induced by various mutations will have differential effects on individual NC activities.
Acknowledgments
We thank Yasumasa Iwatani and Karin Musier-Forsyth for valuable discussion and Alan Rein for a careful reading of the manuscript. We are also indebted to Elena N. Chertova for expert assistance with analysis of the recombinant CCHC/CCCC NC protein under reducing and nonreducing conditions.
This work was supported in part by funds from the National Institutes of Health AIDS Targeted Antiviral Program awarded to J.G.L. and in part by the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400 with SAIC Frederick, Inc.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J.-L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 4.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, and M. F. Summers. 2000. NMR structure of stem-loop SL2 of the HIV-1 Ψ RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 299:145-156. [DOI] [PubMed] [Google Scholar]
- 5.Auxilien, S., G. Keith, S. F. J. Le Grice, and J.-L. Darlix. 1999. Role of posttranscriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 274:4412-4420. [DOI] [PubMed] [Google Scholar]
- 6.Berg, J. M., and Y. Shih. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081-1085. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 8.Bombarda, E., N. Morellet, H. Cherradi, B. Spiess, S. Bouaziz, E. Grell, B. P. Roques, and Y. Mély. 2001. Determination of the pKa of the four Zn2+-coordinating residues of the distal finger motif of the HIV-1 nucleocapsid protein: consequences on the binding of Zn2+. J. Mol. Biol. 310:659-672. [DOI] [PubMed] [Google Scholar]
- 9.Bowles, N. E., P. Damay, and P.-F. Spahr. 1993. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J. Virol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brulé, F., G. Bec, G. Keith, S. F. J. Le Grice, B. P. Roques, B. Ehresmann, C. Ehresmann, and R. Marquet. 2000. In vitro evidence for the interaction of tRNA3Lys with U3 during the first strand transfer of HIV-1 reverse transcription. Nucleic Acids Res. 28:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champoux, J. J. 1993. Roles of ribonuclease H in reverse transcription, p. 103-117. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Chertova, E. N., B. P. Kane, C. McGrath, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1998. Probing the topography of HIV-1 nucleocapsid protein with the alkylating agent N-ethylmaleimide. Biochemistry 37:17890-17897. [DOI] [PubMed] [Google Scholar]
- 14.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 15.Darlix, J.-L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 16.Darlix, J.-L., A. Vincent, C. Gabus, H. De Rocquigny, and B. Roques. 1993. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV 1 RNA. C. R. Acad. Sci. Paris Life Sci. 316:763-771. [PubMed] [Google Scholar]
- 17.Davis, W. R., S. Gabbara, D. Hupe, and J. A. Peliska. 1998. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry 37:14213-14221. [DOI] [PubMed] [Google Scholar]
- 18.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 19.Déméné, H., C. Z. Dong, M. Ottmann, M. C. Rouyez, N. Jullian, N. Morellet, Y. Mely, J. L. Darlix, M. C. Fournié-Zaluski, S. Saragosti, and B. P. Roques. 1994. 1H NMR structure and biological studies of the His23 → Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry 33:11707-11716. [DOI] [PubMed] [Google Scholar]
- 20.DeStefano, J. J. 1995. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch. Virol. 140:1775-1789. [DOI] [PubMed] [Google Scholar]
- 21.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Göttlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driscoll, M. D., and S. H. Hughes. 2000. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J. Virol. 74:8785-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond, J. E., P. Mounts, R. J. Gorelick, J. R. Casas-Finet, W. J. Bosche, L. E. Henderson, D. J. Waters, and L. O. Arthur. 1997. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res. Hum. Retrovir. 13:533-543. [DOI] [PubMed] [Google Scholar]
- 24.Fisher, R. J., A. Rein, M. Fivash, M. A. Urbaneja, J. R. Casas-Finet, M. Medaglia, and L. E. Henderson. 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 72:1902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelick, R. J., W. Fu, T. D. Gagliardi, W. J. Bosche, A. Rein, L. E. Henderson, and L. O. Arthur. 1999. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J. Virol. 73:8185-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 29.Gorelick, R. J., S. M. Nigida, Jr., J. W. Bess, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green, L. M., and J. M. Berg. 1990. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc. Natl. Acad. Sci. USA 87:6403-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo, J., T. Wu, J. Anderson, B. F. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. G. Levin. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, J., T. Wu, J. Bess, L. E. Henderson, and J. G. Levin. 1998. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J. Virol. 72:6716-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, J., W. Wu, Z. Y. Yuan, K. Post, R. J. Crouch, and J. G. Levin. 1995. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry 34:5018-5029. [DOI] [PubMed] [Google Scholar]
- 35.Hargittai, M. R. S., A. T. Mangla, R. J. Gorelick, and K. Musier-Forsyth. 2001. HIV-1 nucleocapsid protein zinc finger structures induce tRNALys,3 structural changes but are not critical for primer/template annealing. J. Mol. Biol. 312:985-997. [DOI] [PubMed] [Google Scholar]
- 36.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. A. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson, L. E., T. D. Copeland, R. C. Sowder, G. W. Smythers, and S. Oroszlan. 1981. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 256:8400-8406. [PubMed] [Google Scholar]
- 38.Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270:20871-20874. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, P. E., R. B. Turner, Z. R. Wu, L. Hairston, J. Guo, J. G. Levin, and M. F. Summers. 2000. A mechanism for plus-strand transfer enhancement by HIV-1 nucleocapsid protein during reverse transcription. Biochemistry 39:9084-9091. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 41.Lapadat-Tapolsky, M., C. Gabus, M. Rau, and J.-L. Darlix. 1997. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 268:250-260. [DOI] [PubMed] [Google Scholar]
- 42.Lapadat-Tapolsky, M., C. Pernelle, C. Borie, and J.-L. Darlix. 1995. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 23:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, B. M., R. N. De Guzman, B. G. Turner, N. Tjandra, and M. F. Summers. 1998. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 279:633-649. [DOI] [PubMed] [Google Scholar]
- 44.Li, X., Y. Quan, E. J. Arts, Z. Li, B. D. Preston, H. De Rocquigny, B. P. Roques, J.-L. Darlix, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1996. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J. Virol. 70:4996-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mély, Y., H. De Rocquigny, N. Morellet, B. P. Roques, and D. Gérard. 1996. Zinc binding to the HIV-1 nucleocapsid protein: a thermodynamic investigation by fluorescence spectroscopy. Biochemistry 35:5175-5182. [DOI] [PubMed] [Google Scholar]
- 46.Mély, Y., N. Jullian, N. Morellet, H. De Rocquigny, C. Z. Dong, E. Piémont, B. P. Roques, and D. Gérard. 1994. Spatial proximity of the HIV-1 nucleocapsid protein zinc fingers investigated by time-resolved fluorescence and fluorescence resonance energy transfer. Biochemistry 33:12085-12091. [DOI] [PubMed] [Google Scholar]
- 47.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. R. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuno, A., E. Ido, T. Goto, T. Kuwata, M. Nakai, and M. Hayami. 1996. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effects on proteolytic processing of Gag precursors and particle formation. AIDS Res. Hum. Retrovir. 12:793-800. [DOI] [PubMed] [Google Scholar]
- 49.Morellet, N., H. de Rocquigny, Y. Mély, N. Jullian, H. Déméné, M. Ottmann, D. Gérard, J. L. Darlix, M. C. Fournie-Zaluski, and B. P. Roques. 1994. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 235:287-301. [DOI] [PubMed] [Google Scholar]
- 50.Morellet, N., N. Jullian, H. De Rocquigny, B. Maigret, J.-L. Darlix, and B. P. Roques. 1992. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 11:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omichinski, J. G., G. M. Clore, K. Sakaguchi, E. Appella, and A. M. Gronenborn. 1991. Structural characterization of a 39-residue synthetic peptide containing the two zinc binding domains from the HIV-1 p7 nucleocapsid protein by CD and NMR spectroscopy. FEBS Lett. 292:25-30. [DOI] [PubMed] [Google Scholar]
- 52.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817-13823. [DOI] [PubMed] [Google Scholar]
- 53.Ramboarina, S., N. Morellet, M.-C. Fournié-Zaluski, and B. P. Roques. 1999. Structural investigation on the requirement of CCHH zinc finger type in nucleocapsid protein of human immunodeficiency virus 1. Biochemistry 38:9600-9607. [DOI] [PubMed] [Google Scholar]
- 54.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 55.Rodríguez-Rodríguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 56.Rong, L., C. Liang, M. Hsu, L. Kleiman, P. Petitjean, H. De Rocquigny, B. P. Roques, and M. A. Wainberg. 1998. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J. Virol. 72:9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, C. M., J. S. Smith, and M. J. Roth. 1999. RNase H requirements for the second strand transfer reaction of human immunodeficiency virus type 1 reverse transcription. J. Virol. 73:6573-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.South, T. L., P. R. Blake, D. R. Hare, and M. F. Summers. 1991. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry 30:6342-6349. [DOI] [PubMed] [Google Scholar]
- 59.South, T. L., P. R. Blake, R. C. Sowder III, L. O. Arthur, L. E. Henderson, and M. F. Summers. 1990. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry 29:7786-7789. [DOI] [PubMed] [Google Scholar]
- 60.Summers, M. F., L. E. Henderson, M. R. Chance, J. W. Bess, Jr., T. L. South, P. R. Blake, I. Sagi, G. Perez-Alvarado, R. C. Sowder III, D. R. Hare, and L. O. Arthur. 1992. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Summers, M. F., T. L. South, B. Kim, and D. R. Hare. 1990. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 29:329-340. [DOI] [PubMed] [Google Scholar]
- 62.Surovoy, A., J. Dannull, K. Moelling, and G. Jung. 1993. Conformational and nucleic acid binding studies on the synthetic nucleocapsid protein of HIV-1. J. Mol. Biol. 229:94-104. [DOI] [PubMed] [Google Scholar]
- 63.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 64.Tanchou, V., D. Decimo, C. Péchoux, D. Lener, V. Rogemond, L. Berthoux, M. Ottmann, and J.-L. Darlix. 1998. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 72:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urbaneja, M. A., B. P. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. R. Casas-Finet. 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 287:59-75. [DOI] [PubMed] [Google Scholar]
- 67.Werner, S., K. Vogel-Bachmayr, B. Hollinderbäumer, and B. M. Wöhrl. 2001. Requirements for minus-strand transfer catalyzed by Rous sarcoma virus reverse transcriptase. J. Virol. 75:10132-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams, M. C., I. Rouzina, J. R. Wenner, R. J. Gorelick, K. Musier-Forsyth, and V. A. Bloomfield. 2001. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. USA 98:6121-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, T., J. Guo, J. Bess, L. E. Henderson, and J. G. Levin. 1999. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 73:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, W., L. E. Henderson, T. D. Copeland, R. J. Gorelick, W. J. Bosche, A. Rein, and J. G. Levin. 1996. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 70:7132-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269:31491-31495. [PubMed] [Google Scholar]
- 72.Yovandich, J. L., E. N. Chertova, B. F. Kane, T. D. Gagliardi, J. W. Bess, Jr., R. C. Sowder II, L. E. Henderson, and R. J. Gorelick. 2001. Alteration of zinc-binding residues of simian immunodeficiency virus p8NC results in subtle differences in Gag processing and virion maturation associated with degradative loss of mutant NC. J. Virol. 75:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu, Q., and J.-L. Darlix. 1996. The zinc finger of nucleocapsid protein of Friend murine leukemia virus is critical for proviral DNA synthesis in vivo. J. Virol. 70:5791-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]